3.1. Physico-Chemical Analysis
The physicochemical analysis results of the 1st, 3rd, 7th, and 21st days of olive leaves fermented with
L. plantarum ATCC 14917 and ATCC-BAA 793 are presented in
Figure 1. In parallel with a slight decrease in pH values, an increase was observed in total acidity values in fermented olive leaves. While the pH value of samples fermented with
L. plantarum ATCC-14917 and
L. plantarum ATCC-BAA 793 was determined as 5.15 and 5.20 on the first day of fermentation, they were found as 4.59 and 4.76, respectively without much change at the end of the 21-day fermentation process. Likewise, the acidity values, which were 0.07 and 0.06% on the 1st day of fermentation, increased at the end of the fermentation process and were found to be 0.40 and 0.31%. In all samples in which LAB were added, the total acidity value was found to be higher than the control samples (non-inoculated) (0.20%) at the end of the 21-day fermentation time. The results show that the use of LAB strain has a significant effect on acidification when compared with non-inoculated samples. The increase in acidity could be explained by the microbial activity of microorganisms, mainly LAB that metabolize sugar and other nutritional contents in the brine leading to the formation of acids and a decrease in pH [
18]. In a study, oleuropein biodegradation by
L. plantarum FSO175 strain was investigated during 7-day fermentation at 30 °C, and the final values of pH and free acidity were 5.7 and 0.42%, respectively [
19]. In another study, olives inoculated with
L. plantarum S175 showed a rapid decrease in pH from 6 to 4.5 during the first 5 days of fermentation, followed by a slight decrease to around pH 4 until the end of the fermentation process. In addition, the free acidity increased continuously and was found to be 1% at the end of the fermentation process [
18]. The same behavior of pH change was observed in the present study. Olives inoculated with
L. plantarum have shown more rapid acidification than spontaneous fermentation during 25 days of fermentation in a previous study [
20]. A pH value of olives inoculated with the
