Discovery of Novel FGFR1 Inhibitors via Pharmacophore Modeling and Scaffold Hopping: A Screening and Optimization Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Compound Preparation
2.2. Protein Preparation
2.3. Pharmacophore Construction
2.4. Pharmacophore Validation
2.5. Virtual Screening Based on Pharmacophore
2.6. Hierarchical Docking
2.7. Scaffold Hopping
2.8. ADMET
2.9. Molecular Dynamics
2.10. MM-PBSA
3. Results
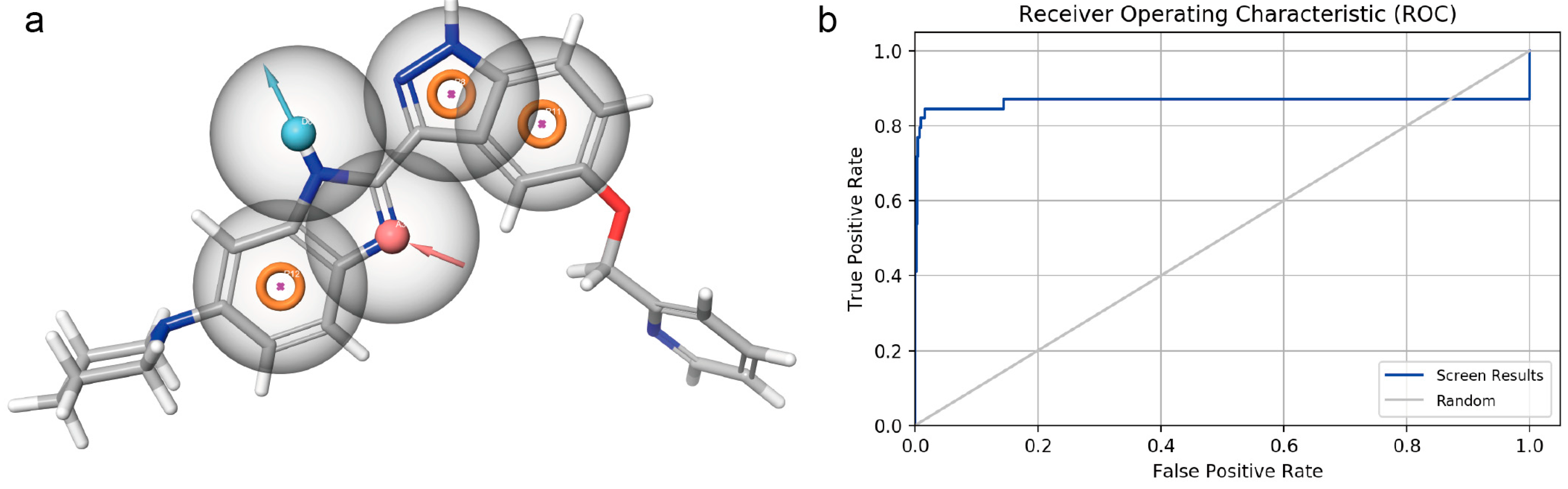
3.1. Pharmacophore Model Establishment
3.2. Pharmacophore Model Verification
3.3. Virtual Screening Based on the Pharmacophore Model
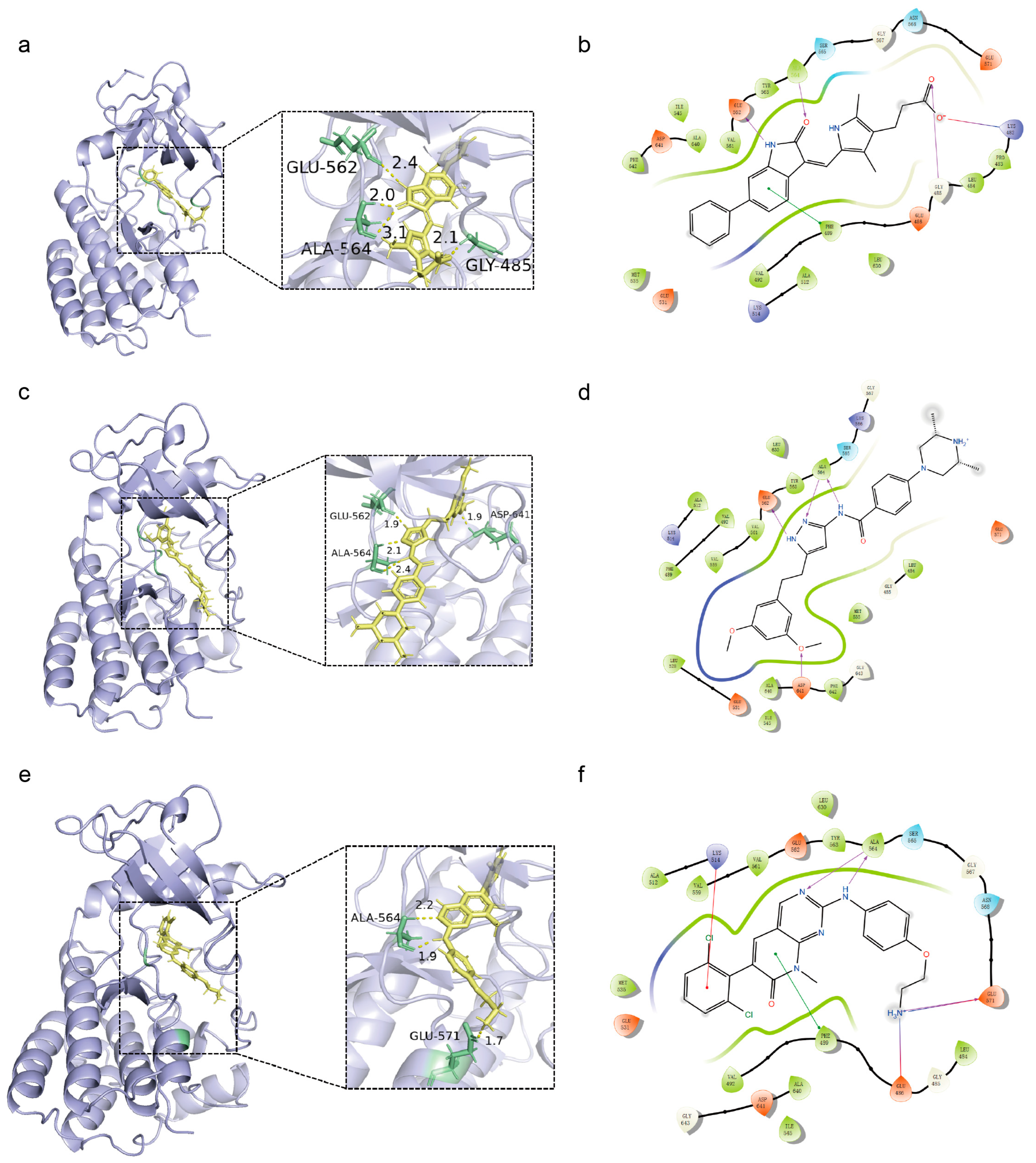
3.4. Molecular Docking
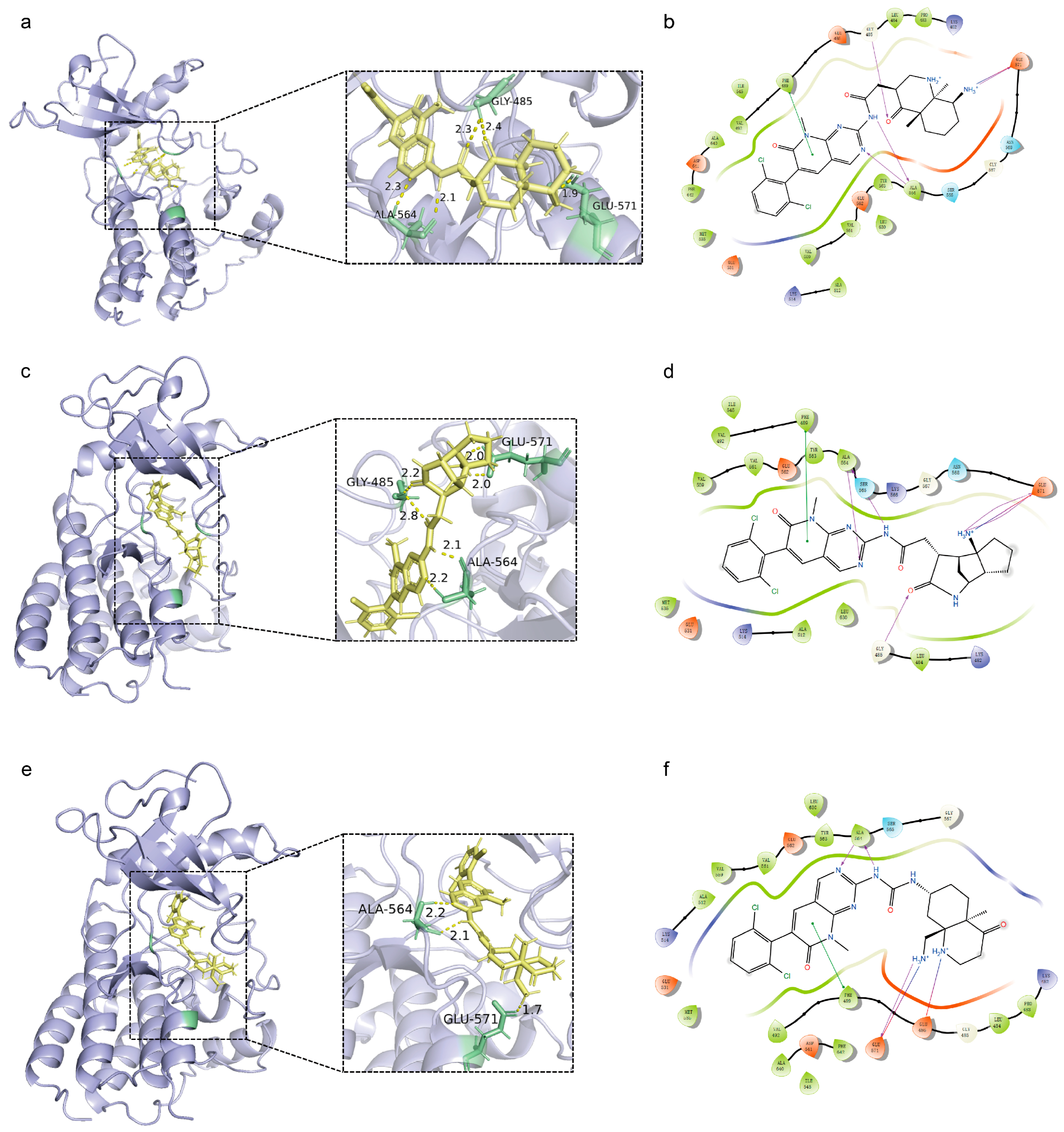
3.5. Scaffold Hopping
3.5.1. Implementation of Scaffold Hopping Strategies for Lead Compounds
3.5.2. Insights into the Scaffold Hopping Results of Compound 20357
3.5.3. Insights into the Scaffold Hopping Results of Compound 18149 and Compound 21769
3.6. ADMET Property Analysis
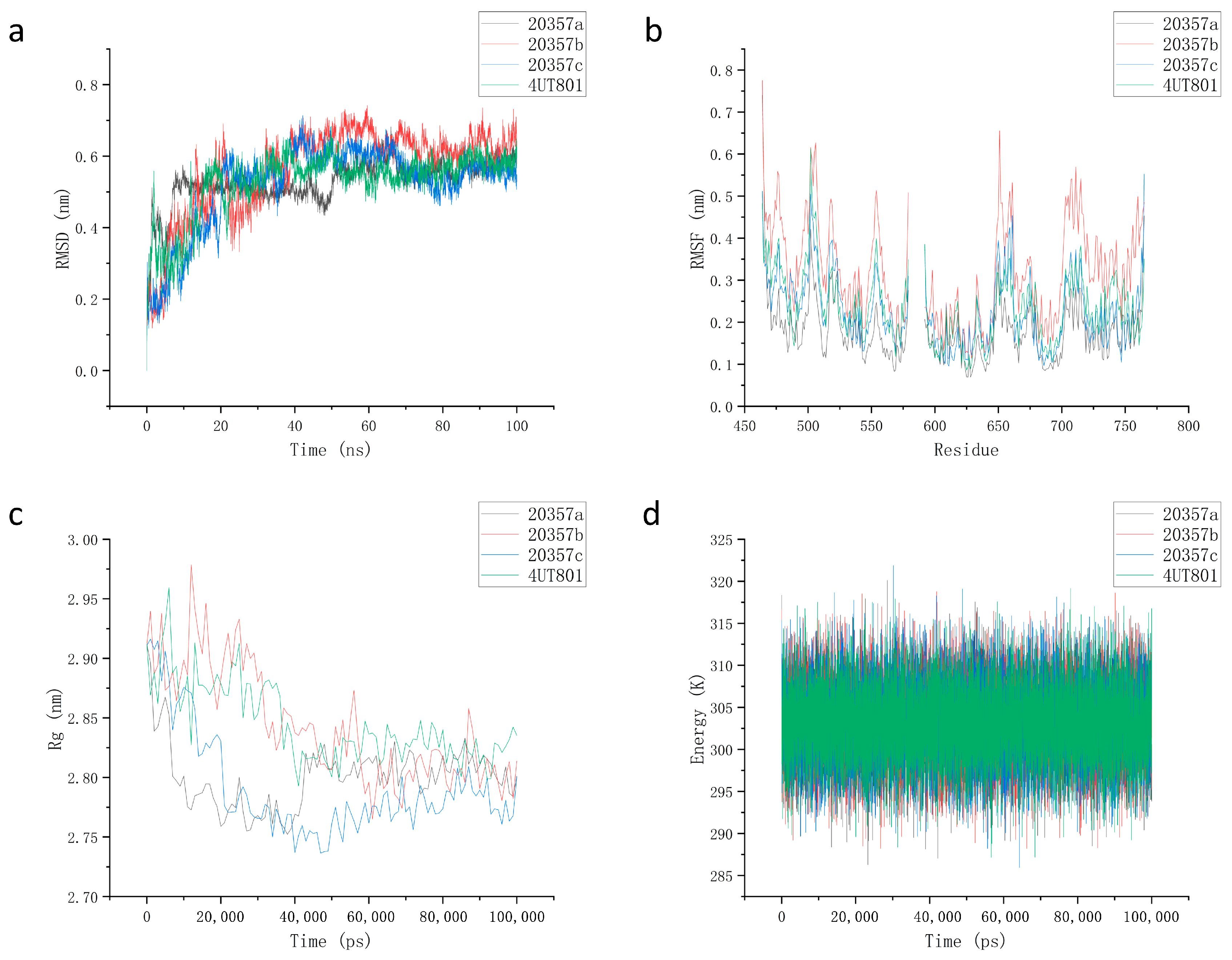
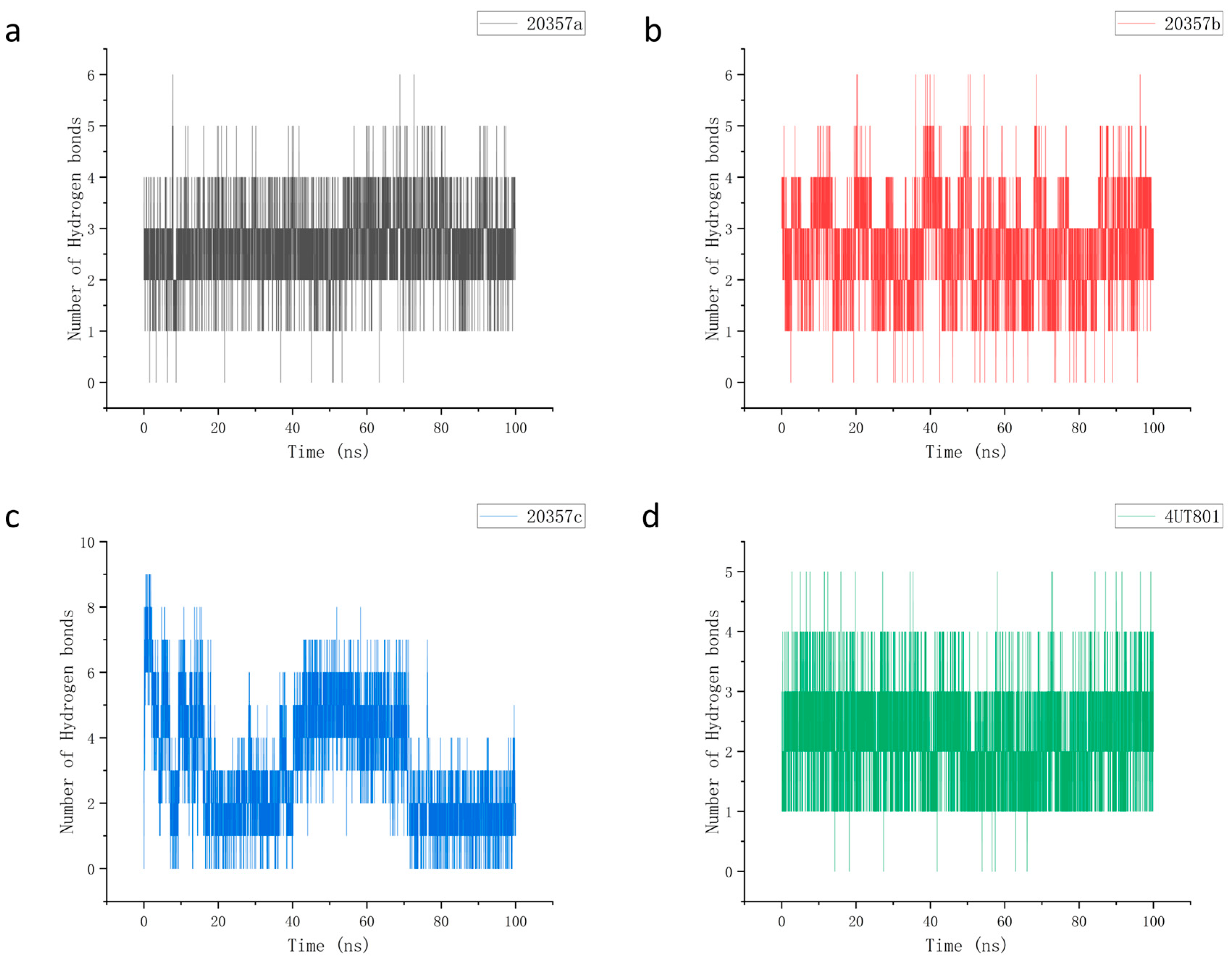
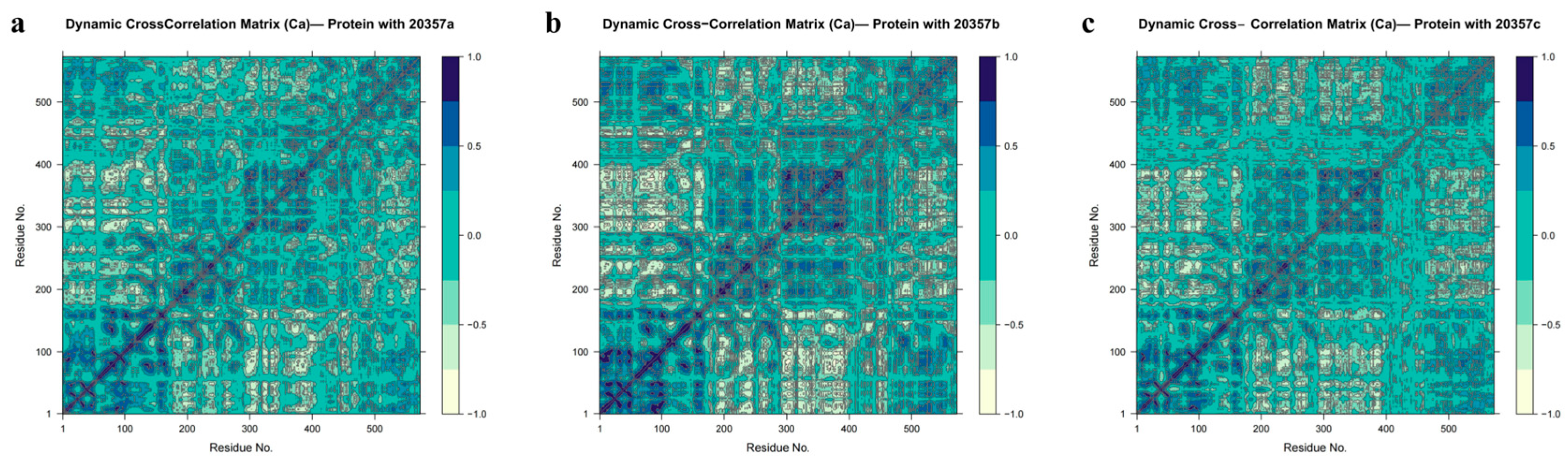
3.7. Molecular Dynamics
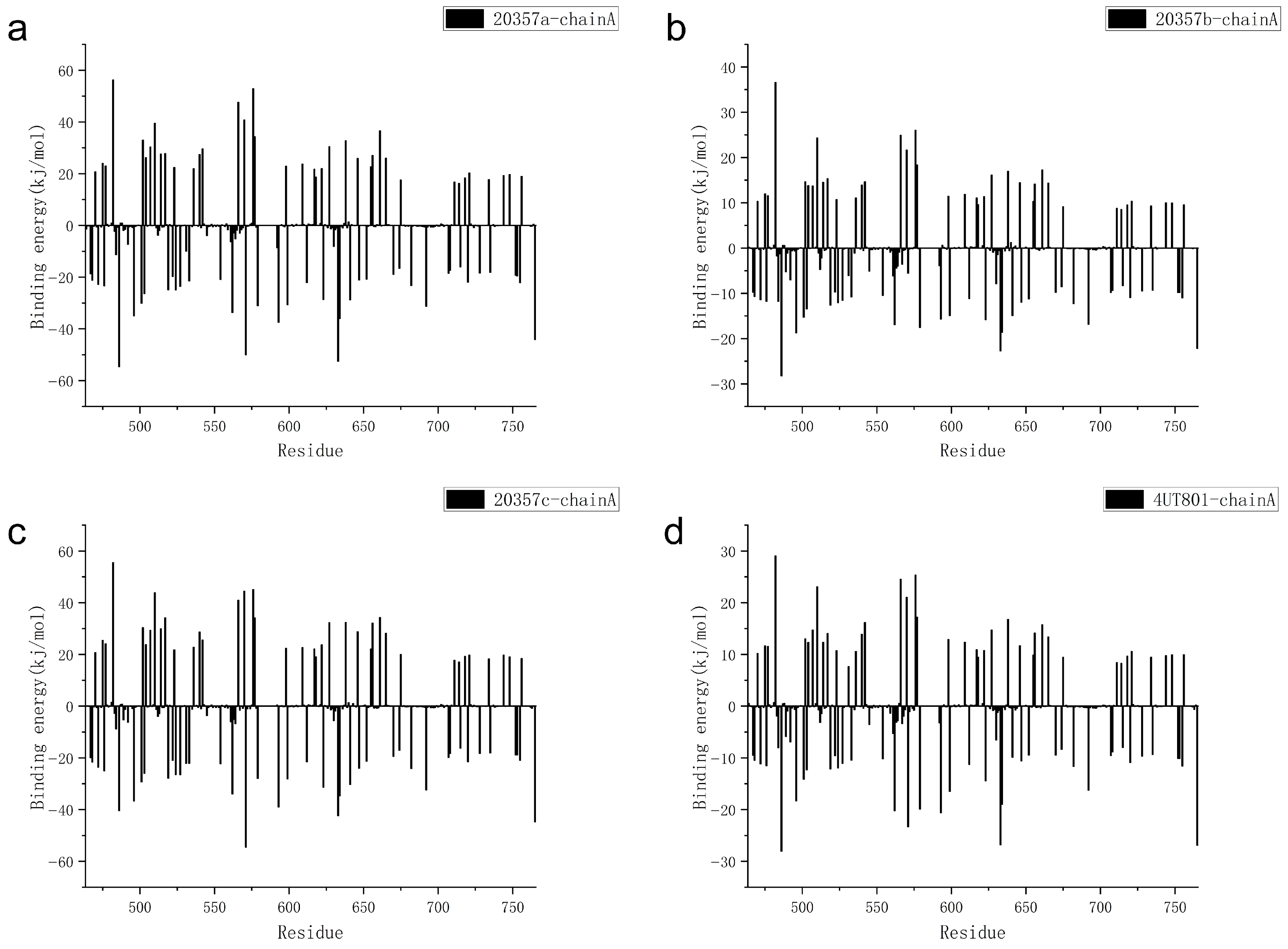
3.8. MM-PBSA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FGFR1 | Fibroblast growth factor receptor 1 |
| FGFs | Fibroblast growth factors |
| HSPG | Heparan sulfate proteoglycan |
| NSCLC | Non-small cell lung cancer |
| TNBC | Triple-negative breast cancer |
| CADD | Computer-aided drug design |
| ROC | Receiver operating characteristic |
| AUC | Area under the curve |
| HTVS | High-throughput virtual screening |
| SP | Standard precision |
| XP | Extra precision |
| MM-GBSA | Molecular Mechanics/Generalized Born Surface Area |
| MM-PBSA | Molecular Mechanics/Poisson–Boltzmann Surface Area |
| ADMET | Absorption, distribution, metabolism, excretion, and toxicity |
| HIA | Human intestinal absorption |
| VD | Volume of distribution |
| DILI | Drug-induced liver injury |
| MD | Molecular dynamics |
| RMSD | Root mean square deviation |
| RMSF | Root-mean-square fluctuation |
| Rg | Radius of gyration |
| DCCM | Dynamic cross-correlation matrix |
| Src | Steroid receptor coactivator |
| EGFR | Epidermal growth factor receptor |
| PDB | Protein data bank |
| FPR | False-positive rate |
| TPR | True-positive rate |
| NVT | Constant temperature-volume |
| NPT | Constant temperature-pressure |
References
