Highly Efficient Biosynthesis of γ-Bisabolene with a New Sesquiterpene Synthase AcTPS5 by Dual Cytoplasmic-Peroxisomal Engineering in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis of Bisabolene Synthases
2.2. Medium, Culture Conditions, and Chemicals
2.3. Construction of Plasmids and Strains
2.4. Shake-Flask Fermentation
2.5. Fed-Batch Fermentation for γ-Bisabolene Biosynthesis
2.6. Analytical Methods
3. Results and Discussion
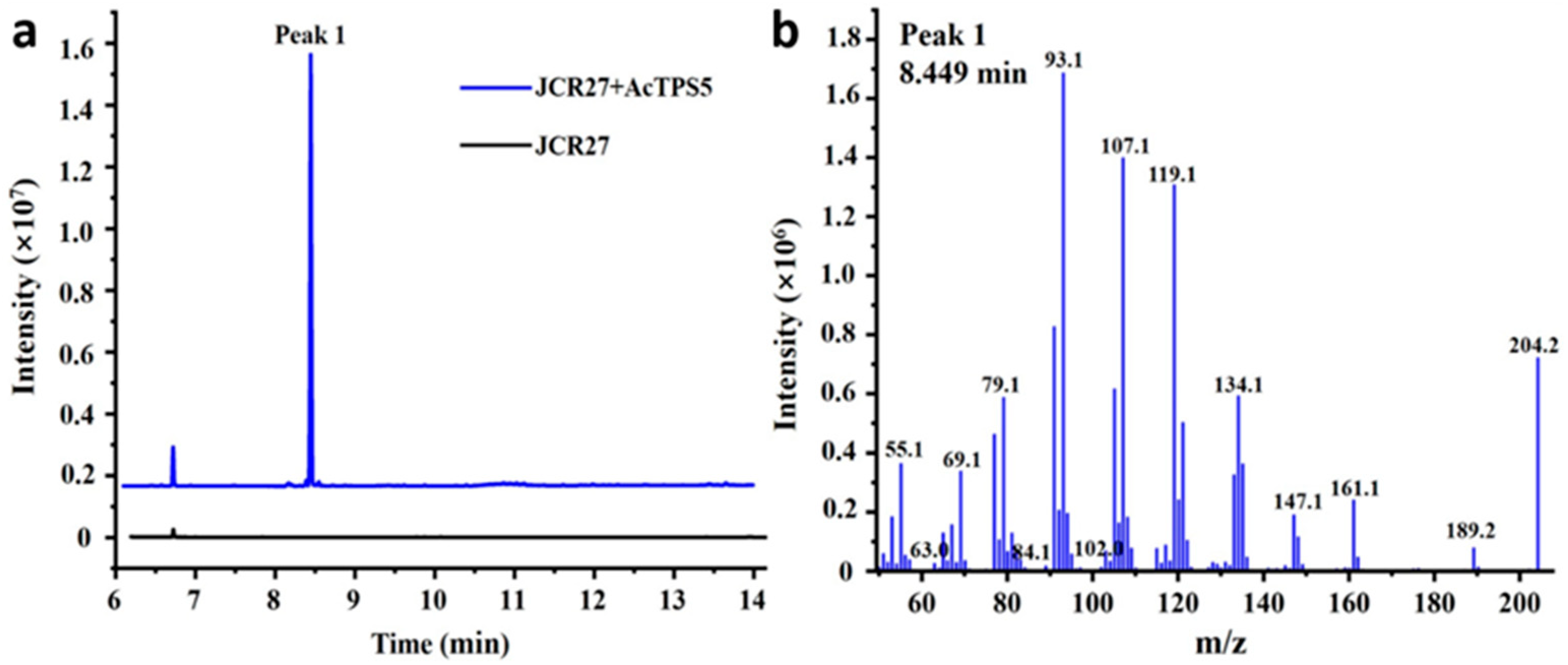
3.1. Functional Characterization and Sequence Analysis of AcTPS5
3.2. Producing γ-Bisabolene in S. cerevisiae Peroxisomes
3.3. Harnessing Peroxisomes to Enhance γ-Bisabolene Production
3.4. Dual Cytoplasmic-Peroxisomal Engineering to Optimize γ-Bisabolene Production
3.5. Engineering Acetyl-CoA Supply to Overproduce Bisabolene
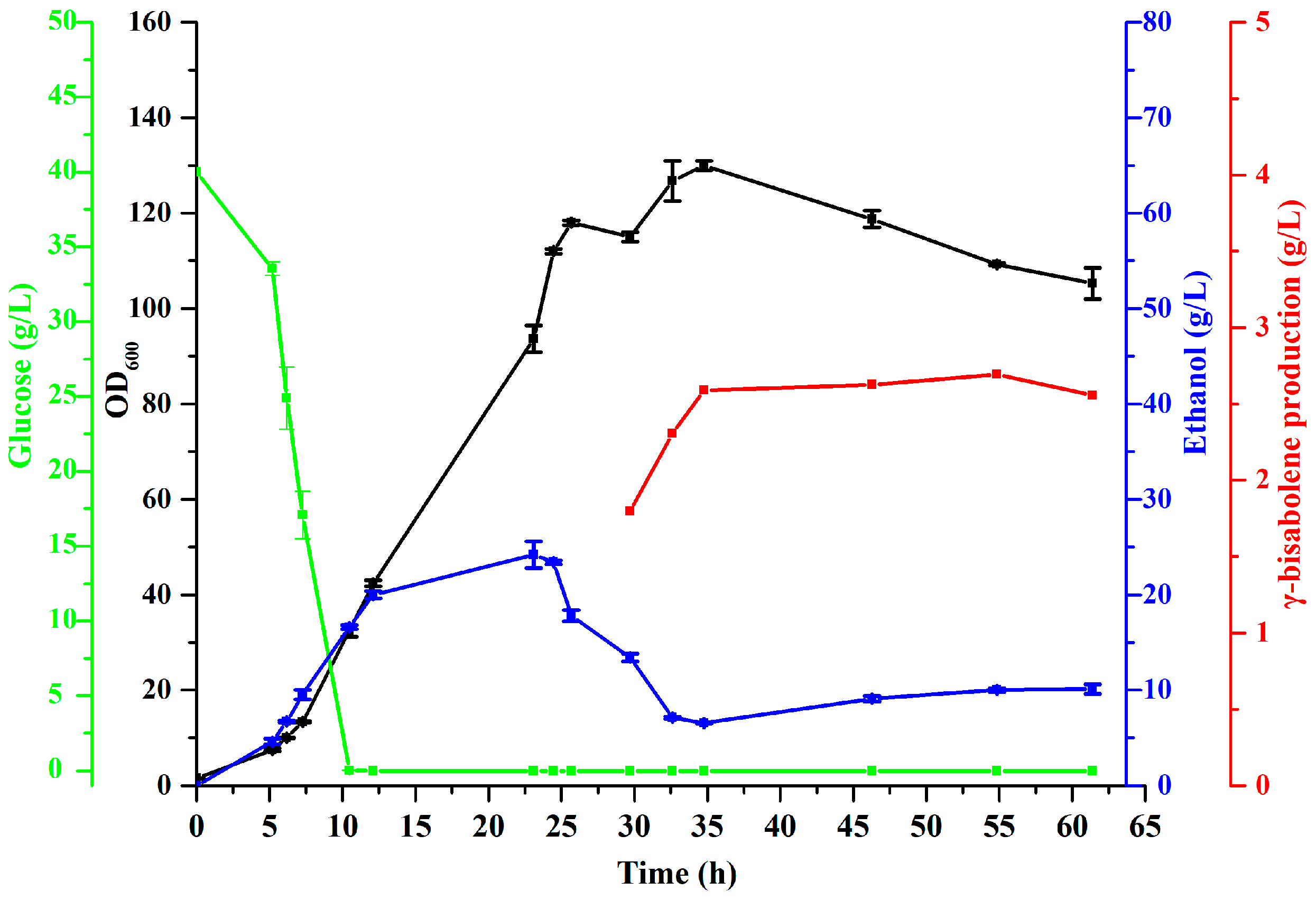
3.6. High-Density Fermentation for γ-Bisabolene Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, C.L.; Xue, K.; Yang, Y.K.; Liu, X.X.; Li, Y.; Lee, T.; Bai, Z.; Tan, T. Metabolic engineering strategies for sesquiterpene production in microorganism. Crit. Rev. Biotechnol. 2022, 42, 3–92. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, X. Biosynthesis of monoterpenoid and sesquiterpenoid as natural flavors and fragrances. Biotechnol. Adv. 2023, 65, 108151. [Google Scholar] [CrossRef] [PubMed]
- Hewage, R.T.; Tseng, C.C.; Liang, S.; Lai, C.Y.; Lin, H.C. Genome mining of cryptic bisabolenes that were biosynthesized by intramembrane terpene synthases from Antrodia cinnamomea. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2023, 378, 20220033. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Yahya, P.P.; Ouellet, M.; Chan, R.; Mukhopadhyay, A.; Keasling, J.D.; Lee, T.S. Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2011, 2, 483. [Google Scholar] [CrossRef]
- Kamatou, G.P.P.; Viljoen, A.M. A review of the application and pharmacological properties of alpha-bisabolol and alpha-bisabolol-rich oils. J. Am. Oil Chem. Soc. 2010, 87, 1–7. [Google Scholar] [CrossRef]
- Yeo, S.K.; Ali, A.Y.; Hayward, O.A.; Turnham, D.; Jackson, T.; Bowen, I.D.; Clarkson, R. β-Bisabolene, a sesquiterpene from the essential oil extract of Opoponax (Commiphora guidottii), exhibits cytotoxicity in breast cancer cell lines. Phytother. Res. 2016, 30, 418–425. [Google Scholar] [CrossRef]
- Nascimento, A.M.; Brandao, M.G.; Oliveira, G.B.; Fortes, I.C.; Chartone-Souza, E. Synergistic bactericidal activity of Eremanthus erythropappus oil or β-bisabolene with ampicillin against Staphylococcus aureus. Antonie Leeuwenhoek. 2007, 92, 95–100. [Google Scholar] [CrossRef]