L. plantarum KLOS 1.0328 strain isolated from the Italian variety was found between 5.0 and 9.0 and the results indicated that
L. plantarum KLDS 1.0328 exhibits stress tolerance under the acid and alkali stress conditions [
21]. In the study comparing the spontaneous and
L. pentosus inoculated fermentation of olives, the average pH value started with 5, and decreased significantly, reaching approximately 4 on the 7th day of fermentation. pH values below 4.5 inhibited the growth of Proteobacteria and other acid-sensitive bacteria [
22].
3.2. Microbiological Analysis
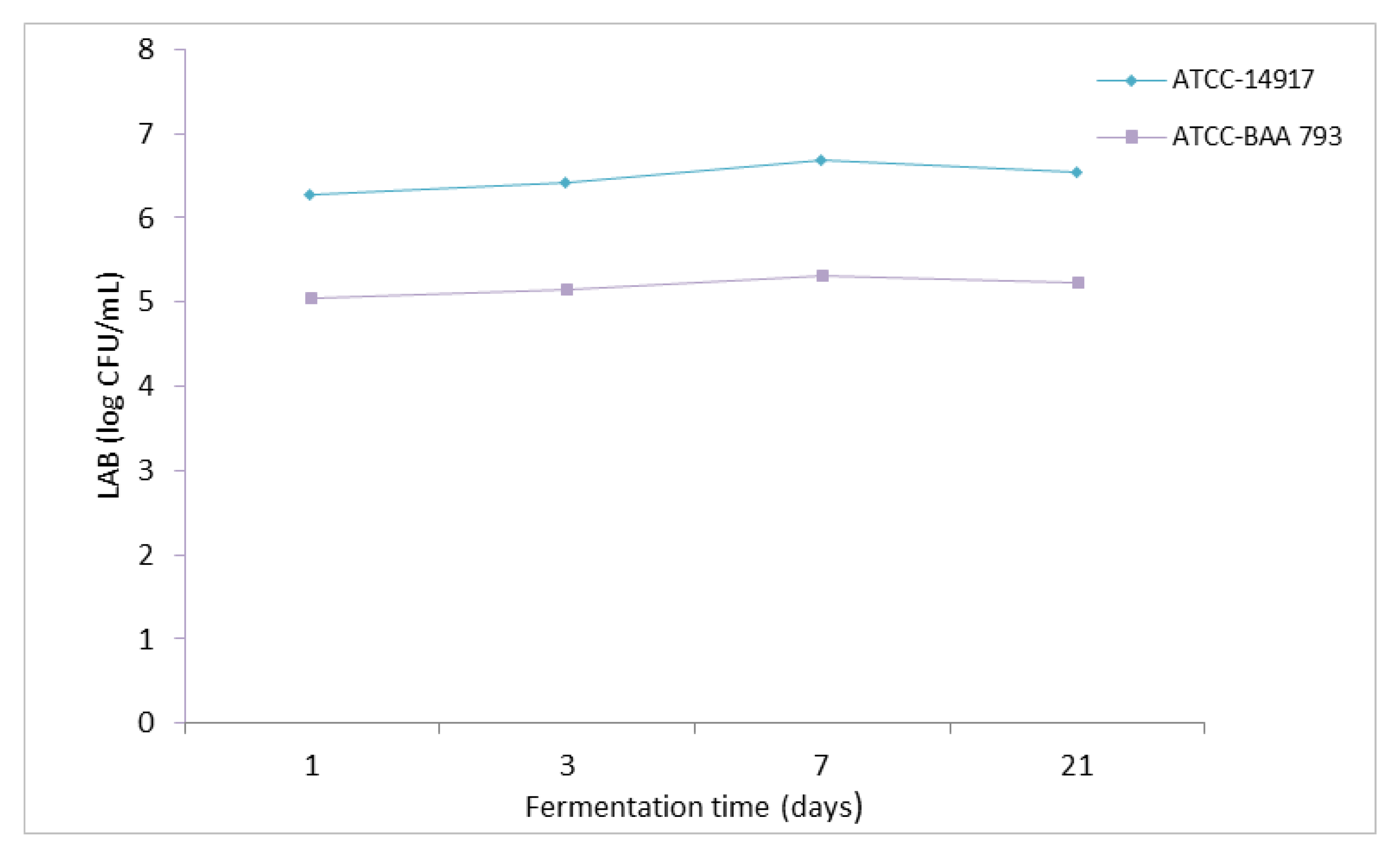
The number of LAB in the brines of fermented olive leaves varied between 5.14 and 6.68 log CFU/mL (
Figure 2). The LAB counts were higher in the samples with
L. plantarum ATCC 14917 compared to the
L. plantarum ATCC-BAA 793. This is consistent when evaluated together with the total acidity results. The total acidity increased in the fermented leaf brine with higher LAB counts. In addition, after the first 7 days of the fermentation process, a slight decrease in the LAB number was observed at the end of the fermentation process in leaves inoculated with both bacterial strains. In another study investigating the effect of stress factors (pH, temperature, and NaCl) associated with olive fermentation on the growth and oleuropein degradation abilities of selected
L. plantarum strains, all tested strains increased cell densities more than 2 log units, starting from an average of 7 logs CFU/mL in the control condition at pH 6.0 and incubation at 32 °C [
23]. In general, all strains were found to be more resistant to both single and multiple stress conditions at 32 °C than at 16 °C [
24]. In a study conducted on
L. plantarum KLOS 10328 strain, it was determined that the strain tended to tolerate acidic stress rather than osmotic stress. It has been reported that pH values between 5.0 and 9.0 do not significantly affect the growth of isolated
L. plantarum strains [
24]. Our results were compatible with a study reported by Bevilacqua et al. [
24] concerning the pH value (4.59–4.76) which led to less stress for the growth of