- Zhou, R.; Tang, X.; Wang, Y. Emerging strategies to investigate the biology of early cancer. Nat. Rev. Cancer 2024, 24, 850–866. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Wang, Y.; Zhang, N.; Xing, J.; Wang, T.; Bao, Y.; Hou, Y.; Zhao, Y. Small-molecule FGFR-targeted medicinal chemistry: Advances since 2020 and future perspectives. Eur. J. Med. Chem. 2025, 300, 118117. [Google Scholar] [CrossRef]
- Wiedlocha, A.; Haugsten, E.M.; Zakrzewska, M. Roles of the FGF-FGFR Signaling System in Cancer Development and Inflammation. Cells 2021, 10, 2231. [Google Scholar] [CrossRef]
- Edirisinghe, O.; Ternier, G.; Alraawi, Z.; Suresh Kumar, T.K. Decoding FGF/FGFR Signaling: Insights into Biological Functions and Disease Relevance. Biomolecules 2024, 14, 1622. [Google Scholar] [CrossRef] [PubMed]
- Korc, M.; Friesel, R.E. The Role of Fibroblast Growth Factors in Tumor Growth. Curr. Cancer Drug Targets 2009, 9, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J.; Plotnikov, A.N.; Ibrahimi, O.A.; Eliseenkova, A.V.; Yeh, B.K.; Yayon, A.; Linhardt, R.J.; Mohammadi, M. Crystal Structure of a Ternary FGF-FGFR-Heparin Complex Reveals a Dual Role for Heparin in FGFR Binding and Dimerization. Mol. Cell 2000, 6, 743–750. [Google Scholar] [CrossRef]
- Ong, S.H.; Guy, G.R.; Hadari, Y.R.; Laks, S.; Gotoh, N.; Schlessinger, J.; Lax, I. FRS2 Proteins Recruit Intracellular Signaling Pathways by Binding to Diverse Targets on Fibroblast Growth Factor and Nerve Growth Factor Receptors. Mol. Cell. Biol. 2000, 20, 979–989. [Google Scholar] [CrossRef]
- Belov, A.A.; Mohammadi, M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb. Perspect. Biol. 2013, 5, a015958. [Google Scholar] [CrossRef]
- Xue, W.J.; Li, M.T.; Chen, L.; Sun, L.P.; Li, Y.Y. Recent developments and advances of FGFR as a potential target in cancer. Future Med. Chem. 2018, 10, 2109–2126. [Google Scholar] [CrossRef]
- Turner, N.; Pearson, A.; Sharpe, R.; Lambros, M.; Geyer, F.; Lopez-Garcia, M.A.; Natrajan, R.; Marchio, C.; Iorns, E.; Mackay, A.; et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010, 70, 2085–2094. [Google Scholar] [CrossRef]
- Ji, W.; Yu, Y.; Li, Z.; Wang, G.; Li, F.; Xia, W.; Lu, S. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget 2016, 7, 15118–15134. [Google Scholar] [CrossRef] [PubMed]
- Maehara, O.; Suda, G.; Natsuizaka, M.; Ohnishi, S.; Komatsu, Y.; Sato, F.; Nakai, M.; Sho, T.; Morikawa, K.; Ogawa, K.; et al. Fibroblast growth factor-2-mediated FGFR/Erk signaling supports maintenance of cancer stem-like cells in esophageal squamous cell carcinoma. Carcinogenesis 2017, 38, 1073–1083. [Google Scholar] [CrossRef]
