Abstract
Pecorino Siciliano PDO is a semi-hard cheese that is produced in wooden vats using raw sheep’s milk and its associated autochthonous microbial community. In the present study, we evaluated the microbial ecology of the milk, curd and whey from five Pecorino Siciliano PDO-producing farms in Sicily using a combination of metagenomic and microbiological approaches. We present an overview of the species and strain-level diversity of dairy lactococcal and streptococcal isolates using established genotyping tools and compare the lactic acid bacterial populations present in samples from these farms. Whole genome sequences of representative isolates of Lactococcus spp. and Streptococcus thermophilus were elucidated and the genetic diversity of the strains was established through analysis of predicted phage-resistance systems and prophage-associated regions. The analysis revealed farm-specific dairy lactococcal and streptococcal isolates that possess diverse genotypic features including newly described phage-resistance systems.
1. Introduction
Lactic acid bacteria (LAB) are a large group of Gram-positive bacterial genera that include starter and non-starter bacteria that are applied in food fermentations and associated with a variety of food products. Among these, Lactococcus lactis/cremoris, Streptococcus thermophilus and certain lactobacilli and Leuconostoc spp. are predominantly associated with the production of fermented dairy products. In industrial fermentation where defined starter cultures are employed, L. lactis/cremoris and S. thermophilus are the dominantly applied species for mesophilic and thermophilic production systems, respectively [1]. However, the starter culture composition of artisanally produced cheeses is only recently being studied in detail to establish its ecology and diversity. Studies of artisanal cheeses associated with different geographical locations and employing distinct milk substrates (bovine, ovine, caprine milk) have described their distinct and diverse microbiota using metagenomic and/or microbiological approaches [2,3,4]. The microbiota analysis of such cheeses has typically been performed using the final product (often ripened) while limited research attention has been devoted to defining the microbiota of such cheeses during the production process, and thus the dominant starter microbiota. Being a traditional Sicilian product, Pecorino Siciliano PDO is currently under-promoted beyond the island of Sicily and information on the microbial composition and starter potential are essential in order to protect and develop this cheese. Pecorino Siciliano PDO is of considerable value for producers, also in view of territory valorization and to slow down land abandonment of internal Sicilian areas. Until recent changes were implemented [5], the production regulations of Pecorino Siciliano PDO required a minimum ripening period of four months. This medium-term production system entails direct costs related to the handling and storage of the cheeses. Thus, detailed microbiological information during production and ripening of this cheese is of paramount importance to allow certification of quality for fresh (20–30 days) and semi-mature (60–90 days) Pecorino Siciliano cheese, in compliance with traditional processing techniques. This would support a defining improvement of the economic performance for the producers, with predictable positive effects in the socio-economically less favored rural areas where the associated farms are located [6].
Pecorino Siciliano PDO cheese was awarded the EU-protected designation of origin (PDO) status in 1996 and is a semi-hard white cheese produced from raw ovine milk without the deliberate inoculation of starter cultures, thereby adhering to the traditional cheese-making protocol [7]. Fermenting microbiota may either originate from the raw milk, animal rennet added for curdling, the cheese-making environment and, in particular, the microbial biofilm established in the wooden vats used for milk coagulation [8]. The cheese-making process includes preheating milk to 38–40 °C, transferring it into a wooden vat and the addition of animal-rennet paste. The curd is broken to reduce the size of the coagulum to the dimension of rice grains, and syneresis is facilitated by the addition of hot water at a temperature of 70–90 °C. The curd is then placed in moulding rattan baskets where it is pressed firmly to facilitate draining of the whey. The rattan baskets containing the curds are placed in the wooden vat or in a steel tank and coated with hot deproteinized whey (~80 °C) for cooking for approximately 3–4 h [9,10]. The curd is then removed from the rattan baskets, dried at ambient temperature for 24–48 h and salted in saturated brine or by manual dispersion of salt onto the cheese surfaces. Ripening occurs on wooden shelves [11] at a temperature between 14 and 18 °C, with 75–80% relative humidity for a variable period, from 20 to more than 120 days, depending on the cheese type [7] (For a detailed review of the production process, see [10]). Italy is leading producer of sheep milk cheeses within the EU and among these Pecorino (Siciliano) PDO is one of the most significant in terms of production volume and export value [6]. Traditionally, the microbiota of cheeses was evaluated microbiologically using presumptive Lactobacillus and Lactococcus/S. thermophilus counts on MRS or M17, respectively at 30, 37/42 °C [12,13]. In many instances, microbiological analysis was paired with 16S rRNA gene sequencing or genotyping using tools such as RAPD-PCR [13]. Previous microbiological analysis of 4-month ripened Pecorino Siciliano identified a dominance of presumptive mesophilic lactobacilli (approximately 108 cfu/g) based on growth on MRS agar at mesophilic temperatures [3]. Since raw milk is used in the production process, it is perhaps unsurprising that pathogenic and spoilage microorganisms including Staphylococcus, Enterococcus and Listeria spp. have also been identified in Pecorino Siciliano PDO cheeses; however, during the ripening process, these organisms are reported to be eliminated [14].
