Abstract
The popular ornamental plant Petunia is also a valuable model plant in tissue culture. Cellular conversions during differentiation and regeneration have been investigated using various combinations of phytohormones; however, studies on genes for reprogramming toward desired tissue identities have been limited. In this study, we isolated Petunia protoplasts and cultured them in the callus, rooting, or shooting stages, which were used to establish the optimal protoplast culture conditions and to identify genes that epigenetically function as tissue identifiers. The optimal conditions for plasmolysis and enzyme digestion to obtain healthy protoplasts were compared, in which combinations of Viscozyme, Celluclast, and Pectinex (VCP) enzymes were more efficient in isolating protoplasts when followed by 21 to 25% sucrose purification and washing processes. The filtered and washed protoplasts started to divide at 1 day and developed into colonies after 3 weeks of culture, which showed higher efficiency in the Murashige and Skoog (MS) salt culture media compared to that in the Kao and Michayluk (KM) salt media. The pluripotent colonies formed calli on the solid medium supplemented with 3% sucrose after 4 weeks, and were destined to the same cell mass, rooting, or shooting on the regeneration medium. Three epigenetic controllers, ATXR2, ATX4A, and ATX4B, were highly expressed in calli, shoots, and organs of shoots and roots, respectively, confirming that dedifferentiation and regeneration of tissue identity is plastic.
1. Introduction
A protoplast is a plant cell lacking a cell wall, which is removed using fungal enzymes such as cellulase [1,2]. Under the influence of various hormones, almost all plant cells have the potential to regenerate through cell dedifferentiation and differentiation [1]. Leaf mesophyll is a common source of protoplasts and a universal system for the transient expression of plant genes and plant regeneration [2,3]; however, protoplast isolation depends on the explant and plant species [4,5]. Therefore, it is necessary to establish the conditions for protoplast culture and shoot regeneration when different plant cultivars or species are used. Initially, the protoplast was used for symmetric or asymmetric fusion of cells from the same or different species [6]. Recently, however, protoplast culture has drawn attention owing to its role in creating DNA-free plants using a CRISPR-related genome editing technology [7].
Research on Petunia protoplasts was started in the late 1970s to facilitate their isolation and culture. Petunia protoplasts were a suitable model system to characterize the effects of explants, genotypes, and digestion enzymes [5,8,9]. Efficient protoplast isolation from leaf mesophyll cells and its transient gene expression protocols have already been determined for the cultivar Madness Midnight by our group [10]; the yields of mesophyll protoplasts have been found to be similar to those previously reported in Petunia genotypes [4]. This suggested that the cultivar and its protoplast system could be helpful in studying the genetics of phenotypic traits. With the development of technology, protoplast culture for the regeneration of various plants has become more efficient [2,11,12]. This efficient culture system can be the foundation for the research of other transient gene transformations, transgenic plants, or target-directed mutagenesis. Using the cultivar Madness Midnight and its protoplast system, the regeneration of genome-edited Petunia plants has also been successfully demonstrated [7].
Genes involved in callus, root, or shoot formation have been reported to play independent roles in corresponding differentiation processes. Generally, callus induction and proliferation are under the genetic control of a set of developmental regulators; the identification of such genes and regulatory elements will help in the development of in vitro systems in Petunia. According to recent studies by Lee et al. [13,14], Arabidopsis trithorax-related 2 (ATXR2) and Arabidopsis trithorax 4 (ATX4) were found to be dynamically upregulated during callus and shoot formation, respectively. Arabidopsis atxr2-defficient mutants showed defects in the formation of calli and adventitious roots, indicating their involvement in epigenetic regulation of cellular dedifferentiation [13]. The ATX4 plays a vital role in regeneration by accumulating H3K4me3, which participates in shoot identity establishment. Arabidopsis atx4-defficient mutants exhibited enhanced callus and repressed shoot identity [15].
Although previously established protoplast isolation and regeneration protocols which are specific to the genome engineering of Petunia [7,10] are available, this study focused on developing simple and efficient protoplast isolation and protoplast-derived cell proliferation and regeneration methods, as well as investigating the molecular events involved in the dedifferentiation and regeneration phases. The results obtained from this study may be helpful in studying the functional analysis of developmental regulators involved in the cellular dedifferentiation of Petunia parental genomes.