- Pichler, R.; van Creij, N.C.H.; Mertens, L.S.; Del Giudice, F.; Koll, F.; Soria, F.; Subiela, J.D.; Plage, H.; Tymoszuk, P.; Mayr, R.; et al. FGFR1/3 Signaling as an Achilles’ Heel of Phenotypic Diversity in Urothelial Carcinoma. Eur. Urol. Oncol. 2025, in press. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Cai, R.; Han, B.; Liu, Y.; Zhang, Y.; Wu, Z.; Cheng, Y.; Lin, Z.; Qin, G.; Xie, X.; et al. FGFR1-amplified colorectal cancer: A distinct prognostic subtype identified through integrated genomic and immunohistochemical profiling. ESMO Open 2025, 10, 105561. [Google Scholar] [CrossRef]
- Weiss, J.; Sos, M.L.; Seidel, D.; Peifer, M.; Zander, T.; Heuckmann, J.M.; Ullrich, R.T.; Menon, R.; Maier, S.; Soltermann, A.; et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2010, 2, 62ra93. [Google Scholar] [CrossRef]
- Siefker-Radtke, A.O.; Necchi, A.; Park, S.H.; García-Donas, J.; Huddart, R.A.; Burgess, E.F.; Fleming, M.T.; Rezazadeh Kalebasty, A.; Mellado, B.; Varlamov, S.; et al. Management of Fibroblast Growth Factor Inhibitor Treatment–emergent Adverse Events of Interest in Patients with Locally Advanced or Metastatic Urothelial Carcinoma. Eur. Urol. Open Sci. 2023, 50, 1–9. [Google Scholar] [CrossRef]
- Gordon, A.; Johnston, E.; Lau, D.K.; Starling, N. Targeting FGFR2 Positive Gastroesophageal Cancer: Current and Clinical Developments. Onco Targets Ther. 2022, 15, 1183–1196. [Google Scholar] [CrossRef]
- Sur, S.; Nimesh, H. Challenges and limitations of computer-aided drug design. Adv. Pharmacol. 2025, 103, 415–428. [Google Scholar] [CrossRef]
- Vemula, D.; Jayasurya, P.; Sushmitha, V.; Kumar, Y.N.; Bhandari, V. CADD, AI and ML in drug discovery: A comprehensive review. Eur. J. Pharm. Sci. 2023, 181, 106324. [Google Scholar] [CrossRef]
- Al-Zaydi, K.M.; Baammi, S.; Moussaoui, M. Multitarget inhibition of CDK2, EGFR, and tubulin by phenylindole derivatives: Insights from 3D-QSAR, molecular docking, and dynamics for cancer therapy. PLoS ONE 2025, 20, e0326245. [Google Scholar] [CrossRef] [PubMed]
- Mohebbi, A. Ligand-based 3D pharmacophore modeling, virtual screening, and molecular dynamic simulation of potential smoothened inhibitors. J. Mol. Model. 2023, 29, 143. [Google Scholar] [CrossRef]
- Baammi, S.; Daoud, R.; Allali, A.E. Computational design of quinazolin-4(3H)-one derivatives as multitarget inhibitors against angiogenesis and metabolic reprogramming to help overcome cancer resistance. Arabian J. Chem. 2025, 18, 1592024. [Google Scholar] [CrossRef]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef]