Recently, multiplex PCR systems have been established for both L. lactis/cremoris and S. thermophilus based on the genetic diversity of gene clusters associated with the biosynthesis of cell wall polysaccharides (CWPS) termed the cwps and rgp loci in Lactococcus and S. thermophilus, respectively [15,16]. The multiplex PCR system for lactococci is capable of distinguishing between three cwps genotypes, termed A, B and C [15]. Furthermore, C-type strains are distinguished into eight C-subtypes (C1 to C8) with primers established to identify strains with a C1 to C5 genotype [17,18]. The S. thermophilus multiplex PCR system is a dual PCR with one multiplex PCR aimed at distinguishing between three “backbone” (Bt) and five “variable” (Vt) genotypes representing the 3′ and 5′ ends of the rgp gene cluster, respectively [16]. These PCR systems incorporate a control species-specific primer pair which is useful in the species-level identification of isolates.
In addition to genotyping based on the above-mentioned PCR systems, the presence and diversity of predicted prophages and phage-resistance systems can be used to distinguish strains of a given species. Lactococci are reported to harbor up to six prophages on their genomes [19,20]. Conversely, prophages are reportedly present in low abundance in dairy streptococcal strains’ genomes by comparison to their lactococcal counterparts [21]. Prophages may excise from the host chromosome under specific conditions resulting in cell lysis. However, their presence may also be beneficial through the provision of defenses against secondary infecting bacteriophages [22,23,24]. Furthermore, in response to bacteriophage pressure in fermentation environments, dairy lactococci and streptococci have evolved and acquired several phage-resistance mechanisms. Five major phage-resistance systems have been described in lactococci and streptococci, i.e., abortive infection (Abi), restriction/modification (R/M), Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) sequences and CRISPR-associated (Cas) genes, adsorption inhibition, and DNA injection blocking mechanisms [25]. Recently, a number of new phage-resistance systems have been identified and functionally characterized in diverse bacterial species with novel modes of action including nucleotide depletion and chemical interference systems [26,27,28,29]. The recent resurgence in the study of prophages and phage-resistance systems has coincided with improved tools to predict the presence of such phage-defense systems in bacterial genomes. Tools such as PHASTER, PADLOC and DefenseFinder have revolutionized the analysis of prokaryotic genomes and facilitated the rapid identification of putative prophages and phage-resistance genes [30,31,32]. Such elements may be used as markers to differentiate strains.
While several studies have described the overall microbiota composition of cheeses at the species level, few studies have evaluated strain-level diversity [33,34]. In the present study, it was aimed to define the LAB species and strain level diversity of milk, curd and whey obtained from five Pecorino Siciliano PDO-producing farms using culture-dependent and independent approaches.
2. Materials and Methods
2.1. Sample Collection and Processing
Milk (ovine milk), curd and whey samples associated with five geographically distinct farms that produce Pecorino Siciliano PDO in Sicily were collected in March 2022 (Figure 1). Farms 1–3 are low hill farms while Farms 4 and 5 are located in mountainous regions of Sicily. All farms possess the same sheep breed and are farms that predominantly allow sheep to feed on pasture. The samples were stored at 4 °C prior to processing; 5 g of curd or 5 mL of milk and whey samples were mixed with 45 mL 2% trisodium citrate (Sigma-Aldrich, Gillingham, UK) and homogenized for 2 min at 300 rpm in a stomacher (Stomacher Circular 400; Seward, UK). Serial dilutions of each sample were prepared in quarter strength Ringers’ solution and plated on LM17 agar [M17 agar (Oxoid, Hampshire, UK) supplemented with 0.5% lactose (Sigma Aldrich, Gillingham, UK)], or MRS agar (Oxoid, Hampshire, UK) incubated overnight at 30 °C aerobically and at 37 and 42 °C anaerobically (Anaerocult A—Merck, NJ, USA). Viable counts on both agars at various temperatures were recorded as CFU/g or mL of the original sample.
Figure 1.
Map indicating the location of the Sicilian farms from which the milk, curd and whey samples were collected in March 2022. Farm 1 (F1): Santa Margherita di Belice; Farm 2 (F2): Contessa Entellina; Farm 3 (F3): Partanna; Farm 4 (F4): Castronovo di Sicilia; Farm 5 (F5): Santo Stefano Quisquina.
