Abstract
The large consumption of non-renewable fossil fuels has brought about energy depletion and environmental pollution, spawning the production of renewable biofuels, an important alternative to alleviate the energy crisis effectively. As one of the ideal types of biofuel, bioethanol synthesis in Saccharomyces cerevisiae has attracted much attention. S. cerevisiae has been developed as essential chassis cells with high efficiency for bioethanol synthesis on account of many advantages. This study systematically summarized the preponderance of S. cerevisiae in biosynthesis. It objectively stated the research strategies of bioethanol synthesis based on S. cerevisiae and the existing bottleneck problems. This study further proposed reasonable prospects for bioethanol synthesis by S. cerevisiae, attempting to provide alternative research strategies.
1. Introduction
The increasing demand for fossil fuels caused by burgeoning anthropogenic activities and rapid economic growth provoked wicked environmental issues and resource depletion [1,2], which is a direct boost to reconstruct the energy structure, develop and industrialize renewable biofuels [3,4,5].
Biofuels produce in response to the proper time and conditions coping with world environmental concerns and the exhaustion of non-renewable fossil-based fuels [6,7,8]. Biofuel refers to the renewable and sustainable fuel obtained through the processing of biomass materials (crop straw, wood, wheat grass, etc.) that can replace traditional fossil fuels [9], among which bioethanol is particularly attractive, having the potential to accelerate sustainable use of resources and change the global economy toward a greener future [10,11,12].
Continuous biotechnology innovation strongly promotes the upgrading and mass production of biofuels represented by bioethanol. Bio-fermentation based on important model microorganisms is a technology with great development potential beyond all doubt for biofuel production at present and in the future [13,14].
With the rapid development of synthetic biology technology, based on the huge market demand for biofuels, Saccharomyces cerevisiae (S. cerevisiae) is increasingly used in the biosynthesis of biofuels due to their superiorities [15,16]. S. cerevisiae is a food-grade budding yeast eukaryote inextricably linked with human production and life [17], which can be effortlessly found in both natural habitats and various environments affected by human activities [18]. Since ancient times, S. cerevisiae has had a long historical standing in human civilization and social development, mainly reflected in food production and fermentation such as bread, beer, and wine [19,20].
Based on numerous studies in this field, this research took bioethanol as an example, reviewed the significant advantages of S. cerevisiae as a quite efficient synthetic cell factory and the main techniques currently used to improve the production of bioethanol. In addition, the bottlenecks existing in the current bioethanol synthesis process using S. cerevisiae as chassis cells will be discussed in this paper, and a rational and feasible future outlook will be proposed. Hopefully, this study could provide the necessary basis for further research on bioethanol synthesis based on S. cerevisiae.
2. Saccharomyces cerevisiae: An Efficient Cell Factory
S. cerevisiae has conceivably received increasing attention in recent years in consideration of its inseparable relations with humankind. Advances in biotechnology, increased demand for synthetic biological products and dynamic environmental changes have enabled the continuous updating and optimization of S. cerevisiae strains [21]. With long-term selection and domestication, S. cerevisiae strains with specific functions have been gradually selected in the expected direction from the wild types [22]. Nowadays, S. cerevisiae is no longer confined to the fermentation of food and drink but has been designed as a cell factory for producing important pharmaceuticals, recombinant proteins and advanced biofuels based on the continuous innovation of different technical approaches [23,24,25,26]. S. cerevisiae has several prominent advantages compared with other prokaryotic and eukaryotic microorganisms, making it one of the most important cell factories today (Figure 1).
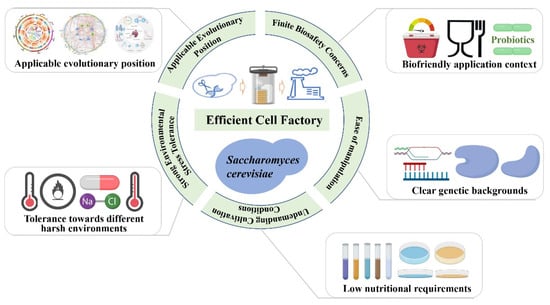
Figure 1.
Main advantages of Saccharomyces cerevisiae as an efficient cell factory.
2.1. Finite Biosafety Concerns
S. cerevisiae is closely related to human diet and life, completely non-pathogenic, and has been confirmed as a food-grade microorganism in the long history [27]. In addition to the function of food fermentation, studies in recent years have expounded that S. cerevisiae also exhibits obvious probiotic properties. A study by Sun et al. showed that the application of engineered S. cerevisiae could attenuate DSS-induced colitis in mice via the suppression of macrophage pyroptosis and modulation of the intestinal microbiota [28]. S. cerevisiae also has significant probiotic effects on farmed animals. The feed digestibility and the number of pathogenic bacteria could be effectively improved by supplementing live S. cerevisiae cells with animal feed [29]. There is also direct evidence about S. cerevisiae-based probiotics as a novel anti-microbial agent for treating bacterial diseases [30,31]. In addition, S. cerevisiae has a significant biological control effect on citrus population disease through nutritional competition and the production of antifungal compounds [32]. Therefore, the high biosafety and probiotic activity are prominent advantage that distinguishes S. cerevisiae from other organisms, displaying great potential and necessary prerequisites as a cell factory.
2.2. Applicable Evolutionary Position
S. cerevisiae is a kind of single-celled eukaryotes, which not only has the characteristics of easy culture, rapid reproduction and toilless genetic manipulation similar to prokaryotes but also has the basic molecular and cell biological characteristics of typical eukaryotes. The applicable evolutionary position of S. cerevisiae has made it an indispensable experimental model for elaborating many regularities in modern genetics, cell biology and biochemistry of both eukaryotic and prokaryotic cells [33]. Also, S. cerevisiae is considered the most promising tool for exogenous gene expression [34,35], essential for remodeling biofuels’ in vivo synthetic pathways. Evidence also indicated that S. cerevisiae is helpful as a host for genetic engineering since it allows the folding and glycosylation of expressed heterologous eukaryotic proteins and can be subjected to many genetic manipulations [36].
2.3. Undemanding Cultivation Conditions
S. cerevisiae is relatively easy to be cultured on different types of media under laboratory conditions, allowing the evaluation of many phenotypes and the construction of different types of cell factories [37]. Generally, S. cerevisiae does not need a complex medium for growth and can be cultured in liquid and solid mediums. The nutrients required by almost all types of S. cerevisiae strains, including nutrient-deficient and gene-deletion mutants, are common, inexpensive, and readily available. The most commonly used media for S. cerevisiae are YPD (also called YPED) media, SC media, minimal media (supplemented with essential amino acids) and sporulation media [38]. Given the certainty of the nutrient requirements of S. cerevisiae, researchers can conduct in-depth research, such as regulating the nutrient metabolic utilization pathway and corresponding metabolites to improve product quality and yield [39,40,41].
2.4. Strong Environmental Stress Tolerance
The tolerance of microorganisms to diverse stresses is very important for practical applications [42,43]. The living habitat of S. cerevisiae is complex and extensive, always facing a variety of adverse environmental perturbations. S. cerevisiae has developed a capacity to cope with harsh environments in long-term natural evolution and artificial application [44,45,46,47]. Based on the physiological and genetic characteristics of strong tolerance of S. cerevisiae, the rational modification could be carried out through genetic engineering to obtain more robust strains to cope with acute environmental changes, including oxidative stress, low temperature, heat shock, nutrient dropout, osmotic stress, antibiotic pressure, acetic acid stress, etc., in practical application [45,48,49,50,51].