- Shu, H.Z.; Peng, C.; Bu, L.; Guo, L.; Liu, F.; Xiong, L. Bisabolane-type sesquiterpenoids: Structural diversity and biological activity. Phytochemistry 2021, 192, 112927. [Google Scholar] [CrossRef]
- Jou, Y.J.; Hua, C.H.; Lin, C.S.; Wang, C.Y.; Wan, L.; Lin, Y.J.; Huang, S.H.; Lin, C.W. Anticancer activity of γ-bisabolene in human neuroblastoma cells via induction of p53-mediated mitochondrial apoptosis. Molecules 2016, 21, 601. [Google Scholar] [CrossRef]
- Govindarajan, M.; Vaseeharan, B.; Alharbi, N.S.; Kadaikunnan, S.; Khaled, J.M.; Al-Anbr, M.N.; Alyahya, S.A.; Maggi, F.; Benelli, G. High efficacy of (Z)-γ-bisabolene from the essential oil of Galinsoga parviflora (Asteraceae) as larvicide and oviposition deterrent against six mosquito vectors. Environ. Sci. Pollut. Res. Int. 2018, 25, 10555–10566. [Google Scholar] [CrossRef]
- Lim, D.S.; Shi, L.L.; Guo, K.; Luo, S.; Liu, Y.; Chen, Y.; Liu, Y.; Li, S.H. A new sesquiterpene synthase catalyzing the formation of (R)-β-bisabolene from medicinal plant Colquhounia coccinea var. mollis and its anti-adipogenic and antibacterial activities. Phytochemistry 2023, 211, 113681. [Google Scholar]
- Chemat, F.; Vian, M.A.; Cravotto, G. Green extraction of natural products: Concept and principles. Int. J. Mol. Sci. 2012, 13, 8615–8627. [Google Scholar] [CrossRef] [PubMed]
- Walls, L.E.; Otoupal, P.; Ledesma-Amaro, R.; Velasquez-Orta, S.B.; Gladden, J.M.; Rios-Solis, L. Bioconversion of cellulose into bisabolene using Ruminococcus flavefaciens and Rhodosporidium toruloides. Bioresour. Technol. 2023, 368, 128216. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhu, K.; Li, J.; Zhao, Y.; Li, S.L.; Zhang, C.; Xiao, D.; Yu, A.Q. High-efficiency production of bisabolene from waste cooking oil by metabolically engineered Yarrowia lipolytica. Microb. Biotechnol. 2021, 14, 2497–2513. [Google Scholar] [CrossRef] [PubMed]
- Kulagina, N.N.; Besseau, S.; Papon, N.; Courdavault, V. Peroxisomes: A New Hub for Metabolic Engineering in Yeast. Front. Bioeng. Biotechnol. 2021, 9, 659431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.B.; Li, M.; Zhao, G.; Lu, W.Y. Harnessing Yeast Peroxisomes and cytosol acetyl-CoA for sesquiterpene α-Humulene production. J. Agric. Food Chem. 2020, 68, 1382–1389. [Google Scholar] [CrossRef]
- Gerke, J.; Frauendorf, H.; Schneider, D.; Wintergoller, M.; Hofmeister, T.; Poehlein, A.; Zebec, Z.; Takano, E.; Scrutton, N.S.; Braus, G.H. Production of the fragrance geraniol in peroxisomes of a product-tolerant baker’s yeast. Front. Bioeng. Biotechnol. 2020, 8, 582052. [Google Scholar] [CrossRef]