2.2. Identification of Presumptive Lactococci and Dairy Streptococci
Twenty-five colonies representing each of the observed morphologies for each sample on MRS and/or LM17 agar were selected for further analysis to identify potential lactococci and/or dairy streptococci. To achieve this, presumptive strains of Lactococcus were grown at 30 °C, while those of S. thermophilus were incubated at 37 °C overnight in LM17 broth or agar, and a multiplex PCR incorporating primers that are conserved among, and specific to, the genomes of lactococcal or dairy streptococcal strains’ genomes was applied. The Lactococcus-specific primers ConF (5′-GTACACTATGTTTATAACAATCATCCAG-3′) and ConR (5′-GCAAACCAGATTCAAAGTCAGTATG-3′) and S. thermophilus-specific primers MSF (5′- GCTGGTCGTAATTACCTCG-3′) and MSR (5′- CAACATCTTCCAAGGTACG-3′) were used to identify isolates belonging to these genera/species [15,16]. PCRs were performed using Phusion Green Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher, Gloucester, UK) employing the following conditions: 98 °C for 10 min followed by 30 cycles of 98 °C for 15 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a final extension step at 72 °C for 10 min. Amplicons were visualized on a 1% agarose gel with UV transillumination. Amplicon sizes of 891 bp and 2724 bp were indicative of presumptive lactococcal and streptococcal isolates, respectively.
2.3. Genotypic Diversity Analysis of Lactococcal and Dairy Streptococcal Isolates
Using an established multiplex PCR system based on distinct regions within the gene cluster associated with cell wall polysaccharide biosynthesis (termed cwps in Lactococcus spp.), the presumptive lactococci were assigned to one of four genotypes (A, B, C or undefined) [15]. Strains identified as belonging to the C genotype were further evaluated using a C-subtyping multiplex PCR system which is capable of distinguishing five distinct C- subtypes (C1 to C5) [17,35]. The presumptive S. thermophilus isolates were genotyped using a dual multiplex PCR system that distinguishes genotypes based on the left and rightward regions of the rgp gene cluster [16]. The primers associated with these multiplex PCR systems are listed in Table S1. The PCRs were performed using the same conditions as described in the previous section.
2.4. Species Identification of Isolates Using 16S rRNA Gene Sequencing
PCR amplification of the 16S rRNA gene was performed on selected isolates that could not be speciated using the described multiplex PCR systems. The 16S rRNA gene was amplified using the LucF and LucR primers (LucF CTTGTTACGACTTCACCC; LucR TGCCTAATACATGCAAGT), and Taq DNA polymerase mastermix (Qiagen) under the following conditions: initial denaturation at 94 °C for 10 min, 30 cycles of 94 °C for 30 s, 40 °C for 30 s, 72 °C for 1 min and 30 s, followed by a final extension at 72 °C for 10 min. The amplicons were purified using the GenElute™ PCR Clean-Up Kit (Sigma Aldrich) according to the manufacturer’s instructions and subjected to Sanger sequencing (Genewiz, Azenta Life Sciences, Leipzig, Germany). The generated sequences were compared using BLASTn against available sequence data on the National Center for Biotechnology Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 7 June 2022).
2.5. Metagenome Extraction and Microbiota Analysis by 16S rRNA Gene Profiling
Total DNA was extracted from the five curd samples using a protocol adapted from Erkus et al. [36]. Briefly, 2.5 g of curd was added to 22.5 mL 2.2% sodium citrate solution at 45 °C and homogenized in a stomacher at 230 bpm for 5 min. The homogenate was centrifuged at 13,750× g for 10 min and the pellet was washed three times in a 2.2% sodium citrate solution. The cell pellet was resuspended in 1 mL of lysis buffer containing 20 mM Tris HCl (pH8.0), 2 mM EDTA (pH8.0) and 2% polyethylene glycol 6000. The suspension was treated with 50 µg/mL lysozyme and 100 U mutanolysin at 37 °C followed by treatment with proteinase K (250 µg/mL) for 1 h at 56 °C; 1 mL of 96% ethanol was added to the lysed solution which was then applied to the Qiagen DNEasy Blood and Tissue kit as per manufacturer’s instructions. Partial 16S rRNA gene sequences were amplified from the DNA extracts and sequenced on an Illumina MiSeq platform as described previously [37]. The resulting fastq files were processed using a customized QIIME2-based script as described previously [38]. Biodiversity within each of the curd samples was calculated through the richness index (alpha diversity) while the similarity between samples (beta diversity) was calculated using Bray–Curtis dissimilarity with similarities reported within a 0 to 1 range [39]. Principal Coordinate analysis (PCoA) to display the beta diversity was performed using QIIME2 [40]. Statistical analysis of the beta diversity of the microbiota was performed using t-test analysis and PERMANOVA analyses to estimate the significance of the differences in PCoA data as described previously [38].
2.6. DNA Extraction, Genome Sequencing and Analysis
Genomic DNA of the Lactococcus and S. thermophilus strains was isolated from a fresh 10 mL overnight culture of each strain using a NucleoBond® DNA extraction kit with Buffer set III (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions with the inclusion of lysozyme (Sigma-Aldrich; to 20 mg/mL), mutanolysin (Sigma-Aldrich; 500 U/mL), and proteinase K (Macherey Nagel; 40 µg/mL) addition to Buffer G3 prior to an extended (16–18 h) incubation at 37 °C. The DNA was stored at −20 °C prior to shipment to the sequencing facility. Genome sequencing of the selected isolates (paired ends 2 × 250 bp) was performed using Illumina MiSeq sequencing technology (GenProbio, Parma, Italy). De novo sequence assemblies and automated gene calling was performed using the MEGAnnotator pipeline [41]. Predicted open reading frames (ORFs) were predicted via Prodigal v2.6 and Genemark.hmm [42]. CRISPR-Cas encoding regions were identified using CRISPRCasFinder and selecting the evidence level 2 to 4 outputs only [43]. A general search for phage-resistance systems (including and beyond R/M and CRISPR-Cas systems) was performed using PADLOC using the default settings [31]. The prediction of prophage-associated regions in the bacterial genomes was performed using PHASTER [30]. Predictions of anti-microbial compound production were performed using BAGEL4 [44]. The outputs of bioinformatic searches were manually inspected for their validity, where appropriate.
