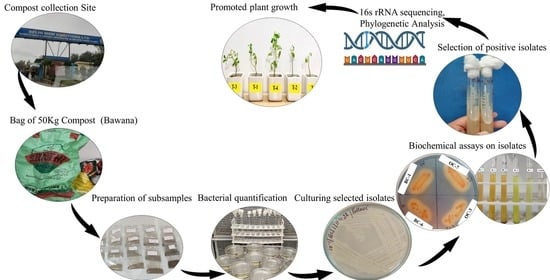
Assessing Growth-Promoting Activity of Bacteria Isolated from Municipal Waste Compost on Solanum lycopersicum L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Compost Sample Collection Sites
- IL&FS Environmental Infrastructure & Services Ltd. (IEISL): It has been a unified waste management organization since 2007. The compost plant at Okhla is operated and maintained by IL & FS Waste Management and Urban Services Ltd. (IWMUSL).
- Delhi MSW Solutions Limited (DMSWSL): DMSWSL is a unit of Hyderabad-based Ramky Enviro Engineers Ltd. and is located in Bawana Industrial Area.
2.2. Preparation of Compost Samples
2.3. Physicochemical Properties of MSWC
2.4. Isolation and Characterization of Culturable Bacteria in the Compost
2.5. Quantitative Estimation of Bacteria
2.6. Inoculum Preparation
2.7. Screening of Pure Bacteria Isolates for Assessment of PGPB Traits
2.7.1. Determining P-Solubilizing Activity of Isolates
2.7.2. Determining K-Solubilizing Activity of Isolates
2.7.3. Estimation of NH3 and HCN Production by Isolates
2.7.4. Estimation of IAA Production by Isolates
2.7.5. Estimation of Siderophore Production by Isolates
2.8. Molecular Characterization of the Isolates
2.8.1. DNA Extraction
2.8.2. 16s rRNA Amplification of Desired Genomic DNA
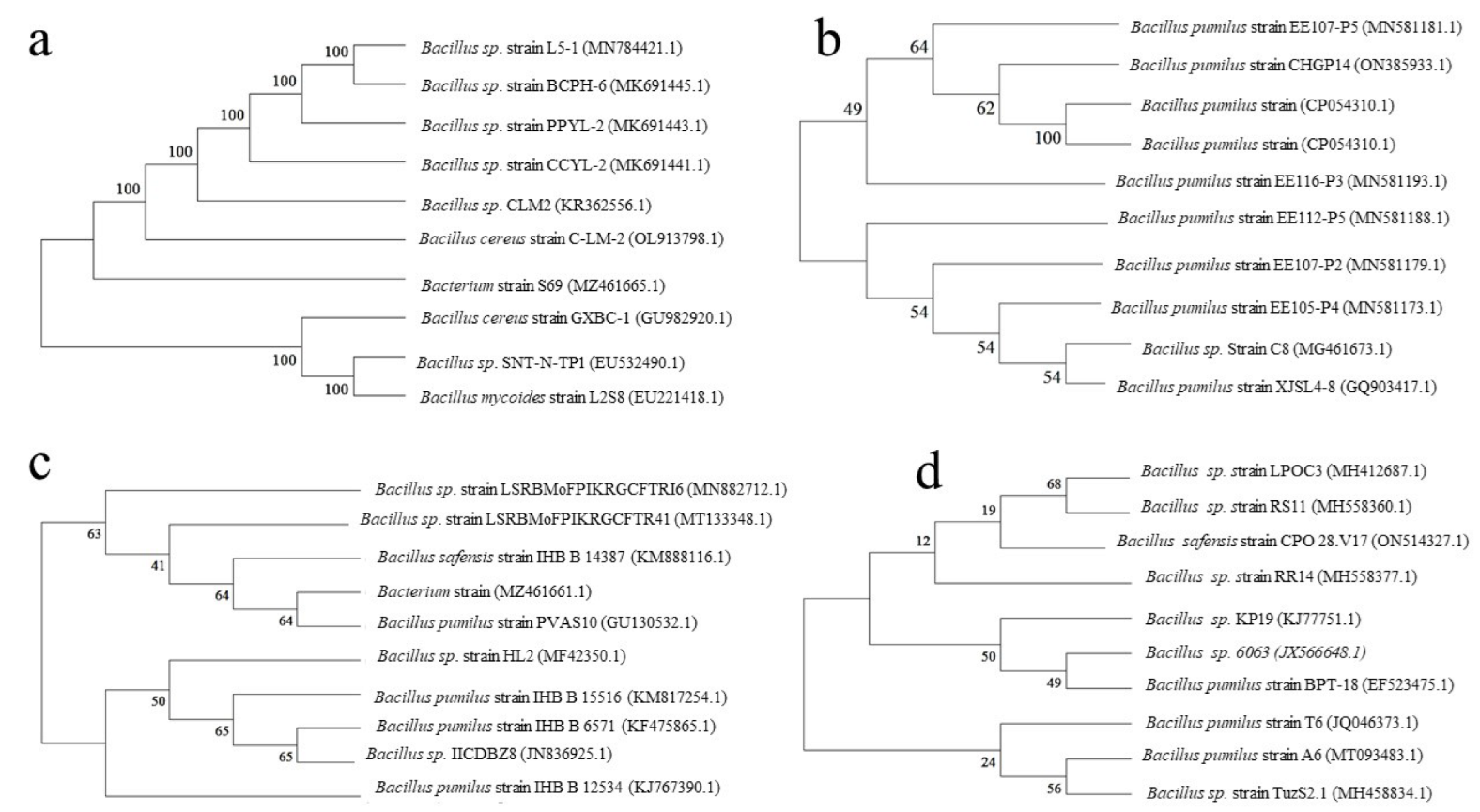
2.8.3. Phylogenetic Analysis
3. Evaluating Potential Effects of Isolates on the Growth of S. lycopersicum
3.1. Seed Procurement
3.2. Effect of Isolates on the Seed Germination Rate
3.3. Plant Material and Growth Conditions
3.4. Inoculation Effects on Growth Parameters
3.5. Harvesting and Plant Analysis
3.6. Statistical Analysis
4. Results
4.1. Compost Physiochemical Analysis
4.2. Quantifying Bacteria
4.3. Screening of Isolates for PGP Traits
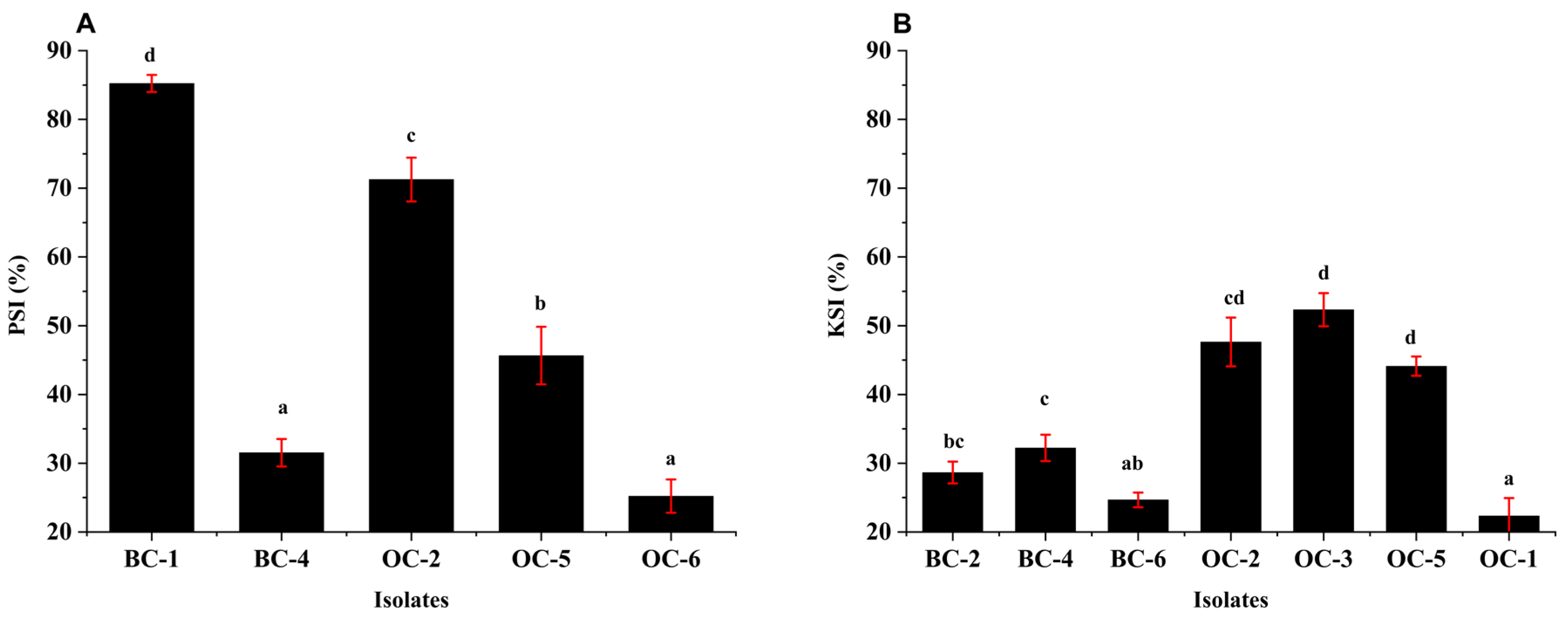
4.3.1. P-Solubilizing Potential of Isolates
4.3.2. K-Solubilizing Potential of Isolates
4.3.3. NH3 and HCN Production Potential of Isolates
4.3.4. IAA Production Potential of Isolates
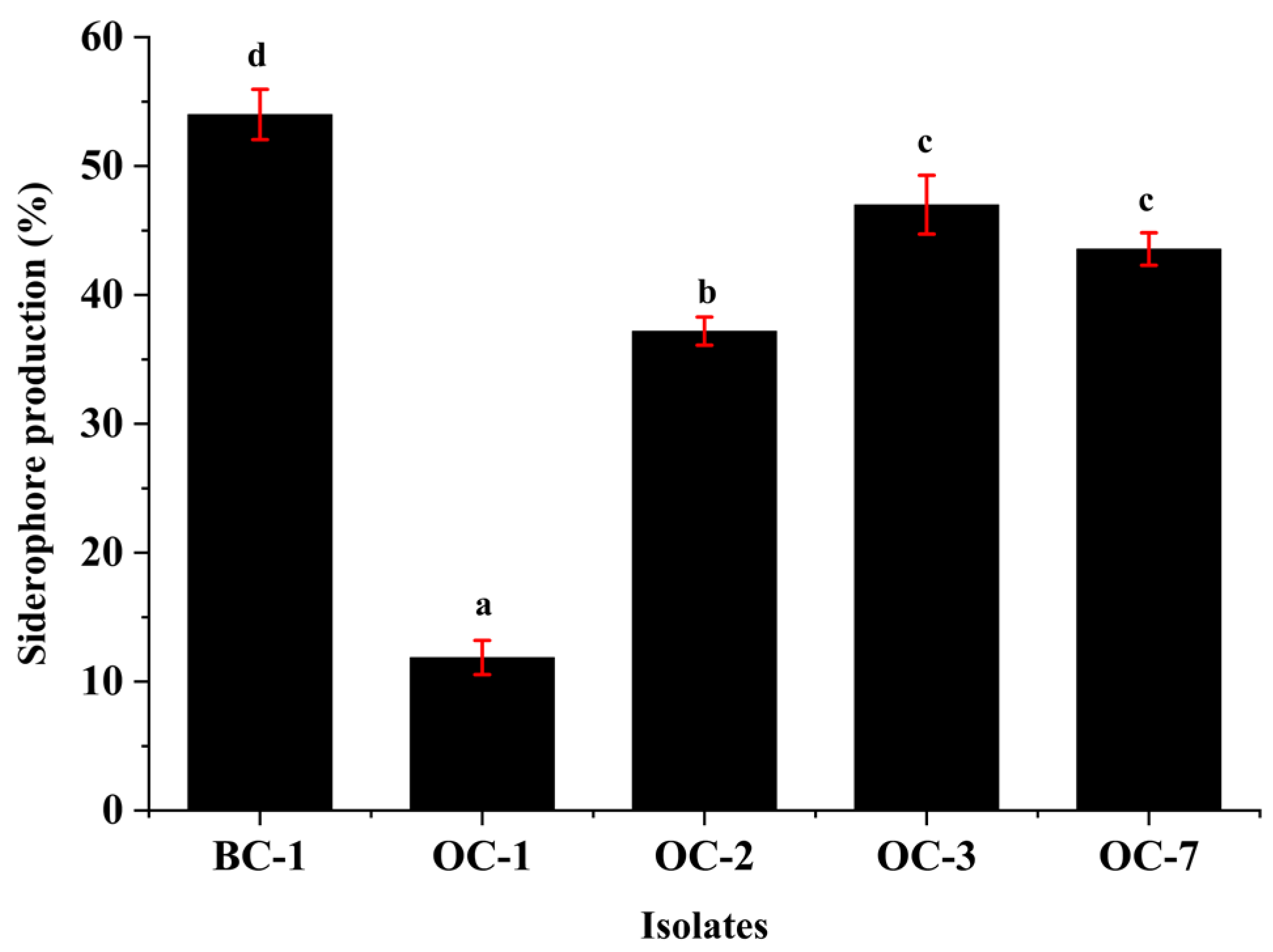
4.3.5. Siderophore Production Potential of Isolates
4.4. Molecular Identification of the Potent PGPB
4.5. Plant Growth Influenced by Inoculation of Isolates
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.M.H.; Soares, H.M.V.M.; Soares, E.V. Promising bacterial genera for agricultural practices: An insight on plant growth-promoting properties and microbial safety aspects. Sci. Total. Environ. 2019, 682, 779–799. [Google Scholar] [CrossRef] [PubMed]