- Liu, G.S.; Li, T.; Zhou, W.; Jiang, M.; Tao, X.Y.; Liu, M.; Zhao, M.; Ren, Y.; Gao, B.; Wang, F.Q.; et al. The yeast peroxisome: A dynamic storage depot and subcellular factory for squalene overproduction. Metab. Eng. 2020, 57, 151–161. [Google Scholar] [CrossRef]
- Hammer, S.K.; Avalos, J.L. Harnessing yeast organelles for metabolic engineering. Nat. Chem. Biol. 2017, 13, 823–832. [Google Scholar] [CrossRef]
- Liu, J.J.; Chen, C.; Wan, X.K.; Yao, G.; Bao, S.H.; Wang, F.L.; Wang, K.; Song, T.Y.; Han, P.G.; Jiang, H. Identification of the sesquiterpene synthase AcTPS1 and high production of (–)-germacrene D in metabolically engineered Saccharomyces cerevisiae. Microb. Cell Fact. 2022, 21, 89. [Google Scholar] [CrossRef]
- Rinkel, J.; Dickschat, J.S. Stereochemical investigations on the biosynthesis of achiral (Z)-γ-bisabolene in Cryptosporangium arvum. Beilstein J. Org. Chem. 2019, 15, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, H.; Ukeba, S.; Kitaoka, T. Latent potentials of the white-rot basidiomycete Phanerochaete chrysosporium responsible for sesquiterpene metabolism: CYP5158A1 and CYP5144C8 decorate (E)-α-bisabolene. Enzym. Microb. Technol. 2022, 158, 110037. [Google Scholar] [CrossRef] [PubMed]
- Siemon, T.; Wang, Z.Q.; Bian, G.K.; Seitz, T.; Ye, Z.L.; Lu, Y.; Cheng, S.; Ding, Y.K.; Huang, Y.L.; Deng, Z.X.; et al. Semisynthesis of plant-derived englerin A enabled by microbe engineering of guaia-6,10(14)-diene as building block. J. Am. Chem. Soc. 2020, 142, 2760–2765. [Google Scholar] [CrossRef]
- Wang, F.; Lv, X.; Xie, W.; Zhou, P.; Zhu, Y.; Yao, Z.; Yang, C.; Yang, X.; Ye, L.; Yu, H. Combining Gal4p-mediated expression enhancement and directed evolution of isoprene synthase to improve isoprene production in Saccharomyces cerevisiae. Metab. Eng. 2017, 39, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.Z.; Yao, M.D.; Wang, Y.; Zhou, L.; Song, T.; Liu, H.; Xiao, W.H.; Yuan, Y.J. Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metab. Eng. 2017, 41, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Apel, A.R.; d’Espaux, L.; Wehrs, M.; Sachs, D.; Li, R.A.; Tong, G.J.; Garber, M.; Nnadi, O.; Zhuang, W.; Hillson, N.; et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2017, 45, 496–508. [Google Scholar]
- Chen, Y.; Wang, Y.; Liu, M.; Qu, J.Z.; Yao, M.D.; Li, B.; Ding, M.Z.; Liu, H.; Xiao, W.H.; Yuan, Y.J. Primary and secondary metabolic effects of a key gene deletion (Δypl062w) in metabolically engineered terpenoid-producing Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2019, 85, e01990-18. [Google Scholar] [CrossRef] [PubMed]