2. Results and Discussion
2.1. Enzyme Combinations for Protoplast Isolation
P × hybrida cv. Madness Midnight leaf protoplast is known to have higher transformation capacity, which allows for efficient protoplast isolation and optimized culture conditions [7]. After 3 h of shaking incubation, protoplasts were released into the enzyme solution. Depending on the enzyme combinations, the yield of protoplasts varied between 1.82 × 106 and 6.9 × 106 cells per gram fresh weight (Figure 1A). For Macerozyme R and Cellulase R-1 (MC) enzyme combinations (EC 1, EC 2, and EC 3), the range of protoplast yield was 1.8 × 106–3.2 × 106 cells per gram fresh weight, while the range of protoplast was 3.8 × 106–6.9 × 106 for combinations of Viscozyme, Celluclast, and Pectinex (VCP) enzymes (EC 4, EC 5, EC 6, and EC 7). More protoplasts were obtained with the combinations of VCP enzymes than with those of MC enzymes, indicating that the VCP enzyme combinations were more efficient in isolating Petunia protoplasts. When the VCP enzyme concentration was increased, protoplast yield increased gradually; however, broken cells were detected at higher enzyme concentrations, as shown at EC 7. The lower number of cells at EC 7 is presumed to be due to damaged protoplasts resulting from the harsh removal of cell walls using higher enzyme concentrations.
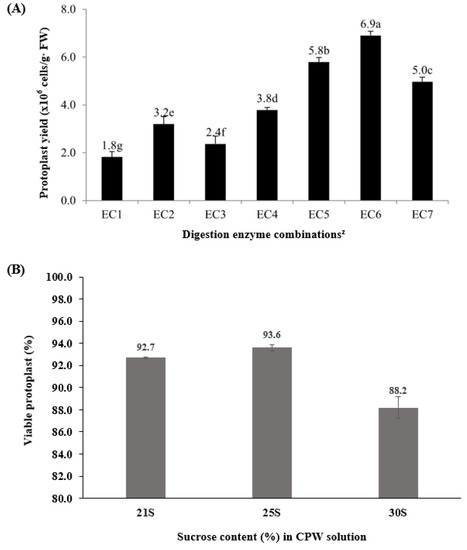
Figure 1.
The calculated protoplast yield after enzyme digestion (A) and sucrose purification (B). The percentage of viable protoplasts was calculated by counting the green-dyed cells after reaction in the 0.01% FDA solution. Error bars indicate standard errors of the means and different letters indicate significant differences at p ≤ 0.05 according to Duncan’s multiple range test (DMRT). z Treatments with digestion enzymes comprised seven groups of enzyme combinations (EC). EC 1: 1.0% Cellulase R-10TM (CR) + 0.2% Macerozyme R-10TM (MR); EC 2: 1.5% CR + 0.5% MR; EC 3: 2.0% CR + 0.8% MR; EC 4: 0.2% CelluclastTM (Ce) + 0.2% PectinexTM (PE) + 0.4% ViscozymeTM (Vz); EC 5: 0.4% Ce + 0.4% PE + 0.8% Vz; EC 6: 0.6% Ce + 0.6% PE + 1.2% Vz; EC 7: 0.8% Ce + 0.8% PE + 1.6% Vz.
Various factors affect the isolation of protoplasts, such as osmotic pressure, enzyme type, digestion time, incubation temperature, shaking, and pH value. To obtain high yields and highly viable protoplasts, a shorter enzyme digestion time was suggested. The currently established method will be applicable to the isolation of Petunia protoplasts, which was further verified by cell wall formation from the Fluorescein diacetate (FDA) test (Supplementary Figure S2) [4,10].
2.2. Protoplast Purification and Protoplast Viability
Following centrifugation at 600 rpm for 5 min, the protoplasts were gathered in the middle of the two different solutions for purification. For the test with different concentrations of sucrose, the percentages of actively dividing protoplasts were determined after 1 day of culture. These were found to be 92.7%, 93.6%, and 88.2% from 21%, 25%, and 30% cell and protoplast washing (CPW) sucrose solutions for purification, respectively (Figure 1A). In addition, microscopic observation showed that the cells from the CPW 25% sucrose solution were round and intact.