2.5. Ease of Manipulation
As a most thoroughly studied eukaryotic model organism, research has been conducted on S. cerevisiae based on the well-annotated genome information. Mature genome manipulation and gene editing techniques for S. cerevisiae led to easy operation of building specific cell factories [52]. S. cerevisiae strain S288c was the first eukaryote to be genetically elucidated in 1996, which for the first time opened the opportunity for the global study of the expression and functioning of the eukaryotic genome [53] and created opportunities for further studies in comparative, functional, and evolutionary genomics, laying a solid foundation of systematically understanding and rationally engineering metabolism pathways [54]. On this basis, S. cerevisiae can be studied in depth and detail at multiple levels in the follow-up study [55]. The remarkable plasticity of the S. cerevisiae genome makes it possible for the elaboration of synthetic pathways and large-scale production of biofuels [18,33].
3. Synthesis of Bioethanol in Saccharomyces cerevisiae
Biofuel has the potential to achieve environmentally sustainable development, reduce reliance on imported resources, and meet the energy demand with economic growth [3,7]. Figure 2A shows the biofuel production levels in petajoules (PJ; 1 PJ = 1015 J) in leading countries in 2021, with relatively noteworthy differences between different countries. Bioethanol is a particularly compelling class of biofuels, considered a prominent alternative to fossil fuels in the 21st century due to its advantages of complete combustion, low exhaust emissions and contribution to the reduction of crude oil consumption and environmental pollution [56]. The current situation of bioethanol production in major countries in the world is shown in Figure 2B.
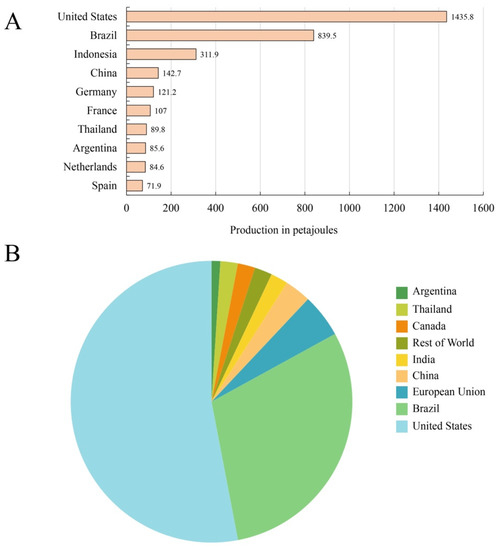
Figure 2.
Leading countries based on biofuel (A) and bioethanol (B) production worldwide in 2021, respectively. (Data source: https://www.statista.com accessed on 25 June 2023).
Bioethanol is mainly produced by microorganisms in a fermentation process that utilizes plant feedstock as raw materials (e.g., corn, sugarcane, sugar beets, sweet sorghum) [57]. According to the different production raw materials, bioethanol production can be divided into three categories: the use of grain, the use of lignocellulose and the use of algae for bioethanol production [58,59]. There are several microbial cell factories to choose from for fermenting the product ethanol based on different raw materials, including Escherichia coli [60,61], Zymomonas mobilis [62,63], Bacillus subtilis [64,65], S. cerevisiae [66,67], etc. Within all fermentative microorganisms, S. cerevisiae is regarded as an excellent industrial ethanologenic organism based on the outstanding advantages mentioned above. The process relies on the ability of S. cerevisiae to efficiently and completely ferment sugars from feedstock biomass into ethanol on a large scale [68].
Based on the latest research results, the main measures taken to produce bioethanol using S. cerevisiae as chassis cells can be summarized as follows (Figure 3).
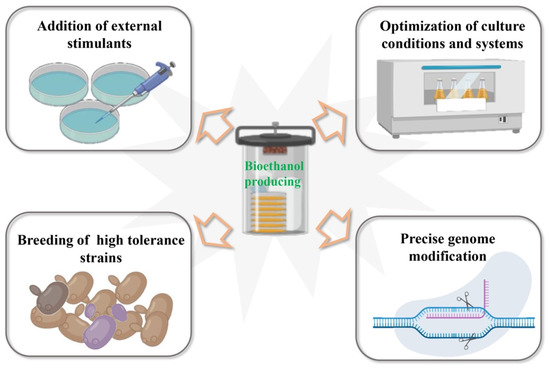
Figure 3.
Major strategies for promoting bioethanol synthesis in Saccharomyces cerevisiae.
3.1. The Addition of External Stimulants to the Medium
The nutritional conditions required by the growth and metabolism of S. cerevisiae are relatively simple. In fermentation to produce ethanol from biomass raw materials, adding exogenous nutrients in the medium is an effective stimulus to increase the ethanol yield. A study by Li et al. showed that adding collagen peptide to the media significantly increased the bioethanol yield under different glucose concentrations and fermentation times compared with the non-added group [69]. In second-generation bioethanol production, the finite tolerance of S. cerevisiae to the inhibitors in lignocellulosic hydrolysates remains a significant challenge. Adding a mixture of pyridoxine, thiamine, and biotin to propagation media could improve cell growth and ethanol yields during fermentation in both corn stover and wheat straw hydrolysate to a large extent [70]. This method is simple and easy and can achieve a significant increase in yield in a short time.
3.2. Optimization of S. cerevisiae Culture Components and Systems
Rational optimization and improvement of culture conditions and systems could effectively increase bioethanol production. A study by Pereira et al. showed an optimized medium based on corn steep liquor and other low-cost nutrient sources that significantly increased the final ethanol titer, yield, and yeast activity, providing valuable insights into cost-effective nutritional supplementation of bioethanol production [71]. In addition, the innovation of the fermentation culture system is also a helpful practice worth trying. By co-culture of S. cerevisiae and Pichia pastoris, the yield of bioethanol can be increased to a great extent [72]. This method of increasing yield requires trial and error to determine the most appropriate medium component and mixed culture system.
3.3. Breeding of High-Tolerance Strains
As a chassis cell, S. cerevisiae faces complex and varied pressures in bioethanol fermentation. Physicochemical conditions, substrate concentration, toxic effects of ethanol and other factors are essential elements affecting the final yield of bioethanol. Therefore, the screening and breeding S. cerevisiae strains with better tolerance is very important.
At present, the breeding of S. cerevisiae strains is mainly focused on improving the thermo-tolerance, glucose-tolerance, and ethanol tolerance, which is the most common type of stress faced by S. cerevisiae cells in the process of bioethanol fermentation [73,74]. The breeding of excellent strains is an essential prerequisite for achieving high bioethanol yield, and that’s why a considerable amount of research is currently focused on this through various physicochemical methods.
3.4. Precise Modification of the S. cerevisiae Genome
To increase ethanol yield, it is a research hotspot to edit and modify the genome of S. cerevisiae accurately from different metabolic pathways based on the transparent genetic background and mature gene operating system. There is a lot of relevant research, and remarkable progress has been made. Modification, knock-out and overexpression of key genes and promoters may be closely related to bioethanol yield. For example, disrupting the alcohol dehydrogenase (ADH2) gene via complete deletion of the gene and introducing a frameshift mutation in the ADH2 locus via CRISPR/Cas9 technology showed that the ethanol yield improved by up to 74.7% compared with the yield obtained using the native strain [75]; the introduction of glucose-proton symporter, fructose-proton symporter and extracellular invertase under the context of deletion of genes encoding hexose transporters, disaccharide transporters and disaccharide hydrolases resulting in a 16.6% increased anaerobic ethanol yield [76]; the knock-in and knock-out of key genes in the glucose metabolic pathway are also crucial for increasing bioethanol production [77,78]. This approach is more precise, more targeted, and more thorough, illustrating the applicability to promote the improvement of bioethanol production in S. cerevisiae via genome engineering.