- Chen, G.; Chen, L.; Li, X.; Mohammadi, M. FGF-based drug discovery: Advances and challenges. Nat. Rev. Drug Discov. 2025, 24, 335–357. [Google Scholar] [CrossRef] [PubMed]
- Anighoro, A.; Bajorath, J.; Rastelli, G. Polypharmacology: Challenges and opportunities in drug discovery. J. Med. Chem. 2014, 57, 7874–7887. [Google Scholar] [CrossRef]
- Azuma, K.; Kawahara, A.; Sonoda, K.; Nakashima, K.; Tashiro, K.; Watari, K.; Izumi, H.; Kage, M.; Kuwano, M.; Ono, M.; et al. FGFR1 activation is an escape mechanism in human lung cancer cells resistant to afatinib, a pan-EGFR family kinase inhibitor. Oncotarget 2014, 5, 5908–5919. [Google Scholar] [CrossRef] [PubMed]
- Hashim, G.M.; Shahgolzari, M.; Hefferon, K.; Yavari, A.; Venkataraman, S. Plant-Derived Anti-Cancer Therapeutics and Biopharmaceuticals. Bioengineering 2024, 12, 7. [Google Scholar] [CrossRef]
- Bell, D.S. Combine and conquer: Advantages and disadvantages of fixed-dose combination therapy. Diabetes Obes. Metab. 2013, 15, 291–300. [Google Scholar] [CrossRef]
- Zhou, N.; Zheng, C.; Tan, H.; Luo, L. Identification of PLK1-PBD Inhibitors from the Library of Marine Natural Products: 3D QSAR Pharmacophore, ADMET, Scaffold Hopping, Molecular Docking, and Molecular Dynamics Study. Mar. Drugs 2024, 22, 83. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Tian, Y.; Cao, L.; Guo, P.; Cai, Z.; Zhou, J. 3D-QSAR, design, molecular docking and dynamics simulation studies of novel 6-hydroxybenzothiazole-2-carboxamides as potentially potent and selective monoamine oxidase B inhibitors. Front. Pharmacol. 2025, 16, 1545791. [Google Scholar] [CrossRef]
- Verburgt, J.; Kihara, D. Benchmarking of structure refinement methods for protein complex models. Proteins 2022, 90, 83–95. [Google Scholar] [CrossRef]
- Bellissent-Funel, M.C.; Hassanali, A.; Havenith, M.; Henchman, R.; Pohl, P.; Sterpone, F.; van der Spoel, D.; Xu, Y.; Garcia, A.E. Water Determines the Structure and Dynamics of Proteins. Chem. Rev. 2016, 116, 7673–7697. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Ignasiak, M.T.; Chan, B.; Croft, A.K.; Radom, L.; Schiesser, C.H.; Pattison, D.I.; Davies, M.J. Reactivity of disulfide bonds is markedly affected by structure and environment: Implications for protein modification and stability. Sci. Rep. 2016, 6, 38572. [Google Scholar] [CrossRef] [PubMed]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Mohan, A.; Krishnamoorthy, S.; Sabanayagam, R.; Schwenk, G.; Feng, E.; Ji, H.F.; Muthusami, S. Pharmacophore based virtual screening for identification of effective inhibitors to combat HPV 16 E6 driven cervical cancer. Eur. J. Pharmacol. 2023, 957, 175961. [Google Scholar] [CrossRef] [PubMed]