This step determined the protoplast quality of the CPW solution containing round and healthy or broken protoplasts, as well as undigested cell debris. Separation of high-quality protoplasts was achieved with 21% to 25% sucrose solution. Previous studies have reported on feasible purification solutions for different plant species, such as 20% sucrose for Petunia [4], 25% sucrose for Pinellia cordata [16], and 30–44% sucrose for Torreya nucifera [17]. In the current work, determination of the optimized isolation enzyme and purification solution for healthy and viable protoplasts was further verified using the FDA method (Supplementary Figure S2).
2.3. Different Culture Media for Colony and Calli Induction
After culturing the protoplast cells for 24 h, the cells appeared to divide and form a cell mass 3 days after culture. After 4 weeks, the cell mass reached the microcolony phase (Figure 2(D1,E1)). The colonies were transferred to callus induction solid media consisting of Murashige and Skoog medium (MS), 2 mg/L 6-benzylaminopurine (6-BAP), 0.5 mg/L naphthalene acetic acid (NAA), 3% sucrose, and 0.6% agar (pH 5.8). In Kalanchoe species, the interaction between BAP and 2.4-D has been found to affect the formation of protoplast-derived calli [18]. In this study, the calli were formed 3 weeks later (Figure 2(D3,E3)), indicating that BAP and NAA had a significant influence on the formation of calli as noted in previous reports on the plant species Nigella damascene [19] and Chrysanthemum [20]. After culturing in shallow-layered liquid media for 3 days, many cell masses formed which stuck to the bottom of the petri dish, and gentle shaking was necessary to separate the calli for subsequent shooting or rooting induction.
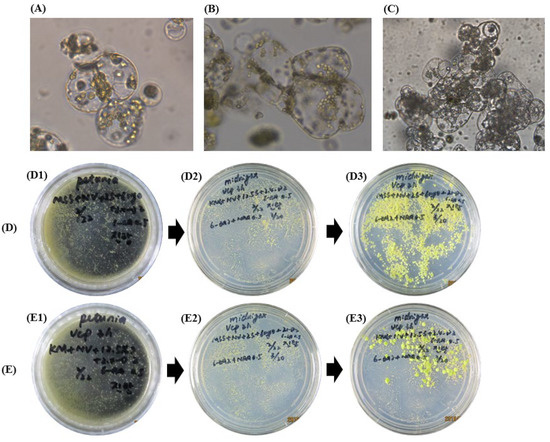
Figure 2.
Protoplast-derived cell division for colony formation and proliferation in two callus induction media. A to C: Isolated protoplasts showing initiation of cell division on colony induction medium for 1 (A), 3 (B), and 7 (C) days after culture. The colonies were moved to callus induction media (D and E, respectively) as indicated. Medium D was composed of MS salt, 6% myo-inositol, 2% sucrose, 2 mg/L 2,4-D, and 0.5 mg/L 6-BAP. Medium E was composed of Kao and Michayluk (KM) salt, NLN vitamin, 12.5% sucrose, 2 mg/L 2,4-D, and 0.5 mg/L 6-BAP. The pictures in (D1,E1) were taken 4 weeks after colony formation. The pictures in (D2,E2) show the colony growing to form callus after transferring to callus induction media. The pictures (D3,E3) were taken 3 weeks after callus formation.
For callus growth, most plant protoplasts were cultured on MS salt [21,22], while protoplasts of Calibrachoa, sweet potato, and Petunia showed improved calli formation on KM salt [4,23]. Rather than the higher concentration of sucrose (medium E), the mixture with a lower concentration of myo-inositol and sucrose (medium D) was more effective for greater callus induction. Similarly, in previous studies, callus proliferation was preferred on MS salt medium with an energy source containing a mixture of myo-inositol and sucrose [17,18,19]. This result demonstrates the varying requirement of media sugar and salts according to plant genotypes and growth stages [19]. It also indicates that carbohydrate type and concentration are important factors in providing energy and adjusting the osmotic potential, which regulates nutrient absorption. Our study suggested that the MS salt medium with a relatively low level of sucrose was preferrable for Petunia callus proliferation, but further studies are needed to elucidate the detailed mechanisms.
