Abstract
The rapid advancement of medical imaging technologies has propelled the development of automated systems for the identification and classification of lung diseases. This study presents the design and implementation of an innovative image retrieval system utilizing artificial neural networks (ANNs) to enhance the accuracy and efficiency of diagnosing lung diseases. The proposed system focuses on addressing the challenges associated with the accurate recognition and classification of lung diseases from medical images, such as X-rays and CT scans. Leveraging the capabilities of ANNs, specifically convolutional neural networks (CNNs), the system aims to capture intricate patterns and features within images that are often imperceptible to human observers. This enables the system to learn discriminative representations of normal lung anatomy and various disease manifestations. The design of the system involves multiple stages. Initially, a robust dataset of annotated lung images is curated, encompassing a diverse range of lung diseases and their corresponding healthy states. Subsequently, a pre-processing pipeline is implemented to standardize the images, ensuring consistent quality and facilitating feature extraction. The CNN architecture is then meticulously constructed, with attention to layer configurations, activation functions, and optimization algorithms to facilitate effective learning and classification. The system also incorporates image retrieval techniques, enabling the efficient searching and retrieval of relevant medical images from the database based on query inputs. This retrieval functionality assists medical practitioners in accessing similar cases for comparative analysis and reference, ultimately supporting accurate diagnosis and treatment planning. To evaluate the system’s performance, comprehensive experiments are conducted using benchmark datasets, and performance metrics such as accuracy, precision, recall, and F1-score are measured. The experimental results demonstrate the system’s capability to distinguish between various lung diseases and healthy states with a high degree of accuracy and reliability. The proposed system exhibits substantial potential in revolutionizing lung disease diagnosis by assisting medical professionals in making informed decisions and improving patient outcomes. This study presents a novel image retrieval system empowered by artificial neural networks for the identification and classification of lung diseases. By leveraging advanced deep learning techniques, the system showcases promising results in automating the diagnosis process, facilitating the efficient retrieval of relevant medical images, and thereby contributing to the advancement of pulmonary healthcare practices.
1. Introduction
The field of medical imaging has witnessed remarkable progress over recent years, ushering in a new era of diagnostic accuracy and efficiency. Within this landscape, the identification and classification of lung diseases have emerged as crucial tasks that demand innovative solutions. In response to this, the present study introduces a cutting-edge image retrieval system underpinned by artificial neural networks (ANNs), particularly focusing on convolutional neural networks (CNNs), to revolutionize the diagnosis and understanding of lung diseases as Figure 1 [1,2,3,4,5].
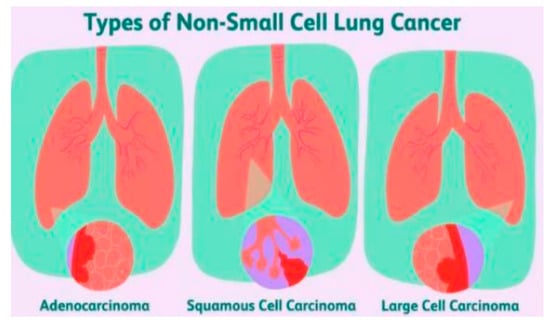
Figure 1.
Different types of non-small-cell lung cancer [5].
Lung diseases present a significant global health challenge, contributing to substantial morbidity and mortality rates. Traditional diagnostic methods often rely on the expertise of radiologists to interpret medical images such as X-rays and CT scans. However, this process can be time-consuming, subjective, and prone to human error. The advent of ANNs, fueled by the power of deep learning, has paved the way for more precise and automated analysis of medical images. CNNs, a subset of ANNs, excel in capturing intricate patterns and features within images, enabling them to identify subtle anomalies indicative of various lung diseases as Figure 2 [6,7,8,9,10,11].
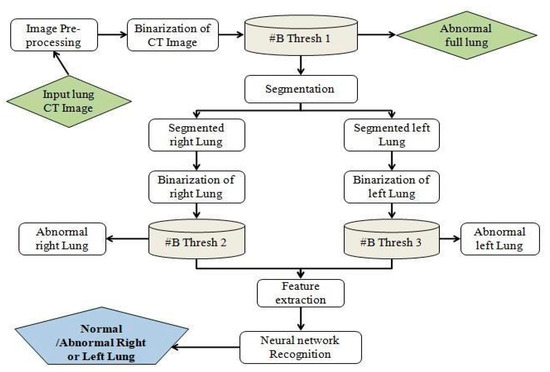
Figure 2.