4. The Bottleneck of Producing Bioethanol by Saccharomyces cerevisiae
Yeasts such as S. cerevisiae have been used in bioethanol production, especially in the brewery and wine industries, thousands of years ago. Numerous efforts have been aimed at comprehending and further improving yeast fermentation. Nevertheless, bioethanol production by fermentation in S. cerevisiae is not without obstacles. There are many external constraints, including environmental conditions, price factors, policy background, scale limitation, and complexity and uncertainty of the fermentation process, and these factors have evolved into the main bottleneck problems, inevitably preventing the increase of ethanol production. Based on the existing research results, some representative bottlenecks are summarized below (Figure 4).
4.1. The Utilization Dilemma of Fermentation Raw Materials
The first generation of bioethanol is produced from food crops (corn, wheat, sweet potato, etc.) with excellent fermentation characteristics [79]. However, food crops are extremely important resources with high production and storage costs, mainly reflected in using valuable and scarce available farmland and irrigation water. Therefore, in terms of cost, producing bioethanol from food crops is not an option based on the lowest cost and highest return.
Lignocellulosic biomass represents the largest resource pool worldwide and is the raw material for producing second-generation bioethanol with low cost and wide sources. One outstanding advantage of lignocellulosic biomass is that it can be obtained for ethanol production without competing for arable land and agricultural inputs with crops for human or livestock consumption [80]. However, despite intensive research exploring lignocellulosic ethanol, this option still accounts for <1% of global ethanol production [68]. The major drawback of these feedstocks is the recalcitrance to degradation of the lignocellulosic matrix, which is comprised of covalently and hydrogen-bonded cellulose and hemicellulose polymers that are further linked to lignin in its natural state [81,82,83]. In addition, S. cerevisiae, the most widely used ethanol-producing species in bioethanol production, has difficulty utilizing β-D-xylose and α-L-arabinose, the main pentoses in hemicellulose polymers [84,85]. This is an important reason that makes it impossible to expand the production of second-generation bioethanol further.
The third generation of bioethanol production is derived from microalgal biomass. Microalgae is currently a promising option for producing new forms of renewable energy with a wide range of sources and rich nutrients. Nevertheless, due to the particularity and complexity of microalgae, the process of using microalgae raw materials to produce bioethanol is relatively complicated, which needs to go through several steps such as cell wall breaking treatment, starch extraction, hydrolysis saccharification and the final fermentation process [86]. Besides, compared with the equipment and process for producing bioethanol from food crops, the production equipment of microalgal biomass for bioethanol synthesis is not mature enough, and many key technologies are still in the theoretical research stage. Also, how to effectively prevent bacterial contamination become another significant constraint in mass cultivation and impedes the industrial process [87].
At present, the largest source of feedstock for the production of bioethanol is still food crops [88]. How to crack the bottleneck of raw material utilization in bioethanol synthesis is a problem that needs to be solved in the future.
4.2. Limitations of Gene Editing Techniques in Saccharomyces cerevisiae
CRISPR-Cas9 gene editing technology to identify the target genome sequence through artificially designed sgRNA (guide RNA). It guides Cas9 protease to cut the DNA double-strand, resulting in double-strand breaks effectively. The CRISPR-Cas9 system of S. cerevisiae is mature enough compared to other chassis cells in synthetic biology, but there are still some problems impeding practical application. For example, chromatin affects the gene-editing efficiency of the Cas9 protein in S. cerevisiae because PAM accessibility in chromatin is a critical factor regulating Cas9 targeting and cleavage [89]. In addition, Cas9-based gene editing can sometimes lead to excessive cutting of DNA, which can lead to some mutations. gRNA is crucial for editing activity, but how to avoid unnecessary side effects of gene editing by rationalizing gRNA modification remains unclear. The low efficiency of the HDR-mediated genome will also significantly affect the efficiency of gene editing. This is undoubtedly a problem that needs to be paid attention to the improving bioethanol production through further genetic modification [90].
In conclusion, a gene-editing system of S. cerevisiae must be better constructed for improved bioethanol production.
4.3. Factors of Yield Constraint
The efficient production of bioethanol is desired for large-scale bioenergy applications, but it is severely challenged by ethanol stress, which is a mutual problem in the global context [91]. Bioethanol accumulates continuously and reaches a higher concentration in fermentation by S. cerevisiae. Although S. cerevisiae has been modified through mutagenic breeding, gene editing and other technologies to improve ethanol tolerance, it is still far from the expected value. It cannot reach the scale fermentation level of higher density.
On the other hand, looking at bioethanol production worldwide, it is distinctly found that the degree of marketization is appreciably high, with fierce competition and development gaps between different countries and regions [92]. In 2020 and 2021, the global bioethanol fermentation industry capacity showed a relatively downward trend compared to 2019 and 2020 (Table 1). Among the major countries producing bioethanol, the overall production of China, Brazil and Thailand has shown a downturn in the past two years. In addition to the impact of macroeconomic regulation policies and the effect of the COVID-19 pandemic, specific agricultural development among countries also significantly affects bioethanol production. Taking China as an example, it is difficult to form a large-scale and stable supply of bioethanol raw materials of first-generation bioethanol in China in terms of the challenge of food security, which is the biggest obstacle to bioethanol industrialization in China at present [93].

Table 1.
Annual world fuel ethanol production (Mil. Gal.) (Data source: https://ethanolrfa.org accessed on 25 June 2023).
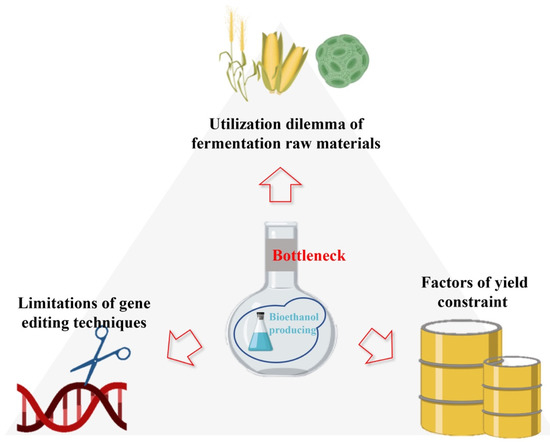
Figure 4.
Main bottlenecks of bioethanol production by Saccharomyces cerevisiae.
5. Future Perspectives
Global concerns about fossil fuel depletion and the environmental effects of greenhouse gas emissions have led to widespread fermentation-based production of bioethanol [94]. S. cerevisiae, the preferred microorganism for bioethanol production, given its convenient cultivation and genetic manipulations, can use biomass raw materials to synthesize bioethanol through specific metabolic pathways more effectively. Until now, bioethanol fermentation in S. cerevisiae represents the predominant product of industrial biotechnology [95,96].
While seeing the remarkable advantages of bioethanol production by S. cerevisiae, it is also urgent to recognize the significant challenges. To make substantial improvements better, the following aspects could be considered and strengthened in future research.