2.7. Anti-Microbial Production Evaluation
The spot-on-lawn assay was adapted from the method of Parlindungan et al. [45] using LM17 agar; 10 µL spots of fresh overnight cultures of the lactococcal and streptococcal isolates were transferred on LM17 agar, incubated overnight at 30 (Lactococcus) or 37 °C (S. thermophilus), UV-treated for 45 min and overlayed with a semi-solid agar containing 200 µL of a fresh overnight culture of the indicator strain Lactococcus cremoris HP cross-sensitivity test was performed for the S. thermophilus isolates using the same approach but with 400 µL of the seven S. thermophilus strains as indicators in place of L. cremoris HP followed by incubation at 37 °C overnight. Inhibition was indicated where zones of clearing were observed and the diameter of the zones of inhibition was recorded (mm).
3. Results
3.1. Metagenome Analysis of Pecorino Siciliano PDO Curd from Five Sicilian Farms
Pecorino Siciliano PDO is a traditional Sicilian cheese produced through spontaneous fermentations of the autochthonous microbiota of wooden vats and using raw ovine milk. The metagenome of the curd associated with Pecorino production from five Sicilian farms was evaluated using 16S rRNA gene profiling. Between 36,000 and 70,000 reads were generated for each sample (Table 1) and after quality filtering between 33,000 and 65,000 reads were applied to subsequent taxonomic analysis.

Table 1.
Number of reads from each farm’s curd sequenced metagenome extract.
Metagenome analysis established that each curd possessed a unique microbiota. A close relationship between those samples collected from Farm 1 and Farm 2 which were observed to group most closely in a 3-Dimensional Principal Coordinate Analysis (PCoA) (Figure 2a,c). The low abundance/absence of lactococci and streptococci in the curd sample from Farm 4 corroborates the observed low microbial richness evaluated by the Chao1 index through its alpha rarefaction curve (Figure 2b). Among the LAB, at genus level, Lactococcus, Streptococcus, Lactobacillus, Leuconostoc and Enterococcus were identified in four of the five curd samples (Farm 4 curd was shown to possess 0.07% reads which mapped to Streptococcus). Farms 3, 1 and 2 curds were found to contain the highest relative abundance levels of Lactococccus reads (29.37, 17.85 and 11.48%, respectively) and the curds from Farms 3 and 2 were observed to contain the highest proportion of Streptococcus reads (47.66 and 6.72%, respectively). Notably, Leuconostoc was shown to be present in high relative abundance in Farm 5 curd (37.47%), while Lactobacillus was shown to be typically present at <1% in any of the assessed samples.
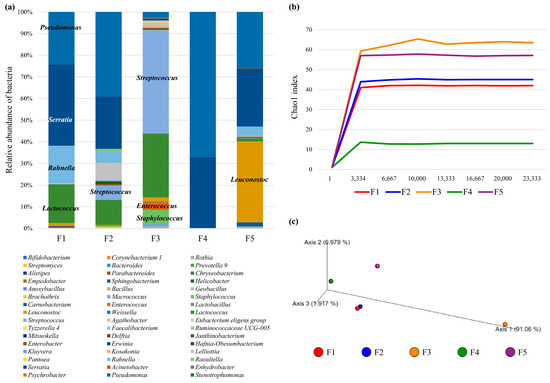
Figure 2.
Bacterial diversity associated with curd samples from each of the five farms (F1–F5). (Panel a) shows the bacterial distribution among cheeses, in which each bacterial genus is color-coded as described in the chart legend. (Panel b) reports the rarefaction curves based on the Chao1 index at increasing sequencing depth of cheeses samples. (Panel c) displays the predicted PCoA through a three-dimensional image.
At the species level, L. lactis was identified at a high relative abundance in the curd from Farm 3 (29.27%) and in low abundance in Farms 1 (0.36%), 2 (0.33%) and 5 (1.18%). Undefined lactococcal species were also observed in high abundance (12.91%) in the curd from Farm 1 (Figure 1). The curd from Farm 5 was determined to contain a high relative abundance of Leuconostoc mesenteroides (32.87%), while lower levels of this species were observed in curd samples from Farms 1 (1.27%), 2 (0.14%) and 3 (1.77%). Interestingly, the curd from Farm 4 appeared to contain very limited LAB. Farm 3 is the only sample with an appreciable abundance of reads relating to streptococcal species although this may include species other than S. thermophilus. Reads relating to lactobacilli were present in <1% of each of the samples except for Farm 4 curd. It is noteworthy that a varying proportion of sequence reads were unassigned in any of the assessed samples (0.3 to 18% of reads).