- Abrar, M.M.; Shah, S.A.A.; Sun, N.; Mehmood, K.; Aziz, T.; Waqas, M.A.; Luo, Y.; Zhou, B.; Ma, X.; Xu, M.; et al. Long-term manure application enhances organic carbon and nitrogen stocks in Mollisol subsoil. Land Degrad. Dev. 2022. [Google Scholar] [CrossRef]
- Zahid, M.; Kaleem Abbasi, M.; Hameed, S.; Rahim, N. Isolation and identification of indigenous plant growth promoting rhizobacteria from Himalayan region of Kashmir and their effect on improving growth and nutrient contents of maize (Zea mays L.). Front. Microbiol. 2015, 6, 207. [Google Scholar] [CrossRef]
- Irshad, A.; Rehman, R.; Abrar, M.; Saeed, Q.; Sharif, R.; Hu, T. Contribution of Rhizobium–Legume Symbiosis in Salt Stress Tolerance in Medicago truncatula Evaluated through Photosynthesis, Antioxidant Enzymes, and Compatible Solutes Accumulation. Sustainability 2021, 13, 3369. [Google Scholar] [CrossRef]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Saeed, Q.; Xiukang, W.; Haider, F.U.; Kučerik, J.; Mumtaz, M.Z.; Holatko, J.; Naseem, M.; Kintl, A.; Ejaz, M.; Naveed, M.; et al. Rhizosphere Bacteria in Plant Growth Promotion, Biocontrol, and Bioremediation of Contaminated Sites: A Comprehensive Review of Effects and Mechanisms. Int. J. Mol. Sci. 2021, 22, 10529. [Google Scholar] [CrossRef] [PubMed]
- Koza, N.A.; Adedayo, A.A.; Babalola, O.O.; Kappo, A.P. Microorganisms in Plant Growth and Development: Roles in Abiotic Stress Tolerance and Secondary Metabolites Secretion. Microorganisms 2022, 10, 1528. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition—Current Knowledge and Future Directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef]
- Me, A.N.P.O.; Ge, A.E.A.N.E.; Farid, B.M.; Adolphe, A.; Lamine, B.M.; Noumavo, P.A.; Agbodjato, N.A.; Baba-Moussa, F.; Adjanohoun, A. Plant growth promoting rhizobacteria: Beneficial effects for healthy and sustainable agriculture. Afr. J. Biotechnol. 2016, 15, 1452–1463. [Google Scholar] [CrossRef]
- Creus, C.M. Inoculantes microbianos: Piezas de un rompecabezas que aún requiere ser ensamblado. Rev. Argent Microbiol. 2017, 49, 207–209. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Aasfar, A.; Bargaz, A.; Yaakoubi, K.; Hilali, A.; Bennis, I.; Zeroual, Y.; Meftah Kadmiri, I. Nitrogen Fixing Azotobacter Species as Potential Soil Biological Enhancers for Crop Nutrition and Yield Stability. Front. Microbiol. 2021, 12, 628379. [Google Scholar] [CrossRef]