2.4. Hormones for Shoot and Root Induction
Shoot regeneration was observed at different hormone combinations and concentrations, resulting in shoot induction efficiencies ranging from 3.7% to 77.8% (Table 1). Treatments with zeatin 1 mg/L, thidiazuron (TDZ) 0.5 mg/L, or TDZ 1 mg/L similarly ensured higher shoot induction efficiency than treatments with NAA or indole acetic acid (IAA) combinations (Table 1 and Figure 3).

Table 1.
Shoot induction efficiency using different hormones.
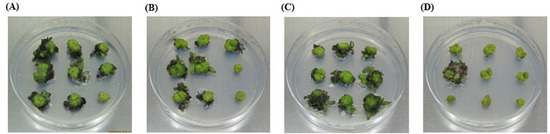
Figure 3.
Shoot induction by cytokinin treatments in Petunia protoplast-derived regeneration, exhibiting relatively higher shooting rates of 74%, 70%, 78%, and 26% at thidiazuron (TDZ) 0.5 mg/L (A), TDZ 1.0 mg/L (B), zeatin 1.0 mg/L (C), and zeatin 2.0 mg/L (D), respectively.
Based on previous reports [4,24,25], various combinations of hormones were tested, but only TDZ or zeatin, as shown in Table 1, were found to be suitable for shoot regeneration in Petunia, demonstrating the species dependence for organogenesis. The direct induction of roots from zeatin supplementation in Kalanchoe species was quite different from our results [18]. Although a combination of 2 mg/L 6-BAP and 0.5 mg/L NAA was reported to show 41% regeneration in Chrysanthemum [20], more than 70% of shoot induction was achieved in Petunia supplemented with TDZ or zeatin alone. Interestingly, further growth of some Petunia calli was either arrested or continued to organogenesis depending on the level and type of phytohormones or protoplast-derived tissues, demonstrating that epigenic factors mediate the phase control of protoplast-based dedifferentiation or regeneration [19].
2.5. Epigenetic Regulation of Callus, Shoot, and Root Development in Petunia
ATX and its related ATXR proteins are generally methyltransferases; these are key regulators of chromatin state and gene expression that are necessary for the vegetative-to-reproductive phase transition [26]. To understand the molecular events surrounding developmental events such as callus, shoot, and root development, the gene expression patterns of ATXR2, ATX4A, and ATX4B were profiled (Figure 4). It was found that PiATXR2 was significantly upregulated in callus and root tissues by 2.3 and 1.9 times, respectively, compared to the control (Figure 4A). The ATXR2 has been found to be involved in cellular dedifferentiation during callus formation in Arabidopsis by inducing the expression of lateral organ boundaries domain (LBD) transcription factors, which is modulated by the histone methylation of H3K36me3 [13]. It has also been noted to play vital roles in de novo root organogenesis [14]. In contrast, both PaATX4A and PiATX4B were upregulated in shoot tissues (Figure 4A,B). The PaATX4A was highly repressed in both callus and root tissues (Figure 4A), while PiATX4B showed significant repression only in the callus (Figure 4B). This is consistent with the Arabidopsis expression study, where ATX4-deficient mutants showed enhanced callus formation. This suggested that the chromatin modifier ATX4 protein facilitates shoot formation during the plant regeneration process by activating shoot identity genes via H3K4me3 modification at these gene loci [15]. Analysis of the mRNA expression results demonstrated that Petunia ATX and ATXR genes could be involved in the epigenetic regulation of molecular events surrounding the acquisition of competence and early developmental events of callus, shoot, and root development.
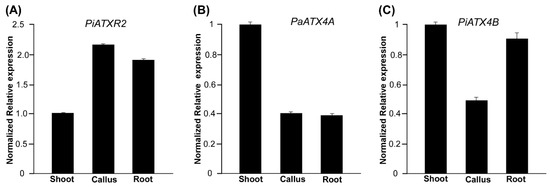
Figure 4.
Transcriptional mRNA expression profiles of genes that act as molecular signatures of the different developmental processes of P. × hybrida cv. Madness Midnight. Expression of genes (A) ATXR2, (B) PaATX4A from P. axillaris, and (C) PiATX4B from P. inflata in developmental stages of callus, shoot, and root in Petunia protoplast-derived tissue culture. Expression levels of genes were normalized relative to the elongation factor 1 gene. Transcriptional fold changes in shoot tissues (control) were set to level 1 for determining the fold changes in other tissues.