Serratia spp. were present in all curd samples ranging in abundance from 0.7 (Farm 3) to 37% (Farm 1). Staphylococci were abundant (6.22%) in the curd sample from Farm 3. Pseudomonas spp. were also abundant in all curd samples ranging from 2.32% of reads from Farm 3 to 49.17% of reads from Farm 4.
3.2. Culture-Based Analysis of Milk, Curd and Whey Samples from Five Farms
In addition to the culture-independent analysis, we also wanted to establish the presence and diversity of dairy lactococci and/or streptococci present in raw sheep’s milk, cheese curd and whey samples from the same Pecorino-producing farms employing selective media and growth conditions (see Section 2). All five milk samples were shown to contain both mesophilic and thermophilic (presumptive) LAB. In general, higher viable counts of mesophiles than thermophiles were observed in the majority of samples. Milk samples from Farms 2 and 3 generally had higher viable counts across all temperatures and media compared to the other farms (~106–108 cfu/mL). Among the curd samples, Farm 5 had the highest viable counts (108 cfu/mL at 30 °C and 105–106 cfu/mL at 37 and 42 °C), while Farm 4 had the lowest (growth only on LM17 at 30 °C–107 cfu/mL and no growth on either medium at other temperatures). The whey samples of Farms 4 and 5 did not harbor presumptive and culturable LAB while those from Farms 1 to 3 harbored presumptive LAB at counts of 102–105 cfu/mL (Figure 3).
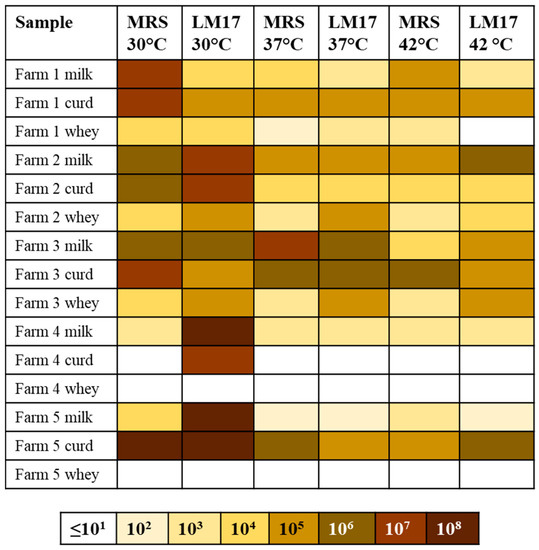
Figure 3.
Overview of the viable cell counts on MRS and LM17 agar at 30, 37 and 42 °C. The scale is color-coded as per the indicator bar beneath the table and is presented as culturable cells/mL or g of sample.
3.3. Diversity of Dairy Lactococci Associated with the Five Farms
Twenty-five representative colonies from each medium and temperature where growth was observed were selected for PCR-based identification of candidate Lactococcus or S. thermophilus strains using species-specific primers. Among the 1800 tested isolates, 206 presumptive lactococci and 27 S. thermophilus isolates were identified. The vast majority of isolates (approximately 77% of isolates) did not type according to the Lactococcus and S. thermophilus PCR typing system indicating that diverse LAB and/or additional species were present in the samples. No lactococci were identified in the milk from Farm 1 while curd and whey from this farm contained very low numbers of lactococcal isolates (Figure 3, Table S2). Similarly low numbers of lactococcal isolates (<10) were found in Farm 2 samples. From curd samples, lactococci were only identified in samples originating from Farms 4 and 5 (11 and 5 isolates, respectively). Conversely, all samples (milk, whey and curd) from Farm 3 contained lactococcal isolates retrieved from either of the selective media (Table 2).

Table 2.
Numbers of lactococcal isolates associated with each sample representing the A, B, C or undefined (X) lactococcal genotypes.
To discern the diversity among the 206 presumptive dairy lactococcal strains, PCR-based genotyping based on the cwps gene cluster was performed. This PCR distinguishes between strains possessing an A, B or C cwps genotype. Using this approach, four, 62 and 25 isolates were identified as possessing cwps genotype A, B or C, respectively; 115 of the presumptive lactococcal isolates could not be (sub-)typed according to this PCR system although the presence of the control PCR band indicates that they are indeed lactococcal strains. The 25 C-type isolates were further analyzed using the C-subtyping multiplex PCR, which may identify and distinguish diverse C-type strains belonging to the C1 to C5 cwps genotypes. C1 and C5 cwps genotypes were dominant among the C-type isolates with 10 and 7 of the 25 confirmed C-type lactococcal isolates exhibiting these genotypes, respectively (Table 2). C2 (six isolates), C3 (one isolate) and C4 (one isolate) type strains were less abundant.