- Nahm, F.S. Receiver operating characteristic curve: Overview and practical use for clinicians. Korean J. Anesthesiol. 2022, 75, 25–36. [Google Scholar] [CrossRef]
- Huang, Q.; Lai, T.; Wang, Q.; Luo, L. mPGES-1 Inhibitor Discovery Based on Computer-Aided Screening: Pharmacophore Models, Molecular Docking, ADMET, and MD Simulations. Molecules 2023, 28, 6059. [Google Scholar] [CrossRef]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021, 60, 273–343. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Osunnaya, S.A.; Ndarawit, W.K.; Aderibigbe, I.; Omolopo, I.A.; Aribisala, P.O.; Elekan, A.O.; Adeyemo, A.S.; Amoo, S.A.; Olamiposi, O.S.; Kimani, N.M.; et al. Identification and exploration of novel FGFR-1 inhibitors in the Lotus database for Cholangiocarcinoma (CCA) treatment. Asp. Mol. Med. 2025, 5, 100085. [Google Scholar] [CrossRef]
- Hu, Y.; Stumpfe, D.; Bajorath, J. Computational Exploration of Molecular Scaffolds in Medicinal Chemistry. J. Med. Chem. 2016, 59, 4062–4076. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.L.G.; Andricopulo, A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today 2019, 24, 1157–1165. [Google Scholar] [CrossRef]
- Tan, H.; Li, C.; Lai, T.; Luo, L. In Silico Analysis of USP7 Inhibitors Based on Building QSAR Models and Fragment Design for Screening Marine Compound Libraries. Mar. Drugs 2024, 22, 1. [Google Scholar] [CrossRef]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef]
- Kagami, L.; Wilter, A.; Diaz, A.; Vranken, W. The ACPYPE web server for small-molecule MD topology generation. Bioinformatics 2023, 39, btad350. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Grant, B.J.; Skjaerven, L.; Yao, X.Q. The Bio3D packages for structural bioinformatics. Protein Sci. 2021, 30, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Hongmao, S. Pharmacophore-Based Virtual Screening. Curr. Med. Chem. 2008, 15, 1018–1024. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, J.L.; Ang, L.; Li, M.C.; Zhao, M.; Wang, Y.; Wu, Q. Proposing a novel molecular subtyping scheme for predicting distant recurrence-free survival in breast cancer post-neoadjuvant chemotherapy with close correlation to metabolism and senescence. Front. Endocrinol. 2023, 14, 1265520. [Google Scholar] [CrossRef]
- Böhm, H.J.; Flohr, A.; Stahl, M. Scaffold hopping. Drug Discov. Today Technol. 2004, 1, 217–224. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Deng, J.; Li, X. Clinical advances and challenges in targeting FGF/FGFR signaling in lung cancer. Mol. Cancer 2024, 23, 256. [Google Scholar] [CrossRef]