In summary, we obtained the optimized culture conditions by performing Petunia protoplast culture for regeneration. For protoplast isolation, we obtained the highest yield using the VCP enzyme combination of Viscozyme, Celluclast, and Pectinex at 1.2%, 0.6%, and 0.6%, respectively. Using a CPW 25% sucrose solution to purify protoplasts, we obtained more dynamic cells. After the addition of culture media, the protoplasts started to divide 1 d and 3 d later, and the cells became a cell mass. Approximately 3–4 weeks later, we observed cell colonies. After comparing two kinds of culture media, MS salt + 6% myo-inositol + 2% sucrose + 2 mg/L 2,4-D + 0.5 mg/L 6-BAP was chosen for colony induction to obtain more colonies. After 3–4 weeks, the produced colonies were transferred to callus induction media (MS + 2 mg/L 6-BAP + 0.5 mg/L NAA + 3% sucrose + 0.6% agar, pH 5.8), and small calli were formed 2 weeks later. Single green calli were picked up to MS + 1 mg/L zeatin + 3% sucrose + 0.7% Agar 1 mg/L for shoot induction. The small calli slowly started to grow larger about 2–3 weeks later, and small shoot tissue was produced around the callus. Shoot induction efficiency reached a maximum of 77.8%. After the regenerated shoots grew to 1–2 cm, the shoots were cut and cultured in MS + 3% sucrose + 0.7% agar media, and roots were formed one week later. Thus, the regenerated plant was successfully obtained. The duration of the entire process was less than three months. Some protoplasts proceeded to the dedifferentiation callus stage through successful cell proliferation, which was explained by the upregulation of ATXR2. ATXR2 is a general regulator for callus and de novo root organogenesis, which was reported by stimulating the lateral organ boundaries domain (LBD) transcription factors [13]. Some of the pluripotent calli further transited to shoot organogenesis, which was confirmed by the enhanced ATX4 gene that was reported to confer H3K4me3 deposition at shoot identity genes [15].
3. Materials and Methods
3.1. Plant Material
P. × hybrida cv. Madness Midnight seeds were sterilized in 2% NaClO solution for 20 min and washed 5 times with sterile distilled water for germination. The seedlings were further grown on MS basic medium containing 30 g/L sucrose and 0.8% agar, and the pH was adjusted to 5.8 [27]. In vitro plants were grown in culture room environments of 16/8 h light (140 µmol m−2 s−1) and dark photoperiod at 25 °C. Young, fully expanded leaves of the in vitro plants were selected and cut into 1–2 mm strips for protoplast isolation, as described in a previous report [10].
3.2. Enzyme Digestion and Purification of Protoplasts
Seven enzyme combinations (ECs) were prepared for enzyme digestion. Three treatments (MC enzyme) using Macerozyme R-10TM (MR; YAKULT Co. Ltd., Tokyo, Japan) and Cellulase R-10TM (CR; YAKULT Co. Ltd., Tokyo, Japan) were EC 1 (0.2% MR + 1.0% CR), EC 2 (0.5% MR + 1.5% CR), and EC 3 (0.8% MR + 2.0% CR). The other four treatments (VCP enzyme) using CelluclastTM (Ce; Novozyme Co. Ltd., Copenhagen, Denmark), PectinexTM (PE; Novozyme Co. Ltd., Copenhagen, Denmark), and ViscozymeTM (Vz; Novozyme Co. Ltd., Copenhagen, Denmark) were EC 4 (0.2% Ce + 0.2% PE + 0.4% Vz), EC 5 (0.4% Ce + 0.4% PE + 0.8% Vz), EC 6 (0.6% Ce + 0.6% PE + 1.2% Vz), and EC 7 (0.8% Ce + 0.8% PE + 1.6% Vz).
The combined enzymes were dissolved in CPW 9% solution containing 27.2 mg/L KH2PO4, 101.0 mg/L KNO3, 1480.0 mg/L CaCl2·2H2O, 246.0 mg/L MgSO4·7H2O, 0.16 mg/L KI, and 0.025 mg/L CuSO4·5H2O [24], which was then combined with 9% mannitol (W/V) and 5 mM MES, and the pH was adjusted to 5.8. Leaf mesophyll cells were digested in each of the enzyme combinations with shaking at 40 rpm in three replicates at 25 °C under dark conditions for 3 h, and then the protoplast cells were isolated. The yield of protoplasts per gram of fresh weight leaves was calculated.