The milk, whey and/or curd samples from each farm exhibited a unique microbiota composition with Farm 1 whey containing C4 and B-type strains and the curd containing C1, C2 and C3 isolates. Both milk and whey from Farm 2 harbored B-type strains while C2 and A-type strains were additionally identified in the milk or whey, respectively. In Farm 3 samples, B-type strains were isolated from the milk, whey and curd while C1 and C2 strains were identified in the milk and whey, respectively. The only confirmed and typed lactococci from Farm 4 was a single B-type CWPS isolate in the curd. Farm 5 was associated with the unique presence of C5 isolates in the milk and A-type isolates in the curd.
Beyond the identified 206 lactococcal isolates, there were a significant number of colonies that were not identified using the two multiplex PCR systems applied in the study. Therefore, to establish if the PCR systems were not capable of capturing all lactococcal/dairy streptococcal strains, ten unclassified isolates were randomly selected for speciation based on 16S rRNA gene sequence analysis. The ten isolates were selected to represent isolates from the different farms, growth media (MRS and LM17) and sample type (milk, whey and curd). Accordingly, isolates from one milk, three whey and five curd samples were analyzed. Four of the ten isolates were identified as Lactococcus lactis or Lactococcus cremoris and these were associated with whey samples from Farm 2 and 3 as well as milk and curd samples from Farm 3. The tested curd isolates from Farms 1 and 5 cultured on MRS and LM17 agar, respectively, were identified as Leuconostoc lactis and Leuconostoc mesenteroides strains, based on BLASTn analysis. The curd and whey isolates from Farms 1, 3 and 4 were identified as Enterococcus faecium (Farms 1 and 3) and Enterococcus durans (Farm 4), respectively.
3.4. Diversity of Dairy Streptococci Associated with the Five Pecorino Siciliano PDO Cheese Producing Farms
Through the species-specific PCR described above for the identification of presumptive lactococci and dairy streptococci, 27 presumptive S. thermophilus strains were identified. All S. thermophilus isolates were identified in whey (18 isolates) and curd (nine isolates) from Farms 2 and 3. Using the dual multiplex PCR system for distinguishing the five variable (Vt) and three backbone (Bt) rgp genotypes currently described, four isolates were identified from the Farm 2 whey sample, two of which were demonstrated to exhibit a Bt1 Vt4 genotype and a single isolate with a Bt3 Vt3 and Bt2 Vt1 genotype. From Farm 3, eight of the nine curd isolates were shown to exhibit a Bt1 Vt1 genotype, while one isolate was shown to possess a Bt2 Vt5 genotype. All Farm 3 whey isolates (14 isolates) were shown to display a Bt1 Vt1 genotype. Therefore, while the number of S. thermophilus isolates was low and isolates were only retrieved from Farms 2 and 3, at least two distinct strains from each farm were isolated and each farm possessed distinct lineages of strains (Table 3). Among these, the Bt2 Vt5 genotype has not been reported previously and thus represents a novel combination of the rgp cluster 5′ and 3′ regions.

Table 3.
Summary of S. thermophilus isolates from Farms 2 and 3.
3.5. Genome Characteristics of Dairy Lactococcal and Streptococcal Isolates
To evaluate the genetic diversity of the bacterial isolates, the genomes of seven S. thermophilus and eight lactococcal isolates were sequenced using an Illumina MiSeq platform. The S. thermophilus isolates were selected based on representing the two farms, the sample type (curd or whey in Farm 3) and the rgp/cwps genotypes. Using the MIGA (http://microbial-genomes.org/, accessed on 15 March 2023) genome analysis tool, the genome completeness and general genome characteristics of each strain were evaluated. Furthermore, the lactococcal isolates were identified as either L. lactis or L. cremoris (Table 4). All genomes except those of the S. thermophilus strains STMM4, STMM22 and STMM25 were considered complete based on the presence of 106 essential genes. All three genomes were predicted to lack secE, which encodes a SecE preprotein translocase. The genomes of the S. thermophilus strains range in size from 1.767 to 1.895 Mb and harbor 1874–2013 predicted open reading frames (ORFs). The lactococcal genomes range in size from 2.465 to 2.709 Mb with 2437–2753 predicted ORFs (Table 4). Six of the eight lactococcal isolates were identified as L. lactis while two strains were identified as L. cremoris (Table 5).

Table 4.
Genome characteristics and source farm (F) and type information for each isolate.

Table 5.
Presence of predicted phage-resistance systems and/or prophages in the strains’ genomes.