- Dutilloy, E.; Oni, F.E.; Esmaeel, Q.; Clément, C.; Barka, E.A. Plant Beneficial Bacteria as Bioprotectants against Wheat and Barley Diseases. J. Fungi 2022, 8, 632. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, S.; Yuan, Z. Adoption of solid organic waste composting products: A critical review. J. Clean. Prod. 2020, 272, 122712. [Google Scholar] [CrossRef]
- Milinković, M.; Lalević, B.; Jovičić-Petrović, J.; Golubović-Ćurguz, V.; Kljujev, I.; Raičević, V. Biopotential of compost and compost products derived from horticultural waste—Effect on plant growth and plant pathogens’ suppression. Process. Saf. Environ. Prot. 2019, 121, 299–306. [Google Scholar] [CrossRef]
- Ho, T.T.K.; Tra, V.T.; Le, T.H.; Nguyen, N.-K.; Tran, C.-S.; Nguyen, P.-T.; Vo, T.-D.; Thai, V.-N.; Bui, X.-T. Compost to improve sustainable soil cultivation and crop productivity. Case Stud. Chem. Environ. Eng. 2022, 6, 100211. [Google Scholar] [CrossRef]
- Rath, P.P.; Das, K.; Pattanaik, S. Microbial Activity during Composting and Plant Growth Impact: A Review. J. Pure Appl. Microbiol. 2022, 16, 63–73. [Google Scholar] [CrossRef]
- Zouari, I.; Masmoudi, F.; Medhioub, K.; Tounsi, S.; Trigui, M. Biocontrol and plant growth-promoting potentiality of bacteria isolated from compost extract. Antonie van Leeuwenhoek 2020, 113, 2107–2122. [Google Scholar] [CrossRef]
- Sherpa, M.T.; Bag, N.; Das, S.; Haokip, P.; Sharma, L. Isolation and characterization of plant growth promoting rhizobacteria isolated from organically grown high yielding pole type native pea (Pisum sativum L.) variety Dentami of Sikkim, India. Curr. Res. Microb. Sci. 2021, 2, 100068. [Google Scholar] [CrossRef]
- Kang, Y.; Shen, M.; Wang, H.; Zhao, Q. A possible mechanism of action of plant growth-promoting rhizobacteria (PGPR) strain Bacillus pumilus WP8 via regulation of soil bacterial community structure. J. Gen. Appl. Microbiol. 2013, 59, 267–277. [Google Scholar] [CrossRef]
- Win, K.T.; Oo, A.Z.; Ohkama-Ohtsu, N.; Yokoyama, T. Bacillus Pumilus Strain TUAT-1 and Nitrogen Application in Nursery Phase Promote Growth of Rice Plants under Field Conditions. Agronomy 2018, 8, 216. [Google Scholar] [CrossRef]