Architecture of detection of lung disease [10].
The cornerstone of the proposed system lies in its ability to learn complex representations from labeled datasets containing a diverse array of lung images. The collected data are meticulously pre-processed to ensure consistency and quality, facilitating subsequent feature extraction. By leveraging the hierarchical structure of CNNs, the system effectively learns and discriminates between normal lung anatomy and various disease manifestations. Additionally, the incorporation of image retrieval techniques enhances the system’s utility by allowing medical practitioners to swiftly access relevant images for comparative analysis and reference [12,13,14,15,16].
This study aims to bridge the gap between technological innovation and medical practice by showcasing the potential of the devised image retrieval system. The implications are profound: the accurate and rapid diagnosis of lung diseases, improved patient care, and optimized treatment planning. The subsequent sections delve into the intricate design and architecture of the system, the methodologies employed for dataset curation and model training, and the comprehensive evaluations that validate its effectiveness [17,18,19,20,21,22].
2. Motivation
The motivation behind this study stems from the pressing need to advance the field of lung disease diagnosis and classification using state-of-the-art technology. Traditional methods reliant on human expertise for interpreting medical images are often time-consuming and subject to human error. As a result, misdiagnosis and delayed treatment can have serious consequences for patients suffering from lung diseases. The advent of artificial neural networks (ANNs) and deep learning has shown remarkable potential in various domains, including image analysis. Convolutional neural networks (CNNs), a subset of ANNs, have exhibited exceptional proficiency in recognizing intricate patterns and features within images. Leveraging these advancements, the proposed image retrieval system aims to significantly improve the accuracy, efficiency, and speed of diagnosing lung diseases.
3. Related Work
The related work section explores existing research and developments in the fields of medical image analysis, lung disease diagnosis, and the application of artificial neural networks (ANNs). The goal is to establish the context for the current study and highlight the gaps and opportunities for further research. Key themes in related work include the following:
- Medical image analysis using ANNs: Numerous studies have demonstrated the efficacy of ANNs, particularly CNNs, in medical image analysis. Researchers have successfully employed CNNs for tasks such as tumor detection, organ segmentation, and disease classification. These studies highlight the potential of deep learning techniques to extract meaningful features from medical images [23,24,25,26].
- Automated lung disease diagnosis: The literature reveals a growing interest in automated methods for diagnosing lung diseases. Researchers have applied machine learning and ANNs to analyze lung images for conditions like pneumonia, tuberculosis, and lung cancer. These studies emphasize the need for accurate and efficient diagnostic tools to alleviate the burden on healthcare professionals [27,28,29].
- CNNs for medical image classification: Prior research showcases the effectiveness of CNN architectures in classifying medical images. Studies have employed transfer learning, data augmentation, and specialized architectures to enhance CNN performance in diagnosing various diseases. This body of work provides insights into optimizing CNNs for specific medical tasks [30,31,32,33].
- Dataset creation and augmentation: The curation of annotated medical image datasets is crucial for training robust models. Research in this area highlights the challenges and strategies for creating diverse and representative datasets. Techniques such as data augmentation, synthetic image generation, and expert annotations have been explored to address dataset limitations [34,35,36].
- Image retrieval in medical imaging: Studies on image retrieval systems within medical imaging focus on improving access to relevant images for healthcare practitioners. These systems assist in diagnosis and treatment planning by retrieving similar cases from databases. Researchers have investigated content-based image retrieval methods and semantic indexing to enhance retrieval accuracy.
- Integration of clinical expertise: Related work also emphasizes the importance of incorporating clinical expertise into automated diagnostic systems. Collaborative efforts between medical professionals and computer scientists are essential for developing tools that align with real-world clinical practices and support healthcare decision making.
- Challenges and future directions: The existing literature acknowledges challenges such as the interpretability of deep learning models, generalization to diverse patient populations, and regulatory approval for clinical use. These challenges offer opportunities for future research, including model explainability techniques, large-scale validation studies, and compliance with medical standards.
The related work demonstrates a growing interest in leveraging ANNs, particularly CNNs, to automate the diagnosis of lung diseases through medical image analysis. The studies underscore the potential of deep learning techniques to enhance diagnostic accuracy and efficiency. However, the need for robust datasets, the integration of expert knowledge, and addressing challenges in model deployment remain critical areas for further exploration. The current study contributes by designing an innovative image retrieval system that leverages ANNs for accurate lung disease identification and classification while addressing these challenges [37,38].