5.1. Redesign of Fermentation Culture System
A good fermentation culture system is critical to increase the production of bioethanol. As mentioned above, microalgae are an essential raw material for third-generation bioethanol production, but there is still a long way to go before their widespread application. Future research on microalgae culture equipment should be strengthened to promote the transformation and upgrading of microalgal raw materials and the construction of a resource-saving and environment-friendly society. For example, to obtain more microalgal biomass, new types of simple photobioreactors suitable for high-density cultivation of microalgae with low cost should be developed, as well as a series of equipment that can be scaled up for harvesting and cell wall breaking to reduce the drying of microalgae and the extraction of carbohydrates as much as possible, saving equipment investment and energy consumption [97].
Furthermore, the symbiotic system of microorganisms and microalgae has become a research hotspot in recent years due to the strong carbon sequestration capacity and rich nutrient factors of microalgae, which can provide most of the essential carbon sources as well as microelements required for microbial growth. There is some research on the symbiotic system of bacteria and algae [98,99], but there are few reports on its application to bioethanol fermentation. A novel symbiotic system between S. cerevisiae and microalgae could be constructed to optimise culture conditions and increase bioethanol yield effectively.
5.2. Targeted Regulation of Fermentation Pretreatment Process
It has been shown that the efficient pretreatment step of raw material fermentation is essential for converting renewable biomass into fuels and chemicals. Therefore, it is a practical scheme to modify fermentation raw materials by directional control of the pretreatment process. The direction design optimization based on machine learning can effectively regulate the pretreatment parameters [100]. The organic coupling of this technology with the pretreatment process of fermentation raw materials could be considered to improve the bioethanol yield continuously. For example, xylose is one of the most abundant sugars in cellulosic biomass but cannot be utilized by wild-type S. cerevisiae [100,101,102]. In the future, based on machine learning and reverse design optimization strategy, the fermentation pretreatment process of raw materials could be continuously optimized and improved to enhance the xylose utilization and conversion efficiency.
5.3. In-Depth Exploration of Important Regulatory Factors
Nowadays, the genetic manipulation of S. cerevisiae strains is very mature, and some metabolic pathways related to ethanol synthesis have been analyzed in depth and detail. Nevertheless, there is still a lack of mining and identification of some important regulatory elements, such as non-coding RNA (ncRNA). ncRNAs cannot be translated into functional proteins but confer important regulatory functions [103]. Many studies have shown that ncRNAs play crucial regulatory roles in all microbial growth and metabolism stages, and S. cerevisiae is no exception [104]. However, there are no relevant studies on whether ncRNA plays an important regulatory role in the bioethanol synthesis pathways of S. cerevisiae. Future studies could emphasize identifying key regulatory elements like ncRNAs closely related to bioethanol metabolism and synthesis in S. cerevisiae genome through high-throughput sequencing and -omics technology so that they could be flexibly used as crucial regulatory biological elements.
5.4. Construction of Cell-Free Synthetic Biological System Based on Saccharomyces cerevisiae Cell Extracts
Due to the complexity of the living cell system, the irrevocability of cell growth, the interference of intracellular noise and the obstruction of the cell membrane, the transformation of biological elements is greatly limited. Hence, the current bioethanol synthesis based on S. cerevisiae still faces problems such as unpredictability, incompatibility, and high complexity that cannot be underestimated.
In recent years, cell-free synthetic biological (CFSB) systems referring to the engineering science that implements the central principles of biology in vitro have come into sight, instructively complementing the study of synthetic biology. The core of this system is to break the shackles of cells, reintegrate cell resources in vitro, and focus on synthesising customized target products [105]. CFSB systems show significant advantages in toxicity tolerance, economic cost, synthetic efficiency, etc. CFSB systems based on cell extracts are widely used technical means at present, among which the CFSB systems based on E. coli cell extracts and wheat germ cell extracts are the most mature among prokaryotic and eukaryotic systems, respectively [106,107]. There is also some research on the cell-free synthesis system of S. cerevisiae, but they are just starting and not deep enough [108,109]. In the future, efficient CFSB systems based on S. cerevisiae cell extracts can be further explored and constructed to lay the specific foundation for providing maximum synthesis efficiency of bioethanol.
5.5. Exploration of Quorum Sensing in Bioethanol Fermentation
Nongenetic approaches to alter metabolism may have the advantages of general applicability and simple control. Quorum sensing is an essential way for bacteria to carry out intra-species or inter-species communication, which uses the secretion and transmission of signal molecules to make bacteria respond to changes in cell density and flora composition in the environment [110,111,112]. Because of its relatively simple structure and transparent mechanism, the system is often used as a gene module to characterize the complex intracellular response mechanism and the interaction of different bacteria, which has far-reaching significance for synthetic biology research [113]. Quorum sensing also plays an essential role in the process of bioethanol synthesis by S. cerevisiae. Studies showed that the ethanol yield could be improved by adding quorum-sensing molecules to inhibit the cell growth of S. cerevisiae [114]. However, it is unknown whether quorum sensing plays an important role and how it works in the mixed fermentation system of microorganisms and microalgae. In the future, it is worth further exploring and elaborating on whether quorum sensing could realize the interactive regulation of the two communities (e.g., S. cerevisiae and microalgae) in the fermentation system while regulating the spatiotemporal behavior of the S. cerevisiae community in the process of bioethanol fermentation, to provide new insights for improving the bioethanol production.