- Aung, H.P.; Djedidi, S.; Oo, A.Z.; Aye, Y.S.; Yokoyama, T.; Suzuki, S.; Sekimoto, H.; Bellingrath-Kimura, S.D. Growth and 137Cs uptake of four Brassica species influenced by inoculation with a plant growth-promoting rhizobacterium Bacillus pumilus in three contaminated farmlands in Fukushima prefecture, Japan. Sci. Total. Environ. 2015, 521–522, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Kang, Y.J.; Wang, H.L.; Zhang, X.S.; Zhao, Q.X. Effect of Plant Growth-promoting Rhizobacteria (PGPRs) on plant growth, yield, and quality of tomato (Lycopersicon esculentum Mill.) under simulated seawater irrigation. J. Gen. Appl. Microbiol. 2012, 58, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Venancio, W.S.; Gomes, J.M.; Nakatani, A.S.; Hungria, M.; Araujo, R.S. Lettuce Production under Reduced Levels of N-fertilizer in the Presence of Plant Growth-promoting Bacillus spp. Bacteria. J. Pure Appl. Microbiol. 2019, 13, 1941–1952. [Google Scholar] [CrossRef]
- Schwarz, D.; Thompson, A.J.; Klã¤Ring, H.-P. Guidelines to use tomato in experiments with a controlled environment. Front. Plant Sci. 2014, 5, 625. [Google Scholar] [CrossRef]
- Walldey, A.; Black, I.A. Estimation of soil organic carbon by the chromic acid titration method. Soil. Sci. 1934, 37, 29–38. [Google Scholar]
- Asija, G.L.; Subbiah, B.V.; Asija, G.L. A Rapid Procedure for the Estimation of Available Nitrogen in Soils. Curr. Sci. 1956, 25, 259–260. [Google Scholar]
- Bray, R.H.; Kurtz, L.T. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- Ashworth, J.; Mrazek, K. “Modified Kelowna” test for available phosphorus and potassium in soil. Commun. Soil Sci. Plant Anal. 1995, 26, 731–739. [Google Scholar] [CrossRef]
- Behera, S.K.; Shukla, A.K. Spatial Distribution of Surface Soil Acidity, Electrical Conductivity, Soil Organic Carbon Content and Exchangeable Potassium, Calcium and Magnesium in Some Cropped Acid Soils of India. Land Degrad. Dev. 2015, 26, 71–79. [Google Scholar] [CrossRef]
- Chauhan, A.; Jindal, T. Microbiological Methods for Water, Soil and Air Analysis. In Microbiological Methods for Environment, Food and Pharmaceutical Analysis; Springer International Publishing: Cham, Switzerland, 2020; pp. 93–196. [Google Scholar]
- Mehta, S.; Nautiyal, C.S. An Efficient Method for Qualitative Screening of Phosphate-Solubilizing Bacteria. Curr. Microbiol. 2001, 43, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, J.; Guo, J. Two Phosphate- and Potassium-solubilizing Bacteria Isolated from Tianmu Mountain, Zhejiang, China. World J. Microbiol. Biotechnol. 2006, 22, 983–990. [Google Scholar] [CrossRef]
- Goswami, D.; Dhandhukia, P.; Patel, P.; Thakker, J.N. Screening of PGPR from saline desert of Kutch: Growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 2014, 169, 66–75. [Google Scholar] [CrossRef]