4. Materials and Methods
In its later stages, lung cancer has a rather slow rate of recovery. If accurate early detection could be achieved, lung cancer survival rates might be greatly increased. Finding lung cancer early is essential for human health. Early lung cancer detection is an exciting area of study for experts in lung cancer diagnosis. The proposed method, which comprises two steps, seeks to detect lung cancer at an early stage. Image capture, binarization, pre-processing, thresholding, division, feature extraction, and brain organization identification are only a few of the innovations incorporated into the suggested method. The recovered properties are used to build the framework of the brain, which is subsequently scanned for signs of malignancy or benignity. The proposed technique has a precision of 94%, and the provided framework generates passable outcomes.
5. Results and Discussion
- A.
- Training and Testing
The neural network is then organized using the extracted attributes. The proposed framework aims to establish which lung is impacted, the left or right. In any case, the only values used to produce affirmative examples (cancerous left lung) are 0 and 1. Only one offers positive examples (malignant right lung), whereas the others all do so in a negative way. The framework designs the organization using 20 CT scans of the lungs, ensuring that the system correctly diagnoses lung cancer. The frameworks categorize the malignant CT scan images after processing and identify the afflicted lung. As indicated in Figure 3 the framework has been assessed for both positive and negative outcomes and is deemed to be acceptable. Two criteria are used to determine whether the suggested framework is effective: neural networks and the Binarization Methods Technique. It provides nearly 100% of the ideal results for this framework.
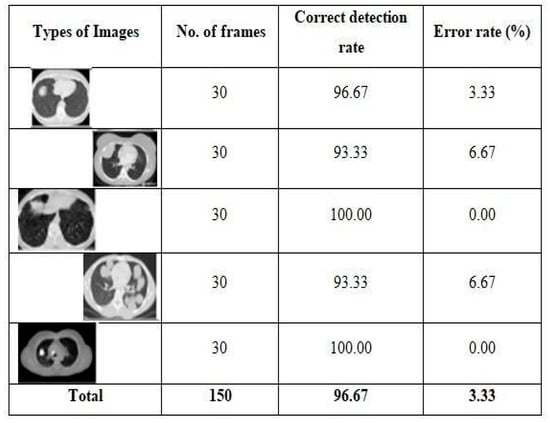
Figure 3.
Testing results on different types of images.
- B.
- Equalization of histograms
Histogram A computation, known as equalization, uses two-dimensional pixel exhibits to address digital images. Each pixel reflects the brightness or shade of the image at any given instant. A picture with a balanced distribution of intelligence levels across the entire range of grandeur is produced using histogram equalization. Programming with MATLAB is a top-notch way to convey technical concepts including computation, visualization, and programming. It enables calculations across many different fields. As demonstrated in Figure 3, the pre-processing method removes noise and obscuration from CT images and yields an unmistakable result.
Salt and pepper noise as well as the median filter are employed. The median filter offers higher noise reduction with less blurring when compared to direct smoothing channels of the same size. From the yield of the pre-handled approach to CT images, the edge is used to turn grayscale images into binary images. In this instance, the histogram equalization technique is applied, and the result is a thresholding yield. Division and pre-processing are two instances of components that cover the entire image. Given the significance of cellular breakdown in the segmentation of the lung picture, many calculations are made. To achieve outstanding outcomes, the calculation’s determination and computations must be made with extreme caution based on the input photos. Even if lung research methods have improved over the past ten years, more can still be done to improve the means of evaluation and distinction. Since lung cancer is a serious condition, early detection is crucial. The most difficult task is without a doubt finding lung cancer. Based on writing audits, a variety of methods are used to detect lung cancer, but each one has unique limitations. The initial stage of our suggested methodology involves balanced thresholding, followed by feature extraction, and, ultimately, the development and testing of the neural structure using the extracted features. As the framework nears completion, we may declare that it has achieved or exceeded all of its original objectives. With a general success rate of 94 percent, the suggested framework assessed 67 different lung CT scans and produced findings that confirmed its predictions.