Author Contributions
H.Z. contributed to the writing and editing of this manuscript. P.Z. and T.W. contributed to the investigation of this study. H.R. contributed to the development and correction of this manuscript. She is also the corresponding author of this paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Tianjin Education Commission Scientific Research Project (grant number 2022KJ004), Tianjin Municipal Science and Technology Bureau (grant number 22ZXJBSN00010) and National Key Research and Development Program (grant number SQ2022YFE013001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Solomon, C.G.; Salas, R.N.; Malina, D.; Sacks, C.A.; Hardin, C.C.; Prewitt, E.; Lee, T.H.; Rubin, E.J. Fossil-Fuel Pollution and Climate Change—A New NEJM Group Series. N. Engl. J. Med. 2022, 386, 2328–2329. [Google Scholar] [CrossRef] [PubMed]
- Thurston, G.D. Fossil Fuel Combustion and PM2.5 Mass Air Pollution Associations with Mortality. Environ. Int. 2022, 160, 107066. [Google Scholar] [CrossRef] [PubMed]
- Eswaran, N.; Parameswaran, S.; Johnson, T.S. Biofuels and Sustainability. Methods Mol. Biol. 2021, 2290, 317–342. [Google Scholar]
- Abid, N.; Ceci, F.; Ikram, M. Green Growth and Sustainable Development: Dynamic Linkage between Technological Innovation, ISO 14001, and Environmental Challenges. Environ. Sci. Pollut. Res. Int. 2022, 29, 25428–25447. [Google Scholar] [CrossRef]
- Golroudbary, S.R.; Makarava, I.; Kraslawski, A.; Repo, E. Global Environmental Cost of Using Rare Earth Elements in Green Energy Technologies. Sci. Total Environ. 2022, 832, 155022. [Google Scholar] [CrossRef]
- Liu, Y.; Cruz-Morales, P.; Zargar, A.; Belcher, M.S.; Pang, B.; Englund, E.; Dan, Q.; Yin, K.; Keasling, J.D. Biofuels for A Sustainable Future. Cell 2021, 184, 1636–1647. [Google Scholar] [CrossRef]
- Hasan, M.; Abedin, M.Z.; Amin, M.B.; Nekmahmud, M.; Oláh, J. Sustainable Biofuel Economy: A Mapping through Bibliometric Research. J. Environ. Manag. 2023, 336, 117644. [Google Scholar] [CrossRef]
- Sharma, S.; Kundu, A.; Basu, S.; Shetti, N.P.; Aminabhavi, T.M. Sustainable Environmental Management and Related Biofuel Technologies. J. Environ. Manag. 2020, 273, 111096. [Google Scholar] [CrossRef]
- Bhattarai, K.; Stalick, W.M.; McKay, S.; Geme, G.; Bhattarai, N. Biofuel: An Alternative to Fossil Fuel for Alleviating World Energy and Economic Crises. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2011, 46, 1424–1442. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.W.; Anderson, W.A.; Moo-Young, M. Ethanol Fermentation Technologies from Sugar and Starch Feedstocks. Biotechnol. Adv. 2008, 26, 89–105. [Google Scholar] [CrossRef]
- Gray, K.A.; Zhao, L.; Emptage, M. Bioethanol. Curr. Opin. Chem. Biol. 2006, 10, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singhania, R.R.; Soam, S.; Chen, C.W.; Haldar, D.; Varjani, S.; Chang, J.S.; Dong, C.D.; Patel, A.K. Production of Bioethanol from Food Waste: Status and Perspectives. Bioresour. Technol. 2022, 360, 127651. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Q.; Liu, X. On the Future Fermentation. Microb. Biotechnol. 2021, 14, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Valle-Rodríguez, J.O.; Siewers, V.; Nielsen, J. Prospects for Microbial Biodiesel Production. Biotechnol. J. 2011, 6, 277–285. [Google Scholar] [CrossRef]
- Hong, K.K.; Nielsen, J. Metabolic Engineering of Saccharomyces cerevisiae: A Key Cell Factory Platform for Future Biorefineries. Cell. Mol. Life Sci. 2012, 69, 2671–2690. [Google Scholar] [CrossRef]
- Buijs, N.A.; Siewers, V.; Nielsen, J. Advanced Biofuel Production by the Yeast Saccharomyces cerevisiae. Curr. Opin. Chem. Biol. 2013, 17, 480–488. [Google Scholar] [CrossRef]
- Landry, C.R.; Townsend, J.P.; Hartl, D.L.; Cavalieri, D. Ecological and Evolutionary Genomics of Saccharomyces cerevisiae. Mol. Ecol. 2006, 15, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.L.; Galeote, V.; Bigey, F.; Camarasa, C.; Marsit, S.; Nidelet, T.; Sanchez, I.; Couloux, A.; Guy, J.; Franco-Duarte, R.; et al. Adaptation of S. cerevisiae to Fermented Food Environments Reveals Remarkable Genome Plasticity and the Footprints of Domestication. Mol. Biol. Evol. 2018, 35, 1712–1727. [Google Scholar] [CrossRef]
- Arranz-Otaegui, A.; Gonzalez Carretero, L.; Ramsey, M.N.; Fuller, D.Q.; Richter, T. Archaeobotanical Evidence Reveals the Origins of Bread 14,400 Years Ago in Northeastern Jordan. Proc. Natl. Acad. Sci. USA 2018, 115, 7925–7930. [Google Scholar] [CrossRef]
- Ting, T.Y.; Li, Y.; Bunawan, H.; Ramzi, A.B.; Goh, H.H. Current Advancements in Systems and Synthetic Biology Studies of Saccharomyces cerevisiae. J. Biosci. Bioeng. 2023, 135, 259–265. [Google Scholar] [CrossRef]
- Eldarov, M.A.; Kishkovskaia, S.A.; Tanaschuk, T.N.; Mardanov, A.V. Genomics and Biochemistry of Saccharomyces cerevisiae Wine Yeast Strains. Biochemistry 2016, 81, 1650–1668. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Ruiz, J.; Santos, A.; Van Wyk, N.; Pretorius, I.S. Saccharomyces cerevisiae. Trends Genet. 2019, 35, 956–957. [Google Scholar] [CrossRef]
- Chen, Y.; Li, F.; Nielsen, J. Genome-Scale Modeling of Yeast Metabolism: Retrospectives and Perspectives. FEMS Yeast Res. 2022, 22, foac003. [Google Scholar] [CrossRef]
- Wang, G.; Huang, M.; Nielsen, J. Exploring the Potential of Saccharomyces cerevisiae for Biopharmaceutical Protein Production. Curr. Opin. Biotechnol. 2017, 48, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Patra, P.; Das, M.; Kundu, P.; Ghosh, A. Recent Advances in Systems and Synthetic Biology Approaches for Developing Novel Cell-Factories in Non-Conventional Yeasts. Biotechnol. Adv. 2021, 47, 107695. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J. Yeast Systems Biology: Model Organism and Cell Factory. Biotechnol. J. 2019, 14, e1800421. [Google Scholar] [CrossRef]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol Yeasts: Mechanisms and Applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xu, X.; Liang, L.; Wang, X.; Bai, X.; Zhu, L.; He, Q.; Liang, H.; Xin, X.; Wang, L.; et al. Lactic Acid-Producing Probiotic Saccharomyces cerevisiae Attenuates Ulcerative Colitis via Suppressing Macrophage Pyroptosis and Modulating Gut Microbiota. Front. Immunol. 2021, 12, 777665. [Google Scholar] [CrossRef]
- Elghandour, M.M.Y.; Tan, Z.L.; Abu Hafsa, S.H.; Adegbeye, M.J.; Greiner, R.; Ugbogu, E.A.; Cedillo Monroy, J.; Salem, A.Z.M. Saccharomyces cerevisiae as A Probiotic Feed Additive to Non and Pseudo-ruminant Feeding: A Review. J. Appl. Microbiol. 2020, 128, 658–674. [Google Scholar] [CrossRef]
