Abstract
Cheap pasture fodder and a long grazing season are favorable for cattle breeding in the Dagestan Republic of Russia. However, specific natural and geographical conditions, including mountain terrain, hypoxia, and high humidity slow down the intensification of cattle breeding in this region. Thus, the maintenance of genetic diversity of local cattle breeds, which fits well into specific environments, is of special importance for mountain ethnic communities. Dagestan Mountain cattle have valuable biological traits, including strong hooves, stamina, and adaptability to extreme mountain conditions. This sample included 32 individuals of Dagestan Mountain cattle collected from private owners in the mountain villages of Dagestan during a scientific expedition. We observed a high level of genetic diversity in Dagestan Mountain cattle, as revealed by calculations of the mean number of alleles per locus (6.82 compared to 4.79–5.82 in other breeds) and observed heterozygosity indices (Ho = 0.73, which was higher compared to the other breeds (Ho = 0.69) excluding Simmentals and Brown Swiss (Ho = 0.74). Based on STRUCTURE results, the individuals with a low level of admixture with other breeds were found within Dagestan Mountain cattle, which can be considered as candidates for using conservation programs in germplasms.
1. Introduction
Dagestan is a peculiar and interesting region located in the southernmost part of the Russian Federation, on the northeastern slopes of the Greater Caucasus and in the Caspian lowland. Almost half of the territory (48%) is occupied by a mountainous zone. At the same time, large areas of mountain pastures, cheap pasture fodder and a long pasture season favor cattle breeding in the region [1].
In Dagestan, from time immemorial, Great Caucasian and Lesser Caucasian cattle have been bred in the mountains and on the plains. Mountain cattle, which were created by the crossing of local cattle with Swiss, Kostroma, and Lebedin breeds, represent a valuable gene pool for obtaining new breeds of cattle. Thus, the Caucasian Brown breed (Dagestan offspring) is the result of the crossbreeding of mountain cattle with Swiss, Brown Carpathian and Lebedin breeds [2].
One of the main reasons for the conservation of the gene pool of mountain cattle is the predominance and specifics of the mountain terrain, where the intensification of cattle breeding is hampered by natural and geographical conditions. The breeding of highly specialized breeds in the mountain zone is associated with a significant loss of their productivity and premature culling for various reasons [3].
In this regard, the aim of our study is to characterize the modern allele pool and to assess the genetic diversity of Dagestan Mountain cattle using STR markers.
2. Materials and Methods
For the present study, samples of mountain cattle were collected in the Republic of Dagestan during a specially organized scientific expedition in 2019.
Polymorphisms of 11 STR loci recommended by ISAG for population genetic studies of cattle were evaluated using a 16-channel capillary genetic analyzer ABI3130xl (Applied Biosystems, Waltham, MA, USA). Raw allele length data were obtained using the Gene Mapper v.4 software (Applied Biosystems).
To compare genetic diversity levels and to establish genetic links, we used STR profiles of different cattle breeds from the Bioresource collection of the L.K. Ernst Federal Research Center for Animal Husbandry, (Unique Scientific Installation (UNU) “Bank of Genetic Materials of Animals and Birds of the L.K. Ernst Federal Science Center for Animal Husbandry, supported by the Russian Ministry of Science and Higher Education). A total of 132 specimens from 5 cattle breeds were analyzed in our study (Table 1).

Table 1.
Sampling information for studied cattle breeds.
3. Results
A total of 117 alleles at 11 loci were found in the studied populations. The total number of alleles was 413, and 90 alleles were identified in the GS group. We found that there was a deficiency of heterozygotes in one locus in the cattle populations bred in the territory of Dagestan (Table 2).

Table 2.
Genetic diversity parameters estimated for 11 microsatellite markers by cattle breeds.
Allelic diversity was the highest in the mountain cattle group compared to other studied breeds.
The level of observed heterozygosity was 0.73 in mountain cattle.
Heterozygote deficiency was found in mountain and in Red Steppe cattle bred in Dagestan.
We identified a total of 29 private alleles (14 in the GS group) with frequencies varying from 0.016 to 0.148 in the studied populations.
Analysis of the Neighbor Net graph (Figure 1) showed a presence of genetic relationships between the GS, BS, and RS groups as well as providing evidence for a common origin of the BS and BS_G groups. Holsteins and Simmentals formed separate branches.