- Lahlali, R.; Mchachti, O.; Radouane, N.; Ezrari, S.; Belabess, Z.; Khayi, S.; Mentag, R.; Tahiri, A.; Barka, E.A. The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits. Agronomy 2020, 10, 1814. [Google Scholar] [CrossRef]
- Ehmann, A. The van URK-Salkowski reagent—A sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. A 1977, 132, 267–276. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Dimkpa, C. Endocytobiosis and Cell Research Microbial siderophores: Production, detection and application in agriculture and environment. Endocytobiosis Cell Res. 2016, 27, 2. [Google Scholar]
- Lau, E.T.; Tani, A.; Khew, C.Y.; Chua, Y.Q.; Hwang, S.S. Plant growth-promoting bacteria as potential bio-inoculants and biocontrol agents to promote black pepper plant cultivation. Microbiol. Res. 2020, 240, 126549. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; Volume 1–3. [Google Scholar]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical Evaluation of Two Primers Commonly Used for Amplification of Bacterial 16S rRNA Genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Martínez-Cano, B.; García-Trejo, J.F.; Sánchez-Gutiérrez, A.E.; Toledano-Ayala, M.; Soto-Zarazúa, G.M. Isolation and Characterization of Plant Growth-Promoting Compost Bacteria That Improved Physiological Characteristics in Tomato and Lettuce Seedlings. Agriculture 2021, 12, 3. [Google Scholar] [CrossRef]
- Islam, S.; Akanda, A.M.; Prova, A.; Islam, M.T.; Hossain, M.M. Isolation and Identification of Plant Growth Promoting Rhizobacteria from Cucumber Rhizosphere and Their Effect on Plant Growth Promotion and Disease Suppression. Front. Microbiol. 2016, 6, 1360. [Google Scholar] [CrossRef]
- Sirajuddin; Khan, A.; Ali, L.; Chaudhary, H.J.; Munis, M.F.H.; Bano, A.; Masood, S. Bacillus pumilus alleviates boron toxicity in tomato (Lycopersicum esculentum L.) due to enhanced antioxidant enzymatic activity. Sci. Hortic. 2016, 200, 178–185. [Google Scholar] [CrossRef]
- Masood, S.; Zhao, X.Q.; Shen, R.F. Bacillus pumilus increases boron uptake and inhibits rapeseed growth under boron supply irrespective of phosphorus fertilization. AoB Plants 2019, 11, plz036. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Tran, Q.N.M.; Mimoto, H.; Koyama, M.; Nakasaki, K. Lactic acid bacteria modulate organic acid production during early stages of food waste composting. Sci. Total. Environ. 2019, 687, 341–347. [Google Scholar] [CrossRef]