- Sabbatini, S.; Monari, C.; Ballet, N.; Mosci, P.; Decherf, A.C.; Pélerin, F.; Perito, S.; Scarpelli, P.; Vecchiarelli, A. Saccharomyces cerevisiae-Based Probiotic As Novel Anti-Microbial Agent for Therapy of Bacterial Vaginosis. Virulence 2018, 9, 954–966. [Google Scholar] [CrossRef]
- Kil, B.J.; Pyung, Y.J.; Park, H.; Kang, J.W.; Yun, C.H.; Huh, C.S. Probiotic Potential of Saccharomyces cerevisiae GILA with Alleviating Intestinal Inflammation in A Dextran Sulfate Sodium Induced Colitis Mouse Model. Sci. Rep. 2023, 13, 6687. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.R.; Klein, M.N.; Ferraz, L.P.; da Silva, A.C.; Kupper, K.C. Saccharomyces cerevisiae: A Novel and Efficient Biological Control Agent for Colletotrichum acutatum during Pre-Harvest. Microbiol. Res. 2015, 175, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Vanderwaeren, L.; Dok, R.; Voordeckers, K.; Nuyts, S.; Verstrepen, K.J. Saccharomyces cerevisiae as a Model System for Eukaryotic Cell Biology, from Cell Cycle Control to DNA Damage Response. Int. J. Mol. Sci. 2022, 23, 11665. [Google Scholar] [CrossRef]
- Bu, X.; Lin, J.Y.; Duan, C.Q.; Koffas, M.A.G.; Yan, G.L. Dual Regulation of Lipid Droplet-Triacylglycerol Metabolism and ERG9 Expression for Improved β-carotene Production in Saccharomyces cerevisiae. Microb. Cell Fact. 2022, 21, 3. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Wang, Q.; Selvaraj, J.N.; Zhou, Y.; Ma, L.; Zhang, G.; Ma, Y. High Copy and Stable Expression of the Xylanase XynHB in Saccharomyces cerevisiae by rDNA-Mediated Integration. Sci. Rep. 2017, 7, 8747. [Google Scholar] [CrossRef]
- Yang, F.; Cao, M.; Jin, Y.; Yang, X.; Tian, S. Development and Application of Saccharomyces cerevisiae Cell-Surface Display for Bioethanol Production. Sheng Wu Gong Cheng Xue Bao 2012, 28, 901–911. [Google Scholar]
- Hanscho, M.; Ruckerbauer, D.E.; Chauhan, N.; Hofbauer, H.F.; Krahulec, S.; Nidetzky, B.; Kohlwein, S.D.; Zanghellini, J.; Natter, K. Nutritional Requirements of the BY Series of Saccharomyces cerevisiae Strains for Optimum Growth. FEMS Yeast Res. 2012, 12, 796–808. [Google Scholar] [CrossRef]
- Dymond, J.S. Saccharomyces cerevisiae Growth Media. Methods Enzymol. 2013, 533, 191–204. [Google Scholar]
- Vallejo, B.; Matallana, E.; Aranda, A. Saccharomyces cerevisiae Nutrient Signaling Pathways Show An Unexpected Early Activation Pattern during Winemaking. Microb. Cell Factories 2020, 19, 124. [Google Scholar] [CrossRef]
- Heitmann, M.; Zannini, E.; Arendt, E. Impact of Saccharomyces cerevisiae Metabolites Produced during Fermentation on Bread Quality Parameters: A Review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1152–1164. [Google Scholar] [CrossRef]
- Rohde, J.R.; Bastidas, R.; Puria, R.; Cardenas, M.E. Nutritional Control via Tor Signaling in Saccharomyces cerevisiae. Curr. Opin. Microbiol. 2008, 11, 153–160. [Google Scholar] [CrossRef]
- Abe, F.; Horikoshi, K. Tryptophan Permease Gene TAT2 Confers High-Pressure Growth in Saccharomyces cerevisiae. Mol. Cell Biol. 2000, 20, 8093–8102. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.C.; Mira, N.P.; Sá-Correia, I. A Genome-Wide Perspective on the Response and Tolerance to Food-Relevant Stresses in Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 2011, 22, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Bravim, F.; Lippman, S.I.; da Silva, L.F.; Souza, D.T.; Fernandes, A.A.; Masuda, C.A.; Broach, J.R.; Fernandes, P.M. High Hydrostatic Pressure Activates Gene Expression that Leads to Ethanol Production Enhancement in A Saccharomyces cerevisiae Distillery Strain. Appl. Microbiol. Biotechnol. 2013, 97, 2093–2107. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takagi, H. Molecular Mechanisms and Highly Functional Development for Stress Tolerance of the Yeast Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2021, 85, 1017–1037. [Google Scholar] [CrossRef]
- Sasano, Y.; Haitani, Y.; Hashida, K.; Oshiro, S.; Shima, J.; Takagi, H. Improvement of Fermentation Ability under Baking-Associated Stress Conditions by Altering the POG1 Gene Expression in Baker’s Yeast. Int. J. Food Microbiol. 2013, 165, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Delling, U.; Raymond, M.; Schurr, E. Identification of Saccharomyces cerevisiae Genes Conferring Resistance to Quinoline Ring-Containing Antimalarial Drugs. Antimicrob. Agents Chemother. 1998, 42, 1034–1041. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, N.; Li, Z.; Wang, J.; Meng, X.; Gao, C.; Chen, J.; Chen, W.; Chen, X.; Liu, L. Mediator Engineering of Saccharomyces cerevisiae to Improve Multidimensional Stress Tolerance. Appl. Environ. Microbiol. 2022, 88, e0162721. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Engineering of Global Regulators and Cell Surface Properties toward Enhancing Stress Tolerance in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2017, 124, 599–605. [Google Scholar] [CrossRef]
- Doğan, A.; Demirci, S.; Aytekin, A.Ö.; Şahin, F. Improvements of Tolerance to Stress Conditions by Genetic Engineering in Saccharomyces cerevisiae during Ethanol Production. Appl. Biochem. Biotechnol. 2014, 174, 28–42. [Google Scholar] [CrossRef]
- Suzuki, T.; Sakamoto, T.; Sugiyama, M.; Ishida, N.; Kambe, H.; Obata, S.; Kaneko, Y.; Takahashi, H.; Harashima, S. Disruption of Multiple Genes Whose Deletion Causes Lactic-Acid Resistance Improves Lactic-Acid Resistance and Productivity in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2013, 115, 467–474. [Google Scholar] [CrossRef]
- Okada, S.; Doi, G.; Nakagawa, S.; Kusumoto, E.; Ito, T. Simple-to-Use CRISPR-SpCas9/SaCas9/AsCas12a Vector Series for Genome Editing in Saccharomyces cerevisiae. G3 2021, 11, jkab304. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, K.; Sherlock, G. High-Throughput Yeast Strain Sequencing. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Engel, S.R.; Weng, S.; Binkley, G.; Paskov, K.; Song, G.; Cherry, J.M. From One to Many: Expanding the Saccharomyces cerevisiae Reference Genome Panel. Database 2016, 2016, baw020. [Google Scholar] [CrossRef][Green Version]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome Evolution across 1,011 Saccharomyces cerevisiae Isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Cunha, J.T.; Soares, P.O.; Baptista, S.L.; Costa, C.E.; Domingues, L. Engineered Saccharomyces cerevisiae for lignocellulosic valorization: A review and perspectives on bioethanol production. Bioengineered 2020, 11, 883–903. [Google Scholar] [CrossRef]