- Akşit, A.; Klei, I.J. Yeast peroxisomes: How are they formed and how do they grow? Int. J. Biochem. Cell Biol. 2018, 105, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Rottensteiner, H.; Stein, K.; Sonnenhol, E.; Erdmann, R. Conserved Function of Pex11p and the Novel Pex25p and Pex27p in Peroxisome Biogenesis. Mol. Biol. Cell 2003, 14, 4316–4328. [Google Scholar] [CrossRef][Green Version]
- Fagarasanu, A.; Fagarasanu, M.; Rachubinski, R.A. Maintaining peroxisome populations: A story of division and inheritance. Annu. Rev. Cell Dev. Biol. 2007, 23, 321–344. [Google Scholar] [CrossRef]
- Motley, A.M.; Nuttall, J.M.; Hettema, E.H. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012, 31, 2852–2868. [Google Scholar] [CrossRef]
- Donald, K.A.G.; Hampton, R.Y.; Fritz, I.B. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997, 63, 3341–3344. [Google Scholar] [CrossRef]
- Lv, X.M.; Wang, F.; Zhou, P.P.; Ye, L.D.; Xie, W.P.; Xu, H.M.; Yu, H.W. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae. Nat. Commun. 2016, 7, 12851. [Google Scholar] [CrossRef]
- Jiang, Y.K.; Xia, L.; Gao, S.; Li, N.; Yu, S.Q.; Zhou, J.W. Engineering Saccharomyces cerevisiae for enhanced (–)-α-bisabolol production. Synth. Syst. Biotechnol. 2023, 8, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.M.; Shi, B.; Ye, Z.L.; Huang, M.; Chen, R.; Cai, Y.S.; Kuang, Z.L.; Sun, X.; Bian, G.K.; Deng, Z.X.; et al. Systematic identification of Ocimum sanctum sesquiterpenoid synthases and (–)-eremophilene overproduction in engineered yeast. Metab. Eng. 2022, 69, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Henry, K.W.; Nickels, J.T.; Edlind, T.D. ROX1 and ERG regulation in Saccharomyces cerevisiae: Implications for antifungal susceptibility. Eukaryot. Cell 2002, 1, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Lv, X.; Ye, L.; Zhou, P.; Yu, H. Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering. Metab. Eng. 2015, 30, 69–78. [Google Scholar] [CrossRef]
- Faulkner, A.; Chen, X.; Rush, J.; Horazdovsky, B.; Waechter, C.J.; Carman, G.M.; Sternweis, P.C. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 14831–14837. [Google Scholar] [CrossRef]
- Wang, H.M.; Yang, Y.; Lin, L.; Zhou, W.L.; Liu, M.Z.; Cheng, K.D.; Wang, W. Engineering Saccharomyces cerevisiae with the deletion of endogenous glucosidases for the production of flavonoid glucosides. Microb. Cell Factories 2016, 15, 134. [Google Scholar] [CrossRef]
- Paramasivan, K.; Mutturi, S. Progress in terpene synthesis strategies through engineering of Saccharomyces cerevisiae. Crit. Rev. Biotechnol. 2017, 37, 974–989. [Google Scholar] [CrossRef]