L. plantarum strain.
Some studies have demonstrated the ability of
L. plantarum strains to tolerate high (>8%) NaCl concentrations [
25,
26,
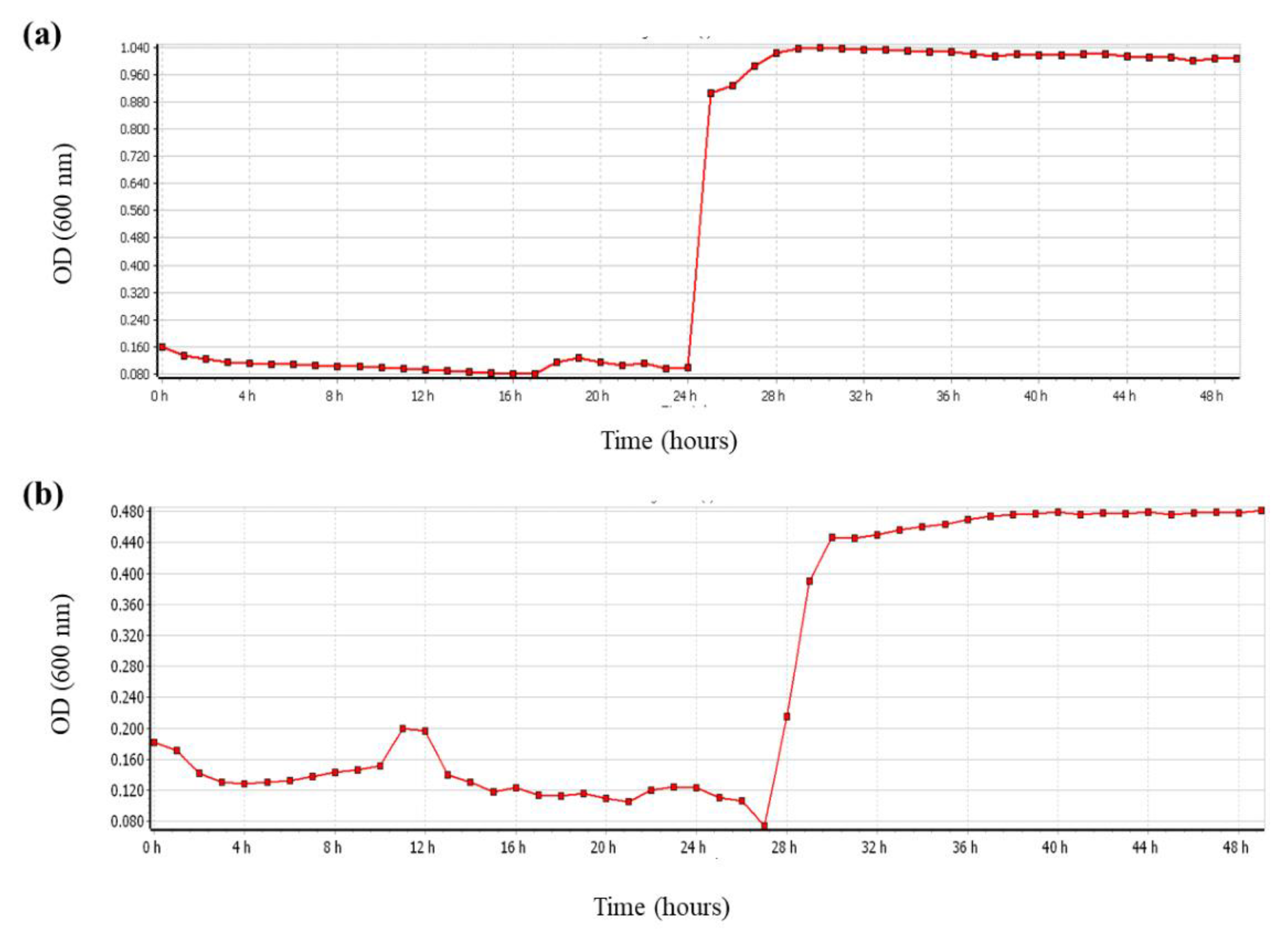
27]. In the present study, the effect of ambient conditions on bacterial growth was determined according to the growth curves of
L. plantarum ATCC 14917 and
L. plantarum ATCC-BAA 793 using a microplate reader. Stress conditions, such as concentration of salt (5%) and incubation temperature (30 °C), did not show a negative effect on the growth and particularly the oleuropein biodegradation capacity of
L. plantarum ATCC 14917 and
L. plantarum ATCC-BAA 793 (
Figure 3). In another study in which the fermentation process of olives was controlled by two bacterial species (
L. plantarum S175 and
L. pentosus), the interaction between cell concentration and time significantly affected the development of the LAB population. This LAB population showed a slight increase from 5.7
−9 × 10
7 cfu/mL to about 10
9 cfu/mL during the first 15 days of the fermentation, then decreased to about 2 × 10
5 cfu/mL at the end of the fermentation [
18].