- Eliodório, K.P.; Cunha, G.C.G.E.; Müller, C.; Lucaroni, A.C.; Giudici, R.; Walker, G.M.; Alves, S.L., Jr.; Basso, T.O. Advances in Yeast Alcoholic Fermentations for the Production of Bioethanol, Beer and Wine. Adv. Appl. Microbiol. 2019, 109, 61–119. [Google Scholar]
- Möllers, K.B.; Cannella, D.; Jørgensen, H.; Frigaard, N.U. Cyanobacterial Biomass as Carbohydrate and Nutrient Feedstock for Bioethanol Production by Yeast Fermentation. Biotechnol. Biofuels 2014, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The Promising Future of Microalgae: Current Status, Challenges, and Optimization of A Sustainable and Renewable Industry for Biofuels, Feed, and Other Products. Microb. Cell Fact. 2018, 17, 36. [Google Scholar] [CrossRef]
- Koppolu, V.; Vasigala, V.K. Role of Escherichia coli in Biofuel Production. Microbiol. Insights 2016, 9, 29–35. [Google Scholar] [CrossRef]
- Tabata, T.; Yoshiba, Y.; Takashina, T.; Hieda, K.; Shimizu, N. Bioethanol Production from Steam-Exploded Rice Husk by Recombinant Escherichia coli KO11. World J. Microbiol. Biotechnol. 2017, 33, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lu, X.; Li, Y.; Jiang, X.; Liu, L.; Wang, H. New Technologies Provide More Metabolic Engineering Strategies for Bioethanol Production in Zymomonas mobilis. Appl. Microbiol. Biotechnol. 2019, 103, 2087–2099. [Google Scholar] [CrossRef] [PubMed]
- Szambelan, K.; Szwengiel, A.; Nowak, J.; Frankowski, J.; Jeleń, H. Bioethanol Production from Sorghum Grain with Zymomonas mobilis: Increasing the Yield and Quality of Raw Distillates. J. Sci. Food Agric. 2023; Epub ahead of print. [Google Scholar] [CrossRef]
- Maleki, F.; Changizian, M.; Zolfaghari, N.; Rajaei, S.; Noghabi, K.A.; Zahiri, H.S. Consolidated Bioprocessing for Bioethanol Production by Metabolically Engineered Bacillus subtilis strains. Sci. Rep. 2021, 11, 13731. [Google Scholar] [CrossRef]
- Rajabi, M.; Nourisanami, F.; Ghadikolaei, K.K.; Changizian, M.; Noghabi, K.A.; Zahiri, H.S. Metagenomic Psychrohalophilic Xylanase from Camel Rumen Investigated for Bioethanol Production from Wheat Bran using Bacillus subtilis AP. Sci. Rep. 2022, 12, 8152. [Google Scholar] [CrossRef] [PubMed]
- Jacobus, A.P.; Gross, J.; Evans, J.H.; Ceccato-Antonini, S.R.; Gombert, A.K. Saccharomyces cerevisiae Strains Used Industrially for Bioethanol Production. Essays Biochem. 2021, 65, 147–161. [Google Scholar] [PubMed]
- Favaro, L.; Jansen, T.; van Zyl, W.H. Exploring Industrial and Natural Saccharomyces cerevisiae Strains for the Bio-Based Economy from Biomass: The Case of Bioethanol. Crit. Rev. Biotechnol. 2019, 39, 800–816. [Google Scholar] [CrossRef]
- Lane, S.; Dong, J.; Jin, Y.S. Value-Added Biotransformation of Cellulosic Sugars by Engineered Saccharomyces cerevisiae. Bioresour. Technol. 2018, 260, 380–394. [Google Scholar] [CrossRef]
- Li, X.; Cen, N.; Liu, L.; Chen, Y.; Yang, X.; Yu, K.; Guo, J.; Liao, X.; Shi, B. Collagen Peptide Provides Saccharomyces cerevisiae with Robust Stress Tolerance for Enhanced Bioethanol Production. ACS Appl. Mater. Interfaces 2020, 12, 53879–53890. [Google Scholar] [CrossRef]
- van Dijk, M.; Mierke, F.; Nygård, Y.; Olsson, L. Nutrient-Supplemented Propagation of Saccharomyces cerevisiae Improves Its Lignocellulose Fermentation Ability. AMB Express 2020, 10, 157. [Google Scholar] [CrossRef]
- Pereira, F.B.; Guimarães, P.M.; Teixeira, J.A.; Domingues, L. Optimization of Low-Cost Medium for very High Gravity Ethanol Fermentations by Saccharomyces cerevisiae Using Statistical Experimental Designs. Bioresour. Technol. 2010, 101, 7856–7863. [Google Scholar] [CrossRef]
- Naseeruddin, S.; Desai, S.; Venkateswar, R.L. Co-Culture of Saccharomyces cerevisiae (VS3) and Pichia stipitis (NCIM 3498) Enhances Bioethanol Yield from Concentrated Prosopis juliflora hydrolysate. 3 Biotech 2021, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Kruasuwan, W.; Puseenam, A.; Am-In, S.; Trakarnpaiboon, S.; Sornlek, W.; Kocharin, K.; Jindamorakot, S.; Tanapongpipat, S.; Bai, F.Y.; Roongsawang, N. Evaluation of Thermotolerant and Ethanol-Tolerant Saccharomyces cerevisiae as An Alternative Strain for Bioethanol Production from Industrial Feedstocks. 3 Biotech 2023, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Zhang, X.; Li, H.X.; Du, X.X.; Liang, S.W.; Zhao, X.H. Screening and Mutation of Saccharomyces cerevisiae UV-20 with A High Yield of Second Generation Bioethanol and High Tolerance of Temperature, Glucose and Ethanol. Indian J. Microbiol. 2018, 58, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Liu, K.; Chen, D.; Yuan, X.; Fang, J.; Yan, H.; Huang, L.; Chen, Y.; He, W. Improved Bioethanol Production Using CRISPR/Cas9 to Disrupt the ADH2 Gene in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2018, 34, 154. [Google Scholar] [CrossRef]
- de Valk, S.C.; Bouwmeester, S.E.; de Hulster, E.; Mans, R. Engineering Proton-Coupled Hexose Uptake in Saccharomyces cerevisiae for Improved Ethanol Yield. Biotechnol. Biofuels Bioprod. 2022, 15, 47. [Google Scholar] [CrossRef]
- Yang, P.; Jiang, S.; Lu, S.; Jiang, S.; Jiang, S.; Deng, Y.; Lu, J.; Wang, H.; Zhou, Y. Ethanol Yield Improvement in Saccharomyces cerevisiae GPD2 Delta FPS1 Delta ADH2 Delta DLD3 Delta Mutant and Molecular Mechanism Exploration Based on the Metabolic Flux and Transcriptomics Approaches. Microb. Cell Fact. 2022, 21, 160. [Google Scholar] [CrossRef]
- Nijland, J.G.; Driessen, A.J.M. Engineering of Pentose Transport in Saccharomyces cerevisiae for Biotechnological Applications. Front. Bioeng. Biotechnol. 2020, 7, 464. [Google Scholar] [CrossRef]
- Shao, X.; DiMarco, K.; Richard, T.L.; Lynd, L.R. Winter Rye as A Bioenergy Feedstock: Impact of Crop Maturity on Composition, Biological Solubilization and Potential Revenue. Biotechnol. Biofuels 2015, 8, 35. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Zi, L.H.; Bai, F.W.; Lin, H.L.; Hao, X.M.; Yue, G.J.; Ho, N.W. Bioethanol from Lignocellulosic Biomass. Adv. Biochem. Eng. Biotechnol. 2012, 128, 25–51. [Google Scholar]
- Park, H.; Jeong, D.; Shin, M.; Kwak, S.; Oh, E.J.; Ko, J.K.; Kim, S.R. Xylose Utilization in Saccharomyces cerevisiae during Conversion of Hydrothermally Pretreated Lignocellulosic Biomass to Ethanol. Appl. Microbiol. Biotechnol. 2020, 104, 3245–3252. [Google Scholar] [CrossRef] [PubMed]