- Chinakwe, E.; Ibekwe, V.; Ofoh, M.; Nwogwugwu, N.; Adeleye, S.; Chinakwe, P.; Nwachukwu, I.; Ihejirika, C. Effect of Temperature Changes on the Bacterial and Fungal Succession Patterns during Composting of Some Organic Wastes in Greenhouse. J. Adv. Microbiol. 2019, 15, 1–10. [Google Scholar] [CrossRef]
- Tondello, A.; Fasolo, A.; Marcato, S.; Treu, L.; Bonato, T.; Zanardi, W.; Concheri, G.; Squartini, A.; Baldan, B. Characterization of bacterial communities isolated from municipal waste compost and screening of their plant-interactive phenotypes. Sci. Total. Environ. 2022, 806, 150592. [Google Scholar] [CrossRef]
- Masood, S.; Zhao, X.Q.; Shen, R.F. Bacillus pumilus promotes the growth and nitrogen uptake of tomato plants under nitrogen fertilization. Sci. Hortic. 2020, 272, 109581. [Google Scholar] [CrossRef]
- Cendales, T.C.; González, C.A.R.; Cuásquer, C.P.V.; Alzate, O.A.T.; Rodríguez, A.H. Bacillus effect on the germination and growth of tomato seedlings (Solanum lycopersicum L.). Acta Biológica Colomb. 2017, 22, 37–44. [Google Scholar] [CrossRef]
- Salehin, A.; Puri, R.; Hafiz, H.R.; Itoh, K. Effect of Co-Inoculation of Bacillus sp. Strain with Bacterial Endophytes on Plant Growth and Colonization in Tomato Plant (Solanum lycopersicum). Microbiol. Res. 2021, 12, 480–490. [Google Scholar] [CrossRef]
- Ricci, E.; Schwinghamer, T.; Fan, D.; Smith, D.L.; Gravel, V. Growth promotion of greenhouse tomatoes with Pseudomonas sp. and Bacillus sp. biofilms and planktonic cells. Appl. Soil Ecol. 2019, 138, 61–68. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Alqarawi, A.; Abd_Allah, E.; Hashem, A. Phytohormones and Beneficial Microbes: Essential Components for Plants to Balance Stress and Fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef]
- Mahapatra, S.; Yadav, R.; Ramakrishna, W. Bacillus subtilis impact on plant growth, soil health and environment: Dr. Jekyll and Mr. Hyde. J. Appl. Microbiol. 2022, 132, 3543–3562. [Google Scholar] [CrossRef]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Hanif, A.; Wu, L.; Colman, M.V.; Gao, X. Plant Growth Promotion by Volatile Organic Compounds Produced by Bacillus subtilis SYST2. Front. Microbiol. 2017, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Saeid, A.; Prochownik, E.; Dobrowolska-Iwanek, J. Phosphorus Solubilization by Bacillus Species. Molecules 2018, 23, 2897. [Google Scholar] [CrossRef]