3.3. Bioconversion of Oleuropein to Hydroxytyrosol
In our study, bioconversion of oleuropein to hydroxytyrosol depending on the activity of β-glucosidase and esterase enzymes of LAB in fermented olive leaves with
L. plantarum ATCC 14917 and
L. plantarum ATCC-BAA 793 strains were investigated and hydroxytyrosol-rich extracts were obtained under optimized conditions. The changes in the amounts of oleuropein and hydroxytyrosol in fermented olive leaves and the brine inoculated with different
L. plantarum strains are given in
Table 1. When the results are examined, hydroxytyrosol is considered to be the main hydrolysis product for oleuropein degradation and this phenol diffused from olive leaves to brine. This diffusion between brine and olive leaves occurs normally due to the osmotic pressure difference [
28]. As a result of this exchange of substances, oleuropein and the water-soluble components diffuse from the olive leaves to the brine. Therefore, concerning fermented olives and olive leaves, brine may be important for the recovery of phenolic compounds and to obtain extracts with high phenolic content [
29].
In the first 5 days of fermentation, the content of oleuropein, which is expected to decrease as a result of biodegradation, increased until the 7th day of fermentation for both bacterial strains (
Table 1). This increase in the amount of oleuropein can be explained by the higher and continuous diffusion of oleuropein by osmosis from olive leaves to the brine [
25]. After the 5th day of fermentation, the oleuropein content decreased from 535.23 to 298.64 mg/mL and from 350.78 to 246.14 mg/L in the brine of fermented olive leaves inoculated with
L. plantarum ATCC-14917 and