- Wu, X.; Dai, M.; Cui, R.; Wang, Y.; Li, C.; Peng, X.; Zhao, J.; Wang, B.; Dai, Y.; Feng, D.; et al. Design, synthesis and biological evaluation of pyrazolo[3,4-d]pyridazinone derivatives as covalent FGFR inhibitors. Acta Pharm. Sin. B 2021, 11, 781–794. [Google Scholar] [CrossRef]
- Hua, H.; Guan, L.; Pan, B.; Gao, J.; Geng, Y.; Niu, M.M.; Li, Z.; Li, J. The identification of potent dual-target monopolar spindle 1 (MPS1) and histone deacetylase 8 (HDAC8) inhibitors through pharmacophore modeling, molecular docking, molecular dynamics simulations, and biological evaluation. Front. Pharmacol. 2024, 15, 1454523. [Google Scholar] [CrossRef]
- Atatreh, N.; Mahgoub, R.E.; Ghemrawi, R.; Sakkal, M.; Sammani, N.; Khair, M.; Ghattas, M.A. The Discovery of Small ERK5 Inhibitors via Structure-Based Virtual Screening, Biological Evaluation and MD Simulations. Molecules 2025, 30, 4181. [Google Scholar] [CrossRef]
- Abdel-Mohsen, H.T.; Ibrahim, M.A.; Nageeb, A.M.; El Kerdawy, A.M. Receptor-based pharmacophore modeling, molecular docking, synthesis and biological evaluation of novel VEGFR-2, FGFR-1, and BRAF multi-kinase inhibitors. BMC Chem. 2024, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Chohan, T.A.; Sarfraz, M.; Chohan, T.A.; Imran Sajid, M.; Tiwari, R.K.; Ansari, S.A.; Alkahtani, H.M.; Yasmeen Ansari, S.; Khurshid, U.; et al. Molecular modeling of pyrrolo-pyrimidine based analogs as potential FGFR1 inhibitors: A scientific approach for therapeutic drugs. J. Biomol. Struct. Dyn. 2023, 41, 14358–14371. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, Y.; Zhang, C.; Zhou, H.; Ma, J.; Vaishnani, D.K.; Zeng, B.; Yu, J.; Mao, H.; Zheng, J. Multi-Omics and Experimental Validation Reveal Anti-HCC Mechanisms of Tibetan Liuwei Muxiang Pill and Quercetin. Pharmaceuticals 2025, 18, 900. [Google Scholar] [CrossRef]
- Tran-Nguyen, V.K.; Randriharimanamizara, U.F.; Taboureau, O. HERGAI: An artificial intelligence tool for structure-based prediction of hERG inhibitors. J. Cheminform. 2025, 17, 110. [Google Scholar] [CrossRef]
- Zhao, J.; Xia, M. Cell-Based hERG Channel Inhibition Assay in High-Throughput Format. In High-Throughput Screening Assays in Toxicology; Methods in Molecular Biology Series; Humana: New York, NY, USA, 2022; Volume 2474, pp. 21–28. [Google Scholar] [CrossRef]



| Hypothesis | Phase Hypo Score | EF1% a | BEDROC160.9 b | ROC | AUAC c |
|---|---|---|---|---|---|
| ADRRR_2 | 1.33 | 26.64 | 0.97 | 0.87 | 0.9 |