After 3 h of enzyme digestion, protoplasts in the enzyme solution were filtered through a 100 µm and 70 µm nylon mesh. The filtrate was centrifuged at 600 rpm for 5 min. The isolated protoplast pellets were washed once with CPW 9% mannitol solution. The obtained pellets were re-suspended in 5 mL CPW 9% mannitol solution and gently added to 5 mL of CPW with different concentrations of sucrose (21%, 25%, and 30%) for purification. The protoplasts were collected in the middle layer of the tube by centrifugation at 600 rpm for 5 min. The protoplasts were purified using different CPW sucrose solutions. The isolated cells in the middle layer were collected and diluted in a CPW 9% mannitol solution. The protoplasts (50 µL) were mixed with FDA solution to maintain a final concentration of 0.01%, which was kept in the dark for 5 min at room temperature. The cells were reacted with FDA solution and then observed using a green fluorescence microscope to count the number of green cells for protoplast viability.
3.3. Induction of Colony and Callus Formation
Finally, the purified protoplasts suspended in CPW 9% mannitol solution were collected by centrifugation at 600 rpm for 5 min. The collected cells were resuspended in the culture media, adjusted to a cell density of 2 × 105 cells mL−1 with a hemocytometer, and incubated at 25 °C in the dark. To compare the colony induction in different culture media, MS salt media consisting of MS salt, 6% myo-inositol, 2% sucrose, 2 mg/L 2,4-D, and 0.5 mg/L 6-BAP at pH 5.8 and KM salt media consisting of KM salt [28], NLN Vitamin [29], 2.5% sucrose, 2 mg/L 2,4-D, and 0.5 mg/L 6-BAP at pH 5.8 were used. After 3–4 weeks of culture, the colonies (micro-calli) were transferred to MS solid medium (pH 5.8) supplemented with 6-BAP (2 mg/L−1), NAA 0.5 mg/L, 3% sucrose, and 0.7% agar at the same culture room conditions of 16 h light and 8 h dark cycles with 120 µmol m−2 s−1 intensity at 25 °C for callus induction.
3.4. Induction of Root and Shoot Generation
Two weeks after culturing in the solid medium, a small callus appeared and was transferred to a different hormone combination media on basic MS media with 3% sucrose in the same culture room conditions as callus induction. Different levels of cytokinins, including 6-BAP (2 mg/L), TDZ (0.5–1 mg/L), and zeatin (1–2 mg/L), a major phytohormone, were prepared with NAA (1 mg/L) or IAA (1 mg/L) as a minor.
3.5. In Silico Identification of Candidate Genes and Estimation of mRNA Expression Using qRT-PCR
To explore the molecular events during the in vitro Petunia plant regeneration, the candidate genes of ATXR2 (AT3G21820.1) and ATX4 (AT4G27910.1) were used to blast (BLASTP and BLASTN) against the parental genomes of P. axillaris (denoted as PaATX4A) and P. inflata (denoted as PiATX4B) (https://solgenomics.net/, accessed on 27 November 2019) and Petunia-specific ATXR2 and ATX4 genes were obtained (Supplementary Table S1), which were successfully amplified (Supplementary Figure S1).
Total RNA was extracted from Petunia tissues exhibiting callus, shooting, or rooting stages using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA synthesis was accomplished using PrimeScript® RT reagent with a gDNA eraser kit (Takara Korea Biomedical, Seoul, Republic of Korea). Primer pairs specific to target genes were designed using Primer Express (version 3.0, Applied Biosystems, Waltham, MA, USA), and are listed in Supplementary Table S1. A qRT-PCR analysis was carried out to estimate the relative mRNA expression level using the SYBR Green-based (iQ™ SYBR® Green Supermix) qRT-PCR method in a CFX Connect™ Real-Time PCR Detection System thermocycler (Bio-Rad, Daejeon, Korea). The qRT-PCR reactions and other parameters were performed according to a previous study [30,31].