L. plantarum BAA-793, respectively.
The highest enzyme activity in both bacterial strains started to occur from the 3rd day of fermentation. Depending on the reduction of oleuropein content, the amount of hydroxytyrosol, which was 28.16 and 11.83 mg/mL on the first day of fermentation increased to 126.89 and 85.93 mg/mL after 21 days of fermentation for the fermented samples with
L. plantarum ATCC-14917 and
L. plantarum BAA-793, respectively (
Table 1). It has also been reported that longer fermentation time can lead to a 40–70% reduction in hydroxytyrosol [
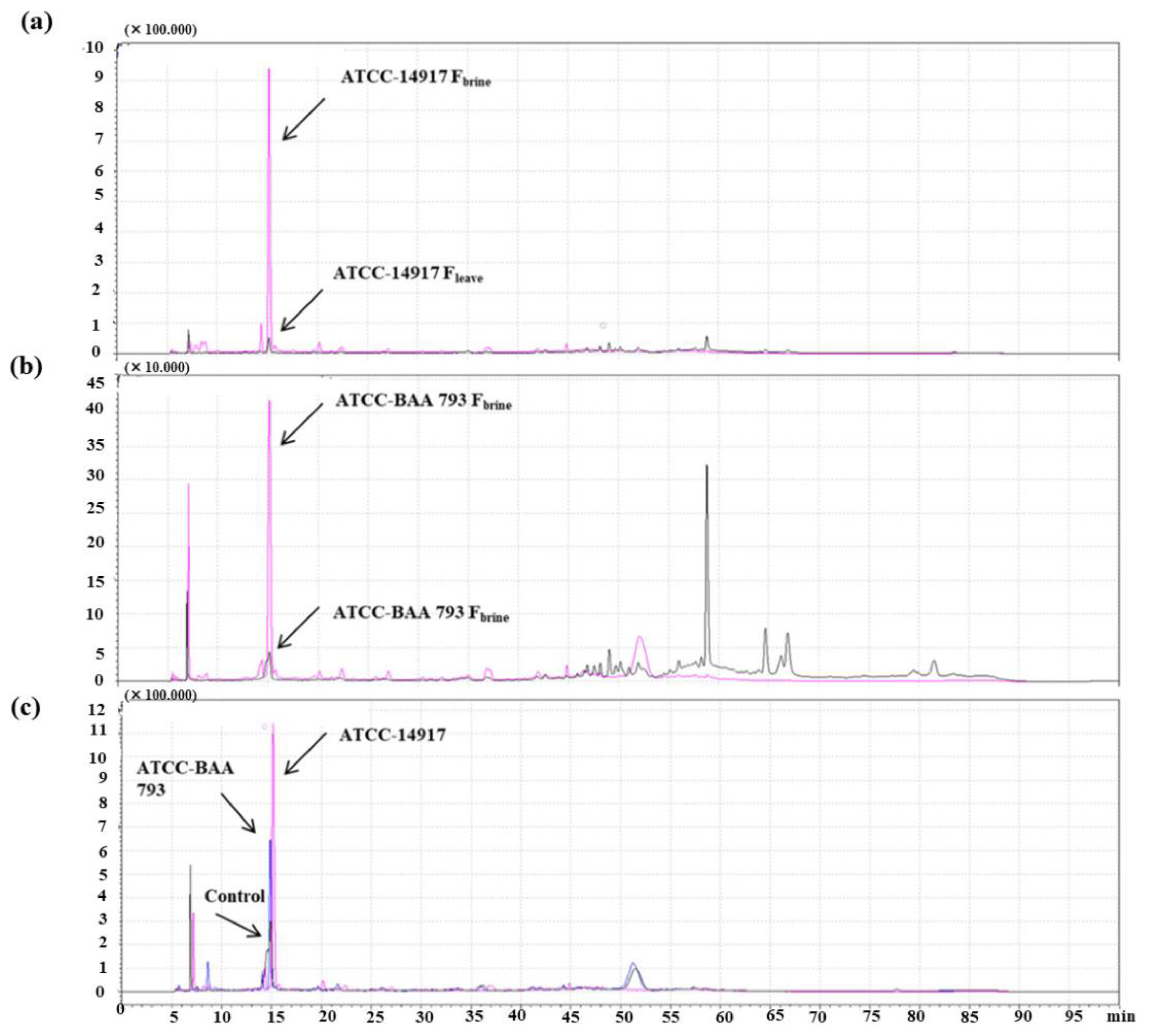
30]. In addition, regarding the oleuropeinolitic activity of both L. plantarun strain, bioconversion of oleuropein to hydroxytyrosol was shown in HPLC chromatograms (
Figure 4) for control (non-inoculated) and brine of fermented olive leaves. When HPLC chromatograms were examined, as a result of the hydrolysis of the initial glycosides during fermentation, a significant difference was observed between the phenolic composition of the control and fermented olive leaf brines. Hydroxytyrosol content in fermented olive leaf brine by adding
L. plantarum strain was considerably higher than the control (9.92 ± 0.09 mg/mL). Moreover, the highest hydroxytyrosol content was determined in the sample with
L. plantarum ATCC 14917 (126.89 mg/mL) after 21 days of fermentation. The degradation of oleuropein was attributed to the β-glucosidase activity of the
L. plantarum strain. However, some studies have shown that oleuropein degradation is not only associated with 𝛽-glucosidase and esterase but also polyphenol oxidase and peroxidase catalyze the conversion of oleuropein [
31,
32].