| ADRRR_1 | 1.32 | 26.64 | 0.97 | 0.86 | 0.9 |
| ARRR_2 | 1.28 | 26.64 | 0.94 | 0.89 | 0.91 |
| DRRR_2 | 1.28 | 26.64 | 0.94 | 0.88 | 0.9 |
| DRRR_1 | 1.28 | 26.64 | 0.93 | 0.9 | 0.91 |
| ARRR_1 | 1.28 | 26.64 | 0.95 | 0.88 | 0.9 |
| DHRRR_1 | 1.27 | 26.64 | 0.97 | 0.84 | 0.9 |
| HRRR_1 | 1.25 | 26.64 | 0.95 | 0.85 | 0.89 |
| ADRR_2 | 1.25 | 26.64 | 0.93 | 0.87 | 0.89 |
| AADRR_1 | 1.25 | 26.64 | 0.97 | 0.68 | 0.78 |
| ADRR_1 | 1.25 | 26.64 | 0.92 | 0.88 | 0.89 |
| DRRR_3 | 1.24 | 26.64 | 0.98 | 0.97 | 0.95 |
| AADRR_2 | 1.24 | 26.64 | 0.98 | 0.69 | 0.8 |
| DHRRR_2 | 1.24 | 26.64 | 0.93 | 0.71 | 0.83 |
| ADRRR_3 | 1.23 | 23.98 | 0.87 | 0.81 | 0.87 |
| DHRR_2 | 1.23 | 26.64 | 0.97 | 0.87 | 0.9 |
| ADRRR_5 | 1.23 | 23.98 | 0.85 | 0.82 | 0.88 |
| ADRRR_4 | 1.23 | 23.98 | 0.85 | 0.84 | 0.88 |
| DHRRR_3 | 1.22 | 26.64 | 0.97 | 0.79 | 0.87 |
| DHRR_1 | 1.2 | 26.64 | 0.93 | 0.93 | 0.93 |
| Ligand | SP Docking Score | XP Docking Score | MM/GBSA_ΔG_Bind |
|---|---|---|---|
| (kcal/mol) | (kcal/mol) | (kcal/mol) | |
| 4UT801 | −8.082 | −13.139 | −69.84 |
| Compound 18149 | −11.405 | −13.527 | −70.69 |
| Compound 20357 | −12.724 | −15.011 | −73.76 |
| Compound 21769 | −11.532 | −13.743 | −92.42 |
| Compound | Structure | Docking Score |
|---|---|---|
| Compound 20357 |  | −15.011 |
| Compound 20357a | 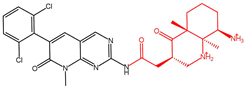 | −14.335 |
| Compound 20357b | 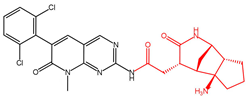 | −15.441 |
| Compound 20357c | 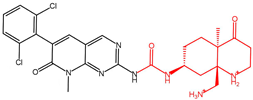 | −13.979 |
| Ligand | Hydrogen Bond | Salt Bridges | Pi-Pi | Sidechain RMSD (Å) | Binding Scores (kcal/mol) |
|---|---|---|---|---|---|
| 4UT801 | GLU562, ALA564 | - | PHE489 | - | −69.84 |
| Compound 20357 | GLU571, ALA564 | GLU571, GLU486 | PHE489 | - | −73.76 |
| Compound 20357a | GLU571, ALA564, GLY485 | GLU571 | PHE489 | 0.674016 | −70.43 |
| Compound 20357b | GLU571, ALA564, GLY485 | GLU571 | PHE489 | 0.466784 | −78.43 |
| Compound 20357c | GLU571, ALA564 | GLU571, GLU486 | PHE489 | 0.239625 | −77.75 |
| 4UT801 | Compound 20357a | Compound 20357b | Compound 20357c | |
|---|---|---|---|---|
| Molecular weight | 455.23 | 556.18 | 540.14 | 557.17 |
| Hydrogen bond acceptors | 7 | 9 | 9 | 10 |
| Hydrogen bond donors | 2 | 4 | 4 | 5 |
| Water solubility (Log S) | −5.121 | −3.561 | −3.456 | −3.264 |
| Lipophilicity (Log P) | 4.492 | 1.556 | 2.322 | 1.993 |
| Human intestinal absorption (HIA) a | 0.038 | 0.014 | 0.035 | 0.021 |
| MDCK Permeability b | 7.63 × 10−6 | 1.23 × 10−5 | 5.89 × 10−6 | 8.57 × 10−6 |