3.6. Statistical Analysis
Statistical analyses were conducted using Statistical Product and Service Solutions for Windows (SPSS; version 12.1, IBM, New York, NY, USA). The data were analyzed using analysis of variance (ANOVA), and differences among the means were tested using Duncan’s multiple range test (p ≤ 0.05).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9020216/s1, Table S1: List of genes, designed primers, and amplicon characteristics. Figure S1: Agarose gel electrophoresis of qRT-PCR amplicons for reference and target genes from Petunia × hybrid. Arrows indicate the bands amplified by designed primers. EF reference gene of elongation factor 1; M DNA ladder; A_ATX4A, I_ATX4B, and I_ATXR2 target genes. Figure S2: Fluorescence microscope images collected from purified protoplast of CPW 25% sucrose solution mixed with 0.01% FDA solution. A: Protoplast stained by FDA in white light. B: Protoplast stained by FDA in green light. C: Combined A and B.
Author Contributions
Conceptualization, G.-J.L.; data curation, L.T., S.S., K.L., Y.J., F.Y. and J.Y.; formal analysis, L.T., Y.-S.K., S.S. and G.-J.L.; funding acquisition, O.-J.K. and G.-J.L.; investigation, L.T., S.S., K.L., Y.J., F.Y., J.Y. and Y.-S.K.; methodology, L.T., S.S., K.L., O.-J.K. and G.-J.L.; project administration, G.-J.L.; resources, G.-J.L.; software, L.T., S.S., K.L., Y.J. and Y.-S.K.; visualization, L.T., K.L. and S.S.; writing—original draft, L.T., S.S., K.L., Y.J., F.Y., J.Y. and Y.-S.K.; writing—review and editing, O.-J.K. and G.-J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a research fund from Chungnam National University.
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Conflicts of Interest
All authors declare that they have no conflict of interest.
References
- Valente, P.; Tao, W.H.; Verbelen, J.P. Auxins and cytokinins control DNA endoreduplication and deduplication in single cells of tobacco. Plant Sci. 1998, 134, 207–215. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Sheen, J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001, 127, 1466–1475. [Google Scholar] [CrossRef]
- Meyer, L.; Serek, M.; Winkelmann, T. Protoplast isolation and plant regeneration of different genotypes of Petunia and Calibrachoa. Plant Cell Tiss. Org. Cult. 2009, 99, 27–34. [Google Scholar] [CrossRef]
- Oh, M.H.; Kim, S.G. Plant-regeneration from petal protoplast culture of Petunia hybrida. Plant Cell Tiss. Org. Cult. 1994, 36, 275–283. [Google Scholar] [CrossRef]
- Bona, C.; Gould, J.; Miller, J.C.; Stelly, D.M.; Louzada, E.S. In vitro regeneration of somatic symmetric and asymmetric hybrid citrus plantlets produced via protoplast fusion. Hortscience 2008, 43, 1206. [Google Scholar]
- Yu, J.; Tu, L.; Subburaj, S.; Bae, S.; Lee, G.J. Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins. Plant Cell Rep. 2021, 40, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Fordlogan, J.; Sink, K.C. Plantlet Regeneration from Protoplasts of Petunia alpicola. Hortscience 1988, 23, 393–395. [Google Scholar] [CrossRef]
- Sangthong, R.; Chin, D.P.; Hayashi, M.; Supaibulwatana, K.; Mii, M. Direct isolation of female germ units from ovules of Petunia hybrida by enzymatic treatment without releasing somatic protoplasts from ovular tissue. Plant Biotechnol. 2009, 26, 369–375. [Google Scholar] [CrossRef]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.M.; Kim, D.H.; Kim, J.S.; Bae, S.; Lee, G.J. Site-directed mutagenesis in Petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef]
- Oberwalder, B.; Schilde-Rentschler, L.; Ruoss, B.; Wittemann, S.; Ninnemann, H. Asymmetric protoplast fusions between wild species and breeding lines of potato—Effect of recipients and genome stability. Theor. Appl. Genet. 1998, 97, 1347–1354. [Google Scholar] [CrossRef]