6. Conclusions
In conclusion, this study has presented a comprehensive approach to addressing the challenges associated with noise reduction in image processing, particularly within the context of the construction domain. The utilization of the two-dimensional median filter, medfilt2, has demonstrated its efficacy in enhancing image quality while preserving essential edge details, a crucial consideration in fields like medical imaging and construction analysis. This study’s findings underscore the superiority of the median filter over convolution-based techniques when the objective is to minimize image disturbance while safeguarding edge integrity. The nonlinear nature of the median filter ensures that extreme noise values are effectively suppressed, resulting in clearer and more reliable images. This property is of paramount importance in applications where accurate interpretation and subsequent analysis are imperative. By applying the median filter within the construction process, this study contributes to the optimization of image quality, setting the stage for improved analysis and decision making. The noise reduction technique employed here prepares images for further investigation, enhancing the accuracy of subsequent measurements and assessments. As a whole, this research sheds light on the significance of noise reduction techniques in image processing and underscores the value of the median filter in scenarios where maintaining edge fidelity is essential. The findings of this study have direct implications for various fields, from medical imaging to construction analysis, where image quality directly impacts the accuracy and reliability of outcomes. In future research, exploring the integration of advanced noise reduction techniques and evaluating their performance across diverse datasets could further enrich our understanding of image enhancement. Additionally, investigating the combination of noise reduction methods with other image processing techniques holds promise for achieving even more robust results.
Author Contributions
Conceptualization, A.P.S.; methodology, A.S.; software; validation, A.K.; formal analysis, A.K.; investigation, A.K.; resources, A.K. and S.Y.; data curation, H.A., S.Y. and M.G. All authors have read and agreed to the published version of the manuscript.
Funding
There is no funding provided for the research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data is itself available in the present study.
Conflicts of Interest
There is no conflict of interest among the authors.
References
- Er, O.; Yumusak, N.; Temurtas, F. Chest diseases diagnosis using artificial neural networks. Expert Syst. Appl. 2010, 37, 7648–7655. [Google Scholar] [CrossRef]
- El-Solh, A.A.; Hsiao, C.-B.; Goodnough, S.; Serghani, J.; Grant, B.J.B. Predicting active pulmonary tuberculosis using an artificial neural network. Chest 1999, 116, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Ashizawa, K.; Ishida, T.; MacMahon, H.; Vyborny, C.J.; Katsuragawa, S.; Doi, K. Artificial neural networks in chest radiography: Application to the differential diagnosis of interstitial lung disease. Acad. Radiol. 2005, 11, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.M.D.; Pereira, B.B.; de Seixas, J.M. Neural networks: An application for predicting smear negative pulmonary tuberculosis. In Proceedings of the Statistics in the Health Sciences, Liège, Belgium, October 2004. [Google Scholar]
- Avni, U.; Greenspan, H.; Konen, E.; Sharon, M.; Goldberger, J. X-ray categorization and retrieval on the organ and pathology level, using patch-based visual words. IEEE Trans. Med. Imaging 2011, 30, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Karargyris, A.; Candemir, S. Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging 2014, 33, 233–245. [Google Scholar] [CrossRef]
- Pattrapisetwong, P.; Chiracharit, W. Automatic lung segmentation in chest radiographs using shadow filter and multilevel thresholding. In Proceedings of the 2016 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Chiang Mai, Thailand, 14–17 December 2016. [Google Scholar]
- Er, O.; Sertkaya, C.; Temurtas, F.; Tanrikulu, A.C. A comparative study on chronic obstructive pulmonary and pneumonia diseases diagnosis using neural networks and artificial immune system. J. Med. Syst. 2009, 33, 485–492. [Google Scholar] [CrossRef]