| PPB c | 92.79% | 63.54% | 91.19% | 59.04% |
| VD d | 2.251 | 1.479 | 0.882 | 2.031 |
| CYP2D6-inhibitor e | 0.85 | 0.187 | 0.268 | 0.278 |
| CL f | 2.543 | 9.084 | 5.671 | 5.7 |
| hERG Blockers a | 0.95 | 0.902 | 0.783 | 0.774 |
| Drug-induced liver injury (DILI) a | 0.954 | 0.301 | 0.705 | 0.913 |
| AMES Toxicity g | 0.902 | 0.13 | 0.178 | 0.336 |
| Rat Oral Acute Toxicity a | 0.677 | 0.347 | 0.232 | 0.303 |
| Skin Sensitization a | 0.645 | 0.499 | 0.362 | 0.587 |
| Eye Corrosion/Irritation a | 0.012 | 0.005 | 0.007 | 0.005 |
| Respiratory Toxicity a | 0.991 | 0.864 | 0.674 | 0.691 |
| Carcinogencity a | 0.078 | 0.753 | 0.199 | 0.11 |
| Lipinski Rule | Accepted | Accepted | Accepted | Accepted |
| Type of Energy | Compound 20357a | Compound 20357b | Compound 20357c | 4UT801 |
|---|---|---|---|---|
| van der Waal energy | −255.379 +/− 9.147 kJ/mol | −252.158 +/− 14.913 kJ/mol | −239.060 +/− 13.270 kJ/mol | −227.125 +/− 12.115 kJ/mol |
| Electrostatic energy | −628.471 +/− 50.228 kJ/mol | −424.579 +/− 34.072 kJ/mol | −622.993 +/− 66.695 kJ/mol | −259.025 +/− 33.853 kJ/mol |
| Polar solvation energy | 401.543 +/− 61.865 kJ/mol | 415.109 +/− 45.852 kJ/mol | 379.525 +/− 103.983 kJ/mol | 218.889 +/− 44.756 kJ/mol |
| SASA energy | −24.168 +/− 0.987 kJ/mol | −23.898 +/− 1.352 kJ/mol | −22.754 +/− 1.278 kJ/mol | −21.418 +/− 1.297 kJ/mol |
| Binding energy | −506.474 +/− 33.056 kJ/mol | −285.526 +/− 28.778 kJ/mol | −505.282 +/− 59.899 kJ/mol | −288.678 +/− 29.135 kJ/mol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, X.; Tao, J.; Zhang, N.; Wang, L.; Zheng, X.; Luo, L. Discovery of Novel FGFR1 Inhibitors via Pharmacophore Modeling and Scaffold Hopping: A Screening and Optimization Approach. Targets 2025, 3, 35. https://doi.org/10.3390/targets3040035
Ji X, Tao J, Zhang N, Wang L, Zheng X, Luo L. Discovery of Novel FGFR1 Inhibitors via Pharmacophore Modeling and Scaffold Hopping: A Screening and Optimization Approach. Targets. 2025; 3(4):35. https://doi.org/10.3390/targets3040035
Chicago/Turabian StyleJi, Xingchen, Jiahua Tao, Na Zhang, Linxin Wang, Xiyi Zheng, and Lianxiang Luo. 2025. "Discovery of Novel FGFR1 Inhibitors via Pharmacophore Modeling and Scaffold Hopping: A Screening and Optimization Approach" Targets 3, no. 4: 35. https://doi.org/10.3390/targets3040035
APA StyleJi, X., Tao, J., Zhang, N., Wang, L., Zheng, X., & Luo, L. (2025). Discovery of Novel FGFR1 Inhibitors via Pharmacophore Modeling and Scaffold Hopping: A Screening and Optimization Approach. Targets, 3(4), 35. https://doi.org/10.3390/targets3040035