- Lian, J.Z.; Si, T.; Nair, N.U.; Zhao, H.M. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab. Eng. 2014, 24, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Krivoruchko, A.; Amatriain, C.S.; Chen, Y.; Siewers, V.; Nielsen, J. Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism. J. Ind. Microbiol. Biotechnol. 2013, 40, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Scalcinati, G.; Knuf, C.; Partow, S.; Chen, Y.; Maury, J.; Schalk, M.; Daviet, L.; Nielsen, J.; Siewers, V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene α-santalene in a fed-batch mode. Metab. Eng. 2012, 14, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Giles, S.S.; Perfect, J.R. Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 60–72. [Google Scholar] [CrossRef]
- Vitolo, M.; Duranti, M.A.; Pellegrim, M.B. Effect of pH, aeration and sucrose feeding on the invertase activity of intact S. cerevisiae cells grown in sugarcane blackstrap molasses. J. Ind. Microbiol. 1995, 15, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, J.; Crock, J.; Jetter, R.; Croteau, R. Terpenoid-based defenses in conifers: cDNA cloning, characterization, and functional expression of wound-inducible (E)-α-bisabolene synthase from grand fir (Abies grandis). Proc. Natl. Acad. Sci. USA 1998, 95, 6756–6761. [Google Scholar] [CrossRef]
- Huber, D.W.; Philippe, R.N.; Godard, K.A.; Sturrock, R.N.; Bohlmann, J. Characterization of four terpene synthase cDNAs from methyl jasmonate-induced Douglas-fir, Pseudotsuga menziesii. Phytochemistry 2005, 66, 1427–1439. [Google Scholar] [CrossRef]
- Fujisawa, M.; Harada, H.; Kenmoku, H.; Mizutani, S.; Misawa, N. Cloning and characterization of a novel gene that encodes (S)-beta-bisabolene synthase from ginger, Zingiber officinale. Planta 2010, 232, 121–130. [Google Scholar] [CrossRef]
- Parveen, I.; Wang, M.; Zhao, J.; Chittiboyina, A.G.; Tabanca, N.; Ali, A.; Baerson, S.R.; Techen, N.; Chappell, J.; Khan, I.A.; et al. Investigating sesquiterpene biosynthesis in Ginkgo biloba: Molecular cloning and functional characterization of (E,E)-farnesol and alpha-bisabolene synthases. Plant Mol. Biol. 2015, 89, 451–462. [Google Scholar] [CrossRef]
- Srivastava, P.L.; Daramwar, P.P.; Krithika, R.; Pandreka, A.; Shankar, S.S.; Thulasiram, H.V. functional characterization of novel sesquiterpene synthases from indian sandalwood, Santalum album. Sci. Rep. 2015, 5, 10095. [Google Scholar] [CrossRef]
- Ro, D.K.; Ehlting, J.; Keeling, C.I.; Lin, R.; Mattheus, N.; Bohlmann, J. Microarray expression profiling and functional characterization of AtTPS genes_ duplicated Arabidopsis thaliana sesquiterpene synthase genes. Arch. Biochem. Biophys. 2006, 448, 104–116. [Google Scholar] [CrossRef]