- Khobragade, S.; Tiwari, A.; Pati, C.Y.; Narke, V. Automatic detection of major lung diseases u sing chest radiographs and classification by feed-forward artificial neural network. In Proceedings of the 1st IEEE International Conference on Power Electronics. Intelligent Control and Energy Systems (ICPEICES-2016), Delhi, India, 4–6 July 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1–5. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, E.B.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Albarqouni, S.; Baur, C.; Achilles, F.; Belagiannis, V.; Demirci, S.; Navab, N. Aggnet: Deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1313–1321. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Bengio, Y.; Lamblin, P.; Popovici, D.; Larochelle, H. Greedy layer-wise training of deep networks. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 3–6 December 2007; pp. 153–160. [Google Scholar]
- Avendi, M.R.; Kheradvar, A.; Jafarkhani, H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med. Image Anal. 2016, 30, 108–119. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roberts, K.; Lu, L.; Demner-Fushman, D.; Yao, J.; Summers, R.M. Learning to read chest X-rays: Recurrent neural cascade model for automated image annotation. arXiv 2016, arXiv:1603.08486. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chest X-ray 8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE CVPR 2017, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Abiyev, R.H.; Altunkaya, K. Neural network based biometric personal identification with fast iris segmentation. Int. J. Control. Autom. Syst. 2009, 7, 17–23. [Google Scholar] [CrossRef]
- Helwan, A.; Tantua, D.P. IKRAI: Intelligent knee rheumatoid arthritis identification. Int. J. Intell. Syst. Appl. 2016, 8, 18. [Google Scholar] [CrossRef][Green Version]
- Helwan, A.; Abiyev, R.H. Shape and texture features for identification of breast cancer. In Proceedings of the International Conference on Computational Biology 2016, San Francisco, CA, USA, 19–21 October 2016. [Google Scholar]
- Cilimkovic, M. Neural Networks and Back Propagation Algorithm; Institute of Technology Blanchardstown: Dublin, Ireland, 2015. [Google Scholar]
- Helwan, A.; Ozsahin, D.U.; Abiyev, R.; Bush, J. One-year survival prediction of myocardial infarction. Int. J. Adv. Comput. Sci. Appl. 2017, 8, 173–178. [Google Scholar] [CrossRef][Green Version]
- Ma’aitah, M.K.S.; Abiyev, R.; Bus, I.J. Intelligent classification of liver disorder using fuzzy neural system. Int. J. Adv. Comput. Sci. Appl. 2017, 8, 25–31. [Google Scholar]
- Cohen, M.A.; Grossberg, S. Absolute stability of global pattern formation and parallel memory storage by competitive neural networks. IEEE Trans. Syst. Man Cybern. 1983, 13, 815–826. [Google Scholar] [CrossRef]
- Abas, A.R. Adaptive competitive learning neural networks. Egypt. Inform. J. 2013, 14, 183–194. [Google Scholar] [CrossRef]
- Barreto, G.A.; Mota, J.C.M.; Souza, L.G.M.; Frota, R.A.; Aguayo, L.; Yamamoto, J.S.; Macedo, P.E.O. Competitive Neural Networks for Fault Detection and Diagnosis in 3G Cellular Systems; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Gao, F.; Yue, Z.; Wang, J.; Sun, J.; Yang, E.; Zhou, H. A novel active semisupervised convolutional neural network algorithm for sar image recognition. Comput. Intell. Neurosci. 2017, 2017, 3105053. [Google Scholar] [CrossRef]
- Rios, A.; Kavuluru, R. Convolutional neural networks for biomedical text classification: Application in indexing biomedical articles. In Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics, Atlanta, GA, USA, 9–12 September 2015; ACM: New York, NY, USA, 2015; pp. 258–267. [Google Scholar]
- Li, J.; Feng, J.; Kuo, C.-C.J. Deep convolutional neural network for latent fingerprint enhancement. Signal Process. Image Commun. 2018, 60, 52–63. [Google Scholar] [CrossRef]
- Bouvrie, J. Notes on Convolutional Neural Networks. Available online: https://cogprints.org/5869/1/cnn_tutorial.pdf (accessed on 5 February 2021).
- Hussain, S.; Anwar, S.M.; Majid, M. Segmentation of glioma tumors in brain using deep convolutional neural network. Neurocomputing 2018, 282, 248–261. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Wijnhoven, R.G.J.; de With, P.H.N. Fast training of object detection using stochastic gradient descent. In Proceedings of the International Conference on Pattern Recognition (ICPR), Tsukuba, Japan, 11–15 November 2010; pp. 424–427. [Google Scholar]
- Abiyev, R.; Altunkaya, K. Iris Recognition for Biometric Personal Identification Using Neural Networks; Lecture Notes in Computer Sciences; Springer-Verlag: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Bar, Y.; Diamant, I.; Wolf, L.; Lieberman, S.; Konen, E.; Greenspan, H. Chest pathology detection using deep learning with non-medical training. In Proceedings of the Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium, Brookly, NY, USA, 16–19 April 2015; pp. 294–297. [Google Scholar]
- Islam, M.T.; Aowal, M.A.; Minhaz, A.T.; Ashraf, K. Abnormality detection and localization in chest X-rays using deep convolutional neural networks. arXiv 2017, arXiv:1705.09850. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).