- Rajawat, M.V.S.; Ansari, W.A.; Singh, D.; Singh, R. Potassium Solubilizing Bacteria (KSB). In Microbial Interventions in Agriculture and Environment; Springer: Singapore, 2019; pp. 189–209. [Google Scholar] [CrossRef]
- Qureshi, S.A.; Qureshi, R.A.; Sodha, A.B.; Tipre, D.R.; Dave, S.R. Bioextraction Dynamics of Potassium from Feldspar by Heterotrophic Microorganisms Isolated from Ceramic and Rhizospheric Soil. Geomicrobiol. J. 2017, 35, 127–131. [Google Scholar] [CrossRef]
- Raji, M.; Thangavelu, M. Isolation and screening of potassium solubilizing bacteria from saxicolous habitat and their impact on tomato growth in different soil types. Arch. Microbiol. 2021, 203, 3147–3161. [Google Scholar] [CrossRef]
- Joseph, B.; Patra, R.R.; Lawrence, R. Characterization of plant growth-promoting rhizobacteria associated with chickpea (Cicer arietinum L.). Int. J. Plant Prod. 2007, 1, 141–152. [Google Scholar] [CrossRef]
- Pathak, E.; Sanjyal, A.; Regmi, C.R.; Paudel, S.; Shrestha, A. Screening of Potential Plant Growth Promoting Properties of Bacillus Species Isolated from Different Regions of Nepal. Nepal J. Biotechnol. 2021, 9, 79–84. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, T.; Lapanje, A. Hydrogen Cyanide in the Rhizosphere: Not Suppressing Plant Pathogens, but Rather Regulating Availability of Phosphate. Front. Microbiol. 2016, 7, 1785. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, N.; El-Nahrawy, S. Siderophore production by Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738 and their efficacy in controlling Cephalosporium maydis in maize plant. Arch. Microbiol. 2021, 203, 1195–1209. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.D.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.-H. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed]
- Kai, M. Diversity and Distribution of Volatile Secondary Metabolites Throughout Bacillus subtilis Isolates. Front. Microbiol. 2020, 11, 559. [Google Scholar] [CrossRef]
- Gautam, S.; Chauhan, A.; Sharma, R.; Sehgal, R.; Shirkot, C. Potential of Bacillus amyloliquefaciens for biocontrol of bacterial canker of tomato incited by Clavibacter michiganensis ssp. michiganensis. Microb. Pathog. 2019, 130, 196–203. [Google Scholar] [CrossRef]
- Lin, C.; Tsai, C.-H.; Chen, P.-Y.; Wu, C.-Y.; Chang, Y.-L.; Yang, Y.-L.; Chen, Y.-L. Biological control of potato common scab by Bacillus amyloliquefaciens Ba01. PLoS ONE 2018, 13, e0196520. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ai, C.; Xin, L.; Zhou, G. The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur. J. Soil Biol. 2011, 47, 138–145. [Google Scholar] [CrossRef]
