- Article
MedROAD V2: An AI-Integrated Electronic Medical Record System with Advanced Clinical Decision Support
- Pierre Boulanger
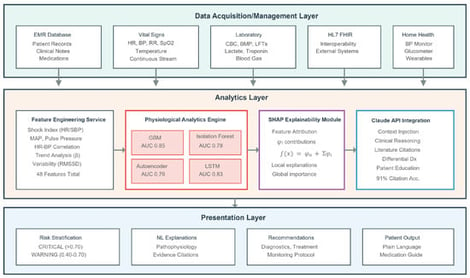
Despite widespread adoption, Electronic Medical Record (EMR) systems remain limited in providing intelligent clinical decision support, particularly for early detection of patient deterioration. We present MedROAD V2 (Medical Records Organization, Analysis, and Display), an open-source EMR that integrates AI-driven physiological analysis with comprehensive patient management. The system combines continuous vital sign monitoring and laboratory data using an ensemble of the following four complementary machine learning models: gradient boosting for supervised prediction, isolation forests for anomaly detection, autoencoders for pattern recognition, and Long Short-Term Memory networks for temporal modeling. A novel framework couples these predictions with a large language model (Claude AI) to generate explainable differential diagnoses grounded in medical literature. Validation on the MIMIC-IV database demonstrated excellent 12 h deterioration prediction. MedROAD demonstrates that combining quantitative prediction with natural language explanation can enhance clinical decision support while extending quality care to populations that would otherwise lack access.
23 January 2026