The genomes of the 15 sequenced dairy isolates were evaluated for the presence of predicted phage-resistance systems using PADLOC. This analysis revealed that all strains except one (L. lactis 74b) harbor at least one predicted phage-resistance system. Among the lactococcal strains, abortive infection (Abi) systems were identified in five of the eight strains and with each such strain possessing distinct systems or combinations of systems (Table 5). Type I restriction/modification (R/M) systems were predicted to be present in three lactococcal strains while two strains were predicted to harbor Type II R/M systems. Interestingly, the Type I R/M system of L. cremoris 71b appears to encompass an AbiJ-encoding gene and may represent a so-called defense island. Furthermore, some lactococcal strains are predicted to harbor more than a single R/M system (L. lactis 67b and L. cremoris 71b). In addition to these well-described phage defense systems, the genomes of some lactococcal strains are predicted to encode recently described novel anti-phage systems including Viperin (six of eight strains), Gabija (one of eight strains) CBass type I (one of eight strains) and AVAST type II systems (one of eight strains) (Table 5). The genomes of the dairy streptococcal strains were similarly analyzed and as expected, all strains were identified to harbor at least one intact CRISPR-Cas system in their genomes. All S. thermophilus genomes were also observed to harbor several predicted R/M systems belonging to Types I (seven strains), III (seven strains) and IV (four of seven strains). Furthermore, three genomes were observed to harbor predicted AbiD-encoding genes while the genomes of two strains were identified as possessing Sirtuin-dependent and Gabija-encoding genes (Table 5). The defense system profiles of the 15 sequenced isolates confirm their unique nature in addition to the cwps/rgp genotyping data. In summary, the milk, curd and/or whey of each Pecorino Siciliano PDO-producing farm presented a unique microbiota with farm-specific strain profiles.
The genomes of the 15 sequenced isolates were also evaluated for the presence of possible prophage-associated regions using PHASTER. This tool classifies the predicted prophage regions as “intact”, “questionable” or “incomplete”. Table 5 provides a summary of the predicted intact and questionable regions reported by PHASTER. Each strain possesses a unique predicted prophage profile based on this analysis. Among the lactococcal isolates, all but one (strain 71b) are predicted to harbor at least one intact prophage, while most also have “questionable” prophage regions and “incomplete” prophage regions in their genomes. Remarkably, strain 463 is predicted to harbor five intact prophages as well as two questionable prophage regions (and five incomplete prophage regions) and represents the highest phage load among all analyzed isolates. The predicted intact prophages range in size from 14.9–76.8 kb while the questionable lactococcal prophage regions range in size from 14.8–53.9 kb. The number of incomplete prophage regions in the lactococcal genomes ranged from 3 to 12. Among the S. thermophilus isolates, one strain was predicted to harbor an intact prophage region (STMM1- 5.7 kb region), while three of the strains’ genomes possessed at least one “questionable” prophage region ranging in size from 6.1–50.7 kb (Table 5). Similarly, all strains were predicted to harbor incomplete prophage regions in their genomes (between two and four regions).
To evaluate the possible production of anti-microbial compounds by the isolates, the genome sequences were interrogated using BAGEL4. All lactococcal isolates were observed to harbor regions of interest with all strains except 76b possessing sactipeptide modification-associated genes and all assessed strains’ genomes possessed lactococcin-associated genes. However, while the lactococcin-associated regions were observed, the majority of these were deemed unlikely to be complete with the exception of the identified region in strain 464. The lactococcin-associated cluster in strain 464′s genome was observed to harbor genes predicted to encode the core peptide (lactococcin B, bit score 145), ABC transport, leader peptide cleavage (LanT) and immunity to lactococcin B. For the dairy streptococcal isolates, regions associated with the predicted biosynthesis of bovicin, streptode, macedovicin, sactipeptide and BplST were common among the analyzed genomes. Among these, the predicted macedovicin clusters of STM22 and STMM25 appeared to represent complete clusters with gene products relating to core peptide biosynthesis (bit score 119), ABC transport systems, modification and leader peptide cleavage identified. Similarly, the genomes of STMM1, STMM2, STMM3, STMM4 and STMM11 harbored seemingly complete BlpST gene clusters with immunity, transport, core peptide and leader peptide cleavage functions readily identified. To validate the bioinformatic predictions of bacteriocin production by these strains, antagonism assays were performed against the bacteriocin-sensitive L. cremoris strain HP. L. lactis 464 was shown to inhibit the growth of L. cremoris HP (8 mm zone of inhibition). Similarly, all S. thermophilus isolates except STMM4 and STMM25 were observed to inhibit L. cremoris HP growth with zones of inhibition ranging from 5 mm (STMM3) to 15 mm (STMM1 and STMM2). Furthermore, the S. thermophilus isolates were applied in cross-sensitivity assays. STMM22 was capable of inhibiting STMM1, STMM2 and STMM11 13–19 mm zone of inhibition) while STMM25 was capable of inhibiting STMM3 (27 mm zone of inhibition).