- Zalma, S.; El-Sharoud, W. Diverse thermophilic Bacillus species with multiple biotechnological activities are associated within the Egyptian soil and compost samples. Sci. Prog. 2021, 104, 003685042110552. [Google Scholar] [CrossRef]
- Sreevidya, M.; Gopalakrishnan, S. Direct and indirect plant growth-promoting abilities of Bacillus species on chickpea, isolated from compost and rhizosphere soils. Org. Agric. 2015, 7, 31–40. [Google Scholar] [CrossRef]
- Selvamani, K.; Annadurai, V.; Soundarapandian, S. Improved co-composting of poultry manure with complementary consortium of indigenous Bacillus spp. 3 Biotech 2019, 9, 215. [Google Scholar] [CrossRef] [PubMed]

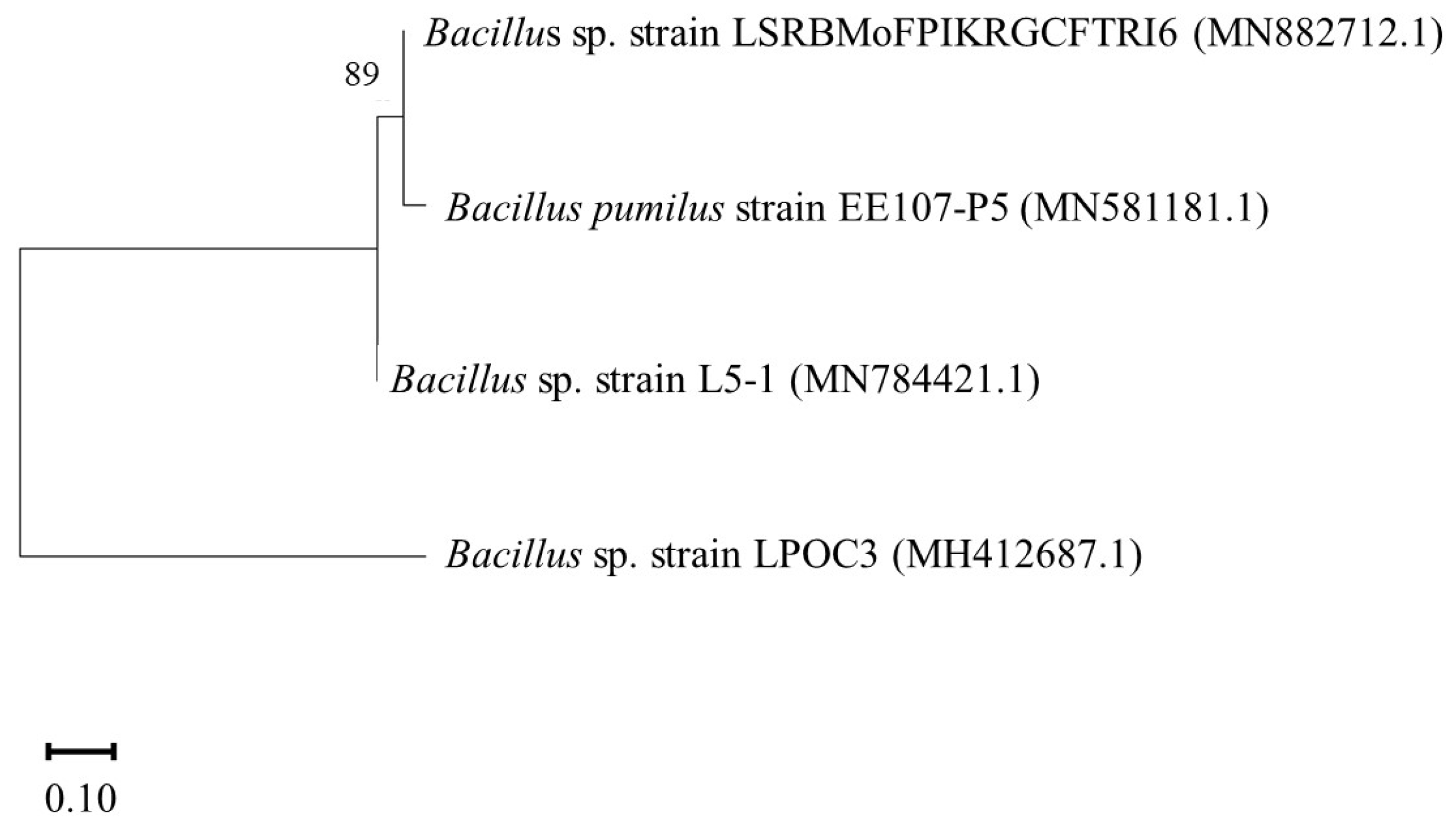

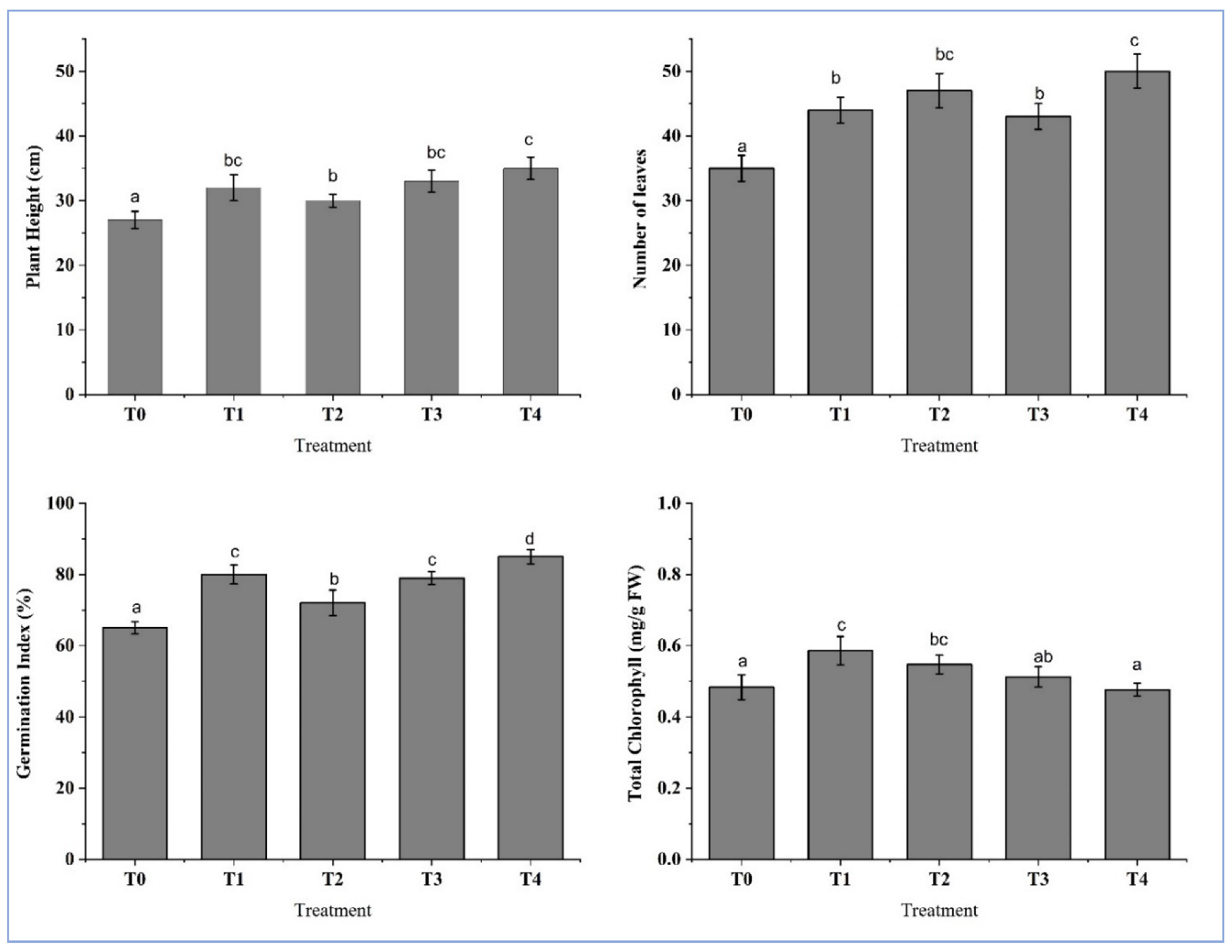
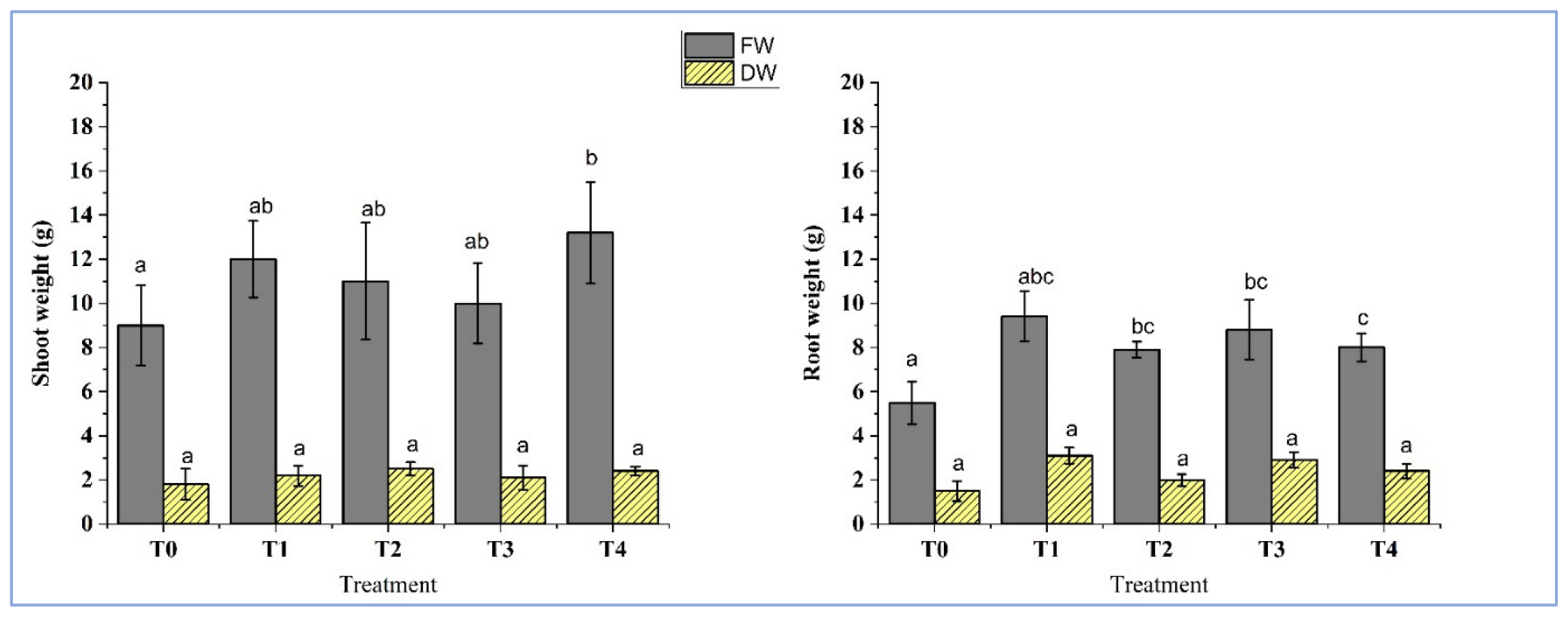
| Treatment | Description |
|---|---|
| T-0 | Control Soil |
| T-1 | Soil inoculated with isolate BC-1 |
| T-2 | Soil inoculated with isolate BC-4 |
| T-3 | Soil inoculated with isolate OC-3 |
| T-4 | Soil inoculated with isolate OC-7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhardwaj, P.; Chauhan, A.; Ranjan, A.; Mandzhieva, S.S.; Minkina, T.; Mina, U.; Rajput, V.D.; Tripathi, A. Assessing Growth-Promoting Activity of Bacteria Isolated from Municipal Waste Compost on Solanum lycopersicum L. Horticulturae 2023, 9, 214. https://doi.org/10.3390/horticulturae9020214
Bhardwaj P, Chauhan A, Ranjan A, Mandzhieva SS, Minkina T, Mina U, Rajput VD, Tripathi A. Assessing Growth-Promoting Activity of Bacteria Isolated from Municipal Waste Compost on Solanum lycopersicum L. Horticulturae. 2023; 9(2):214. https://doi.org/10.3390/horticulturae9020214
Chicago/Turabian StyleBhardwaj, Pallavi, Abhishek Chauhan, Anuj Ranjan, Saglara S. Mandzhieva, Tatiana Minkina, Usha Mina, Vishnu D. Rajput, and Ashutosh Tripathi. 2023. "Assessing Growth-Promoting Activity of Bacteria Isolated from Municipal Waste Compost on Solanum lycopersicum L." Horticulturae 9, no. 2: 214. https://doi.org/10.3390/horticulturae9020214
APA StyleBhardwaj, P., Chauhan, A., Ranjan, A., Mandzhieva, S. S., Minkina, T., Mina, U., Rajput, V. D., & Tripathi, A. (2023). Assessing Growth-Promoting Activity of Bacteria Isolated from Municipal Waste Compost on Solanum lycopersicum L. Horticulturae, 9(2), 214. https://doi.org/10.3390/horticulturae9020214