3.4. Optimization of Ultrasound-Assisted Extraction Conditions of Fermented Olive Leaves
UAE conditions were optimized for highly efficient extraction of hydroxytyrosol, which diffused from leaves to brine after the 21-day fermentation process. In the optimization study, the independent variables were determined as solvent (ethanol/water) concentration, extraction time, ultrasound power, and the dependent variable as the concentration of hydroxytyrosol. Process variables and their levels used in the Box–Behnken design for both bacteria strain are given in
Table 2. It was observed that the model was not significant for the ethanol concentration range from 30–70%
v/v (data not shown). Therefore, the model was reconstituted with the ethanol concentration ranging from 10 to 90% for
L. plantarum ATCC-BAA 793. In the experimental plan designed for
L. plantarum ATCC 14917, the hydroxytyrosol concentration was found to be between 142.24 and 244.02 mg/L, while for
L. plantarum ATCC-BAA 793 it varied between 139.59 and 220.59 mg/L (
Table 3). F-statistic was used to determine significant terms in the ANOVA analysis. Insignificant factors (
p > 0.05) were removed from the model and then, the regression coefficients were recalculated. The adequacy of the model fit was checked for the response according to the lack of fit value. Accordingly, the error caused by the lack of fit value was found insignificant, and the variation resulting from the regression was found significant in the 95% confidence interval, ensuring the model’s suitability for optimization. To determine the optimum extraction conditions, the desirability function method was used, and the response surface plots were obtained to visualize the results clearly [
33].
The sequential model sum of squares obtained for the hydroxytyrosol results of the sample extracts fermented with
L. plantarum ATCC 14917 and ATCC-BAA 793 is given in
Table 4 and
Table 5, respectively. By applying regression analysis, the relationship between factors and response was revealed with a mathematical model. Accordingly, the linear and interaction effect terms of the factors were added to the models. The effects of the factors on the response were evaluated by considering ANOVA.
For
L. plantarum ATCC 14917 fermented sample, the statistical significance of the linear and interaction effects of the factors on response is expressed in
Table 6. While the lack of fit value was determined to be insignificant (
p > 0.05), the model was also found to be insignificant at the 95% confidence interval (
p > 0.05). It was observed that ethanol concentration and time were effective on the hydroxytyrosol content. However, the effect of the amplitude on response was not found significant. Therefore, insignificant factors (
p > 0.05) were removed from the model and the regression coefficients were recalculated (
Table 7). The effect of ethanol concentration and extraction time on the dependent variable was investigated in the new model created by removing the vibration amplitude. The reduced model was found significant at the 95% confidence level, while the lack of fit value was insignificant (
p > 0.05). The interaction (BC) was found effective on the amount of hydroxytyrosol in the fermented samples. Furthermore, the time factor had a greater effect on the response than the ethanol concentration in the samples fermented with
L. plantarum ATCC 14917. In addition, when the terms used in testing the model fit (
Table 8) are examined, the fact that the Pred-R2 and R2 adj values are not close to each other shows that the fit model is not suitable for prediction.
For
L. plantarum ATCC-BAA 793 fermented sample, the statistical significance of the linear and interaction effects of the factors on response is examined in
Table 9. While the lack of fit value was determined to be insignificant (
p > 0.05), the model was insignificant at the 95% confidence interval (
p < 0.05). Moreover, insignificant factors (
p > 0.05) were removed from the model and the regression coefficients were recalculated (
Table 10). Ethanol concentration (
p < 0.0001) was found to be more effective on the response than the time (
p < 0.0244). In terms of the table used in testing the model fit (
Table 11), the Pred-R2 and R2 adj values are found as close to each other and it shows that the fitted model is suitable for prediction for
L. plantarum ATCC-BAA 793 fermented sample. The regression analysis of the data showed a significant effect of linear terms ethanol concentration (B), extraction time (C) and interaction (BC) on the hydroxytyrosol content of fermented extracts. Ethanol concentration and time influenced hydroxytyrosol results of samples with
L. plantarum ATCC 14917 negatively while time affected hydroxytyrosol content positively in samples by adding
L. plantarum ATCC BAA 793. The regression equations for the linear model obtained as a result of the regression analysis for the variables were given in Equations (1) and (2) in terms of actual variables for