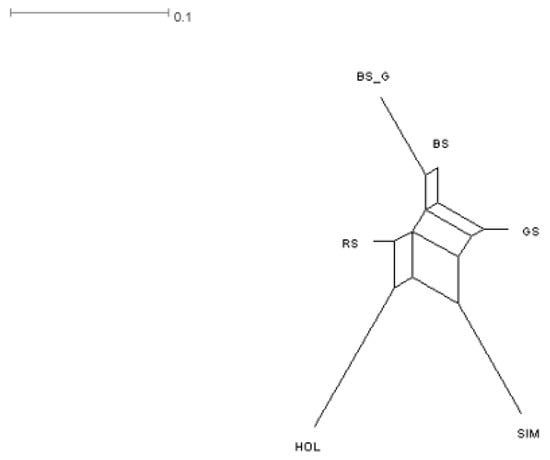
Figure 1.
Dendrogram based on pairwise genetic distances (DJost) plotted using the NeighborNet algorithm.
Based on a cluster analysis, an endemic population of mountain cattle was identified. Most of the identified endemic animals belonged to samples from the most remote area, characterized by rocky and rugged terrain. The remaining cattle groups showed a high degree of membership in their own cluster. In addition, we found differences between Brown Swiss cattle from the Caucasian branch and those who were brought from Germany.
Based on different approaches, we found that GS was characterized by high degrees of polymorphism and genetic diversity. This peculiar pattern might correspond to the following factors. The samples of GS were collected from different private owners, which did not provide any information on whether their animals were crossed with other breeds or not. Moreover, the cattle owners in some villages form a united herd, which is moved to the shared pastures and managed by a single herdsman. The united herd includes cattle from different breeds, which may be admixed. This agrees with a finding that GS and RS share a part of their genome and that the artificial strain BS is already introgressed by the original genome of the region.
However, the recognition of distinguished group of animals within the GS population might indicate the presence of an endemic GS population in the remote isolated areas of the Dagestan region.
4. Conclusions
Thus, our study provides the most complete data on the state of allelic diversity and genetic differentiation of mountain cattle in the Republic of Dagestan compared to other breeds.
The preservation of the valuable gene pool of mountain cattle is necessary for the further development of animal husbandry in the mountains of Dagestan. Comprehensive conservation programs are required to reduce inbreeding and increase the effective population size of this local breed cattle.
Author Contributions
Conceptualization, N.Z.; methodology, V.V. and N.Z.; software, A.A.; investigation, O.R. and V.V.; provision of samples, A.S. and A.K.; writing—original draft preparation, V.V. and T.D.; writing—review and editing, V.V., T.D. and N.Z.; supervision, project administration, and funding acquisition, N.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of the Russian Federation (theme No. FGGN-2022-0002).
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of the L.K. Ernst Federal Re-search Center for Animal Husbandry (protocol № 1B of 21 January 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA genotypes for the Dagestan Mountain breed and other cattle breeds are available on request to the corresponding author.
Acknowledgments
To conduct this study, we used the equipment of the Center for Collective Use “Bioresources and Bioengineering of Farm Animals” of L.K. Ernst Federal Research Center for Animal Husbandry.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study: in the collection, analyses, or interpretation of data; writing of the manuscript; or decision to publish the results.
References
- Ibragimov, R.E.; Dzhalalov, A.P.; Alilov, M.M. Outlook for the development of mountain beef cattle breeding in Dagestan. Sb. Nauchnyh Tr. Sev. Kavk. Nauchno-Issledovatel’skogo Inst. Zhivotnovodstva 2013, 2, 68–74. [Google Scholar]
- Dunin, I.M.; Dankvert, A.G. Directory of Breeds and Types of Farm Animals Bred in the Russian Federation (Spravochnik Porod i Tipov Sel’skohozyajstvennyh Zhivotnyh, Razvodimyh v Rossijskoj Federacii); VNIIPLEM: Moscow, Russia, 2013; pp. 1–560. [Google Scholar]
- Sadykov, M.M. Ways to increase the meat productivity of mountain cattle (Puti povysheniya myasnoj produktivnosti gorskogo skota). Gorskoe Sel’skoe Hozyajstvo 2016, 3, 167–170. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).