- Jeong, Y.Y.; Lee, H.Y.; Kim, S.W.; Noh, Y.S.; Seo, P.J. Optimization of protoplast regeneration in the model plant Arabidopsis thaliana. Plant Methods 2021, 17, 21. [Google Scholar] [CrossRef]
- Lee, K.; Park, O.S.; Seo, P.J. Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci. Signal. 2017, 10, eaan0316. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, O.S.; Seo, P.J. ATXR2 as a core regulator of de novo root organogenesis. Plant Signal. Behav. 2018, 13, e1449543. [Google Scholar] [CrossRef]
- Lee, K.; Park, O.S.; Choi, C.Y.; Seo, P.J. ARABIDOPSIS TRITHORAX 4 Facilitates Shoot Identity Establishment during the Plant Regeneration Process. Plant Cell Physiol. 2019, 60, 826–834. [Google Scholar] [CrossRef]
- Yang, X.; Ma, D.-D.; Jiang, F.-S.; Chen, N.-P.; Ding, B.; Jin, L.-X.; Qian, C.-D.; Ding, Z.-S. Protoplasts isolation, purification and plant regeneration of Pinellia cordata. China J. Chin. Mater. Med. 2014, 39, 4211–4215. [Google Scholar]
- Zhang, Y.; Miao, J.; Xie, M.; Tian, S.; Dong, M.; Jiang, G. Isolation and purification of protoplast cell from the leaves of Torreya nucifera. Biotechnol. Bulletin. 2013, 11, 75. [Google Scholar]
- Cui, J.; Mackenzie, K.K.; Eeckhaut, T.; Muller, R.; Lutken, H. Protoplast isolation and culture from Kalanchoe species: Optimization of plant growth regulator concentration for efficient callus production. Plant Cell Tissue Org. Cult. 2019, 138, 287–297. [Google Scholar] [CrossRef]
- Klimek-Chodacka, M.; Kadluczka, D.; Lukasiewicz, A.; Malec-Pala, A.; Baranski, R.; Grzebelus, E. Effective callus induction and plant regeneration in callus and protoplast cultures of Nigella damascena L. Plant Cell Tissue Org. Cult. 2020, 143, 693–707. [Google Scholar] [CrossRef]
- Adedeji, O.S.; Naing, A.H.; Kim, C.K. Protoplast isolation and shoot regeneration from protoplast-derived calli of Chrysanthemum cv. White ND. Plant Cell Tissue Org. Plant Cell 2020, 141, 571–581. [Google Scholar] [CrossRef]
- Inokuma, C.; Sugiura, K.; Cho, C.; Okawara, R.; Kaneko, S. Plant regeneration from protoplasts of Japanese lawngrass. Plant Cell Rep. 1996, 15, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Mii, M.; Yuzawa, Y.; Suetomi, H.; Motegi, T.; Godo, T. Fertile Plant-Regeneration from Protoplasts of a seed-propagated cultivar of Lilium X Formolongi by utilizing meristematic nodular cell clumps. Plant Sci. 1994, 100, 221–226. [Google Scholar] [CrossRef]
- Sihachakr, D.; Ducreux, G. Plant-regeneration from protoplast culture of sweet-potato (Ipomoea-Batatas Lam). Plant Cell Rep. 1987, 6, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Frearson, E.M.; Power, J.; Cocking, E. The isolation, culture and regeneration of Petunia leaf protoplasts. Dev. Biol. 1973, 33, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Vasil, V.; Vasil, I.K. Regeneration of tobacco and petunia plants from protoplasts an culture of corn protoplasts. In Vitro 1974, 10, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Luo, J.H.; Cui, Z.H.; Xue, M.; Wang, L.; Zhang, X.Y.; Pawlowski, W.P.; He, Y. ATX3, ATX4, and ATX5 encode putative H3K4 methyltransferases and are critical for plant development. Plant Physiol. 2017, 174, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Kao, K. Chromosomal behaviour in somatic hybrids of soybean-Nicotiana glauca. Mol. Gen. Genet. 1977, 150, 225–230. [Google Scholar] [CrossRef]
- Nitsch, C.; Nitsch, J. The induction of flowering in vitro in stem segments of Plumbago indica L. Planta 1967, 72, 355–370. [Google Scholar] [CrossRef]
- Subburaj, S.; Ha, H.J.; Jin, Y.T.; Jeon, Y.; Tu, L.; Kim, J.B.; Kang, S.Y.; Lee, G.J. Identification of gamma-radiation-responsive microRNAs and their target genes in Tradescantia (BNL clone 4430). J. Plant Biol. 2017, 60, 116–128. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).