L. plantarum ATCC 14917 and ATCC-BAA 793, respectively.
Moreover, to visualize the effects of independent variables on hydroxytyrosol content, the response surface plot was obtained (
Figure 5). As shown in
Figure 5a, the hydroxytyrosol concentration increased as the ethanol concentration and extraction time decreased in
L. plantarum ATCC 14917 fermented olive leaf brine. On the contrary, the hydroxytyrosol concentration of the sample fermented with
L. plantarum ATCC-BAA 793 increased as the ethanol concentration decreased and the extraction time increased (
Figure 5b).
Based on the results of the optimization study, the optimum condition was determined as 30% amplitude—30% ethanol concentration—5 min extraction time for
L. plantarum ATCC-14917, while it was found as 45% amplitude—10% ethanol concentration—9 min extraction time for
L. plantarum ATCC-BAA 793 (
Table 12). In addition, the desirability values at the optimum conditions were obtained as 0.746 and 0.952 for
L. plantarum ATCC 14917 and
L. plantarum ATCC-BAA 793, respectively.
Experimental results were obtained at the optimum point to determine the accuracy of the optimum condition. The hydroxytyrosol concentrations of fermented leaves, brine, and control (non-inoculated) extracts under optimum UAE conditions are given in
Table 13. The hydroxytyrosol concentration was higher in the samples fermented with
L. plantarum ATCC 14917 and
L. plantarum ATCC-BAA 793 compared to the control samples (
Table 13). On the other hand, the hydroxytyrosol concentrations of the Fleave-14917 and Fbrine-14917 were determined as 38.97 ± 0.86 and 362.29 ± 2.31 mg/L, respectively. Likewise, it was found to be 25.38 ± 0.53 and 248.79 ± 4.11 mg/L for the Fleave-793 and Fbrine-793. Hydroxytyrosol content in the fermented leaf brine was found to be higher than the leaves due to the diffusion of oleuropein and hydroxytyrosol from the leaves to the brine. In addition,
L. plantarum ATCC 14917 (362.29 ± 2.31 mg/L) strain showed more activity in the bioconversion of oleuropein to hydroxytyrosol than
L. plantarum ATCC-BAA 793 (248.79 ± 4.11) under optimum extraction conditions (
Table 7). Hydroxytyrosol content of extracts obtained at the optimum point is given using HPLC chromatograms in
Figure 6.
In a study in which olives were fermented using two lactobacillus strains (
L. plantarum S175 and
L. pentosus S100), the amount of hydroxytyrosol at the end of the fermentation process was found to be 154.2 mg/100 mL in the samples inoculated with
L. plantarum S175 [
7]. The researchers investigated the oleuropein degradation capacity of
L. plantarum strains, and there was no significant increase in the amount of hydroxytyrosol (approximately less than 0.03 mg/mL in 72 h) in the sample by adding
L. plantarum [
34]. In our study, a notable increase in the amount of hydroxytyrosol (varied between 34.07 and 56.51 mg/mL in 72 h) was detected in samples fermented with
L. plantarum strain when compared to the study of Iorizzo et al. [
34]. In the study examining the conversion of oleuropein to hydroxytyrosol by different bacterial strains, the commercial oleuropein was used and the most effective strain was determined as
L. plantarum 14917 with a hydroxytyrosol efficiency of approximately 30% and oleuropein degradation of 90% under aerobic conditions. After two weeks of fermentation, hydroxytyrosol content was found to be between 216.7 and 476.7 mg/mL for different
L. plantarum strains [
10]. It was seen in the present work,
L. plantarum 14917 has shown less activity in the oleuropein degradation when compared with the study of Zago et al. [
10]. These variations could be attributed to the characteristics of the olive fruit and its by-products, the enzymatic activities of the strains, and other different factors, and so, the degradation of oleuropein in olive leaves may differ from the degradation of commercial oleuropein.