4. Discussion
This study was aimed at evaluating the ecology and diversity of dairy lactococci and streptococci associated with the production of Pecorino Siciliano PDO using samples from five different Sicilian farms. Pecorino Siciliano PDO cheese is a Sicilian historical cheese whose heritage is strictly linked to the production system, especially to the use of wooden tools that have been proven to confer typicality to the final products [46,47]. Wooden vats and equipment that are used in traditional dairy food production practices host complex LAB biofilms representing efficient barriers to the adhesion of the main dairy pathogens [48] and are safe systems [49,50]. Thus, wood cannot be replaced by stainless steel or plastic, because the starter and non-starter cultures adapted to the production systems would be lost [51,52]. Tests conducted on a given bulk milk processed the same day in a single factory by wooden tools (traditional system, with no starter culture addition) and stainless steel equipment (milk inoculated with commercial starter culture preparation) generated Caciocavallo Palermitano cheeses with different quality characteristics imputed exclusively to the different active microbiota during the first phases of production [53] as well as during ripening [53,54]. Using culture-based approaches, we established that the milk, curd and whey from each farm possessed a unique microbiota and that the strains of Lactococcus lactis, Lactococcus cremoris and/or S. thermophilus from each farm are distinct. The genotyping of 206 lactococcal and 27 S. thermophilus isolates highlighted the farm-specific lactococcal and streptococcal populations. Furthermore, among the predicted lactococcal isolates, 115 isolates could not be assigned a cwps genotype and indeed additional lactococcal isolates were identified through 16S rRNA gene sequencing of unclassified isolates. This indicates that there is a likely wealth of diverse lactococcal strains in these samples that may possess unique genetic or functional attributes. The numbers of S. thermophilus isolates was considerably lower than the isolated lactococci. Despite the identified low number, among the seven isolates for which the genomes were sequenced, five distinct rgp genotypes were identified. This is a remarkable level of diversity and is likely a reflection of the adaptation to phages that may be present in the fermentation environment. Most notable among the streptococcal isolates is strain STMM11, which possesses a novel rgp genotype (Bt2 Vt5) among S. thermophilus strains described to date.
The diversity of strains identified in this study highlights the importance of artisanal food production systems as a rich and diverse source of mesophilic and thermophilic strains that could be harnessed in defined starter culture systems. By sequencing the genomes of representative isolates, it was established that these strains harbor an arsenal of defense mechanisms directed against phages including Abi, R/M and in the dairy streptococcal strains, CRISPR-Cas systems (Table 5). Furthermore, several of the recently identified phage-resistance systems are predicted to be encoded in nine of the genomes of the analyzed strains (Table 5). Viperin-like systems were identified in six of eight lactococcal genomes and these systems operate by producing modified ribonucleotides and inhibiting phage polymerase-dependent transcription in coliphage T7 [55]. Gabija, which comprises two genes (GajA and GajB) was identified in the genomes of two S. thermophilus strains (Table 5). Gabija is an endonuclease system that is activated in conditions where NTP and dNTP depletion occurs during phage replication and transcription [56]. CBASS systems synthesize secondary messengers including cyclic di/trinucleotides upon phage infection and induce cell membrane disruption or cleavage of intracellular DNA culminating in cell death [57,58]. The antiviral ATPase/NTPase AVAST systems are also like Abi systems as they are predicted to function through programmed cell death [59]. The presence of these new anti-phage systems dairy lactococci and streptococci has not been widely reported to date and warrants investigation in future studies. The presence of multiple and likely complementary systems reinforces the notion that phages are ever pervasive in fermentation environments whether industrial scale or in artisanal production systems. Indeed, the genomes of the sequenced lactococcal isolates confirms the abundance of (unique) prophage elements in each strain with several strains possessing at least one predicted intact prophage region. The S. thermophilus isolates genomes harbored fewer predicted prophage regions the majority of which are cryptic and incomplete regions. The presence of intact and/or cryptic prophages may offer protection against homologous, secondary infecting phages or in some cases, heterologous infecting phages. Additionally, the presence of diverse prophage regions highlights the diversity of phages that these strains have experienced in their respective environments. The presence of unique predicted prophage profiles combined with each strain possessing diverse anti-phage arsenals and cwps/rgp genotypes indicates that these strains have evolved to survive phages that are resident in the farms and production environments and that there is a farm-specific ecology of lactococci and streptococci.
Metagenomic analysis of the five curd samples revealed the distinct microbiota profile associated with each sample and was largely in agreement with the culture-based analysis. For example, the curd samples with the highest viable counts on M17 and MRS agars correlated well with those samples having the highest counts of reads mapping against lactic acid bacteria. Furthermore, the sample from Farm 4 possessed limited reads corresponding to LAB and this was also reflected in the number of lactococci (and lack of streptococci) retrieved from this sample in microbiological assays. In contrast to a recent metagenomic analysis of industrial and artisanal cheeses, lactococci were more abundant than streptococci [60]. It is noteworthy that the previous study was based on two artisanal cheeses whose origin or production type was not disclosed and these observations suggest that each artisanal and regional cheese has a distinct profile and population dynamic. Previous microbiological studies of ripened Pecorino Siciliano PDO cheeses identified a dominance of mesophilic lactobacilli based on growth on MRS agar. In the present study, both microbiological and metagenomic analyses suggest that mesophilic lactococci are more dominant in the unripened curd and that lactobacilli are present in very low abundance at the early stages of production. It is possible that lactobacilli increase in abundance during the ripening