- Abo, B.O.; Gao, M.; Wang, Y.; Wu, C.; Ma, H.; Wang, Q. Lignocellulosic Biomass for Bioethanol: An Overview on Pretreatment, Hydrolysis and Fermentation Processes. Rev. Environ. Health 2019, 34, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wei, L.; Pan, X.; Liu, C.; Jiang, J.; Wang, K. The Pretreatment of Lignocelluloses with Green Solvent as Biorefinery Preprocess: A Minor Review. Front. Plant Sci. 2021, 12, 670061. [Google Scholar] [CrossRef] [PubMed]
- Moysés, D.N.; Reis, V.C.; de Almeida, J.R.; de Moraes, L.M.; Torres, F.A. Xylose Fermentation by Saccharomyces cerevisiae: Challenges and Prospects. Int. J. Mol. Sci. 2016, 17, 207. [Google Scholar] [CrossRef]
- Cunha, J.T.; Soares, P.O.; Romaní, A.; Thevelein, J.M.; Domingues, L. Xylose Fermentation Efficiency of Industrial Saccharomyces cerevisiae Yeast with Separate or Combined Xylose Reductase/xylitol Dehydrogenase and Xylose Isomerase Pathways. Biotechnol. Biofuels 2019, 12, 20. [Google Scholar] [CrossRef]
- Soni, V.K.; Krishnapriya, R.; Sharma, R.K. Algae: Biomass to Biofuel. Methods Mol. Biol. 2021, 2290, 31–51. [Google Scholar]
- Wang, H.; Zhang, W.; Chen, L.; Wang, J.; Liu, T. The Contamination and Control of Biological Pollutants in Mass Cultivation of Microalgae. Bioresour. Technol. 2013, 128, 745–750. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Dragone, G.; Guimarães, P.M.; Silva, J.P.; Carneiro, L.M.; Roberto, I.C.; Vicente, A.; Domingues, L.; Teixeira, J.A. Technological Trends, Global Market, and Challenges of Bio-ethanol Production. Biotechnol. Adv. 2010, 28, 817–830. [Google Scholar] [CrossRef]
- Hinz, J.M.; Laughery, M.F.; Wyrick, J.J. Nucleosomes Inhibit Cas9 Endonuclease Activity in Vitro. Biochemistry 2015, 54, 7063–7066. [Google Scholar] [CrossRef]
- Jacobus, A.P.; Barreto, J.A.; de Bem, L.S.; Menegon, Y.A.; Fier, Í.; Bueno, J.G.R.; Dos Santos, L.V.; Gross, J. EasyGuide Plasmids Support in Vivo Assembly of gRNAs for CRISPR/Cas9 Applications in Saccharomyces cerevisiae. ACS Synth. Biol. 2022, 11, 3886–3891. [Google Scholar] [CrossRef]
- Dong, S.J.; Yi, C.F.; Li, H. Changes of Saccharomyces cerevisiae Cell Membrane Components and Promotion to Ethanol Tolerance during the Bioethanol Fermentation. Int. J. Biochem. Cell Biol. 2015, 69, 196–203. [Google Scholar] [CrossRef]
- Gnansounou, E. Production and Use of Lignocellulosic Bioethanol in Europe: Current Situation and Perspectives. Bioresour. Technol. 2010, 101, 4842–4850. [Google Scholar] [CrossRef] [PubMed]
- Yinbo, Q.; Zhu, M.; Liu, K.; Bao, X.; Lin, J. Studies on Cellulosic Ethanol Production for Sustainable Supply of Liquid Fuel in China. Biotechnol. J. 2006, 1, 1235–1240. [Google Scholar] [CrossRef]
- Zou, J.; Chang, X. Past, Present, and Future Perspectives on Whey as a Promising Feedstock for Bioethanol Production by Yeast. J. Fungi 2022, 8, 395. [Google Scholar] [CrossRef]
- Jouhten, P.; Ponomarova, O.; Gonzalez, R.; Patil, K.R. Saccharomyces cerevisiae metabolism in ecological context. FEMS Yeast Res. 2016, 16, fow080. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S. The Role of Yeasts in Fermentation Processes. Microorganisms 2020, 8, 1142. [Google Scholar] [CrossRef]
- Huang, K.X.; Vadiveloo, A.; Zhou, J.L.; Yang, L.; Chen, D.Z.; Gao, F. Integrated Culture and Harvest Systems for Improved Microalgal Biomass Production and Wastewater Treatment. Bioresour. Technol. 2023, 376, 128941. [Google Scholar] [CrossRef]
- Wang, H.; Deng, L.; Qi, Z.; Wang, W. Constructed Microalgal-Bacterial Symbiotic (MBS) System: Classification, Performance, Partnerships and Perspectives. Sci. Total Environ. 2022, 803, 150082. [Google Scholar] [CrossRef]
- Kang, Y.; Kim, M.; Shim, C.; Bae, S.; Jang, S. Potential of Algae-Bacteria Synergistic Effects on Vegetable Production. Front. Plant Sci. 2021, 12, 656662. [Google Scholar] [CrossRef] [PubMed]
- Khaleghi, M.K.; Savizi, I.S.P.; Lewis, N.E.; Shojaosadati, S.A. Synergisms of Machine Learning and Constraint-Based Modeling of Metabolism for Analysis and Optimization of Fermentation Parameters. Biotechnol. J. 2021, 16, e2100212. [Google Scholar] [CrossRef]
- Kim, S.R.; Park, Y.C.; Jin, Y.S.; Seo, J.H. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol. Adv. 2013, 31, 851–861. [Google Scholar] [CrossRef]
- Sànchez Nogué, V.; Karhumaa, K. Xylose Fermentation as A Challenge for Commercialization of Lignocellulosic Fuels and Chemicals. Biotechnol. Lett. 2015, 37, 761–772. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Wu, J.; Delneri, D.; O’Keefe, R.T. Non-coding RNAs in Saccharomyces cerevisiae: What Is the Function? Biochem. Soc. Trans. 2012, 40, 907–911. [Google Scholar] [CrossRef]
- Tinafar, A.; Jaenes, K.; Pardee, K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019, 17, 64. [Google Scholar] [CrossRef]
- Worst, E.G.; Finkler, M.; Schenkelberger, M.; Kurt, Ö.; Helms, V.; Noireaux, V.; Ott, A. A Methylation-Directed, Synthetic Pap Switch Based on Self-Complementary Regulatory DNA Reconstituted in an All E. coli Cell-Free Expression System. ACS Synth. Biol. 2021, 10, 2725–2739. [Google Scholar] [CrossRef]
- Fogeron, M.L.; Lecoq, L.; Cole, L.; Harbers, M.; Böckmann, A. Easy Synthesis of Complex Biomolecular Assemblies: Wheat Germ Cell-Free Protein Expression in Structural Biology. Front. Mol. Biosci. 2021, 8, 639587. [Google Scholar] [CrossRef]
- Ares, M., Jr. Preparation of Cell-Free Splicing Extracts from Saccharomyces cerevisiae. Cold Spring Harb. Protoc. 2013, 2013, 978–981. [Google Scholar] [CrossRef]
- Khattak, W.A.; Ullah, M.W.; Ul-Islam, M.; Khan, S.; Kim, M.; Kim, Y.; Park, J.K. Developmental Strategies and Regulation of Cell-Free Enzyme System for Ethanol Production: A Molecular Prospective. Appl. Microbiol. Biotechnol. 2014, 98, 9561–9578. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bassler, B.L. Bacterial Quorum Sensing in Complex and Dynamically Changing Environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Tian, X.; Ding, H.; Ke, W.; Wang, L. Quorum Sensing in Fungal Species. Annu. Rev. Microbiol. 2021, 75, 449–469. [Google Scholar] [CrossRef]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef]
- Stephens, K.; Bentley, W.E. Synthetic Biology for Manipulating Quorum Sensing in Microbial Consortia. Trends Microbiol. 2020, 28, 633–643. [Google Scholar] [CrossRef]
- Huang, X.F.; Reardon, K.F. Quorum-Sensing Molecules Increase Ethanol Yield from Saccharomyces cerevisiae. FEMS Yeast Res. 2021, 21, foab056. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).