- Aschenbrenner, A.K.; Kwon, M.; Conrad, J.; Ro, D.; Spring, O. Identification and characterization of two bisabolene synthases from linear glandular trichomes of sunflower (Helianthus annuus L., Asteraceae). Phytochemistry 2016, 124, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.; Lehner, B.; Khrimian, A.; Muchlinski, A.; Luck, K.; Köllner, T.G.; Weber, D.C.; Gundersen-Rindal, D.; Tholl, D. An IDS-Type sesquiterpene synthase produces the pheromone precursor (Z)-α-Bisabolene in Nezara viridula. J. Chem. Ecol. 2019, 45, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Niu, M.; Silva, J.T.; Zhang, Y.Y.; Yuan, Y.F.; Jia, Y.; Xiao, Y.; Li, Y.; Fang, L.; Zeng, S.; et al. Identification and functional characterization of three new terpene synthase genes involved in chemical defense and abiotic stresses in Santalum album. BMC Plant Biol. 2019, 19, 115. [Google Scholar] [CrossRef] [PubMed]
- Muchlinski, A.; Chen, X.L.; Lovell, J.T.; Köllner, T.G.; Pelot, K.A.; Zerbe, P.; Ruggiero, M.; Callaway, L.; Laliberte, S.; Chen, F.; et al. Biosynthesis and emission of stress-induced volatile terpenes in roots and leaves of switchgrass (Panicum virgatum L.). Front Plant Sci. 2019, 10, 1144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Wang, J.; Wang, Z.B.; Zhang, Y.M.; Shi, S.B.; Nielsen, J.; Liu, Z.H. A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae. Nat. Commun. 2019, 5, 1053. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Yao, G.; Wan, X.; Wang, F.; Han, P.; Bao, S.; Wang, K.; Song, T.; Jiang, H. Highly Efficient Biosynthesis of γ-Bisabolene with a New Sesquiterpene Synthase AcTPS5 by Dual Cytoplasmic-Peroxisomal Engineering in Saccharomyces cerevisiae. Fermentation 2023, 9, 779. https://doi.org/10.3390/fermentation9090779
Liu J, Yao G, Wan X, Wang F, Han P, Bao S, Wang K, Song T, Jiang H. Highly Efficient Biosynthesis of γ-Bisabolene with a New Sesquiterpene Synthase AcTPS5 by Dual Cytoplasmic-Peroxisomal Engineering in Saccharomyces cerevisiae. Fermentation. 2023; 9(9):779. https://doi.org/10.3390/fermentation9090779
Chicago/Turabian StyleLiu, Jiajia, Ge Yao, Xiukun Wan, Fuli Wang, Penggang Han, Shaoheng Bao, Kang Wang, Tianyu Song, and Hui Jiang. 2023. "Highly Efficient Biosynthesis of γ-Bisabolene with a New Sesquiterpene Synthase AcTPS5 by Dual Cytoplasmic-Peroxisomal Engineering in Saccharomyces cerevisiae" Fermentation 9, no. 9: 779. https://doi.org/10.3390/fermentation9090779
APA StyleLiu, J., Yao, G., Wan, X., Wang, F., Han, P., Bao, S., Wang, K., Song, T., & Jiang, H. (2023). Highly Efficient Biosynthesis of γ-Bisabolene with a New Sesquiterpene Synthase AcTPS5 by Dual Cytoplasmic-Peroxisomal Engineering in Saccharomyces cerevisiae. Fermentation, 9(9), 779. https://doi.org/10.3390/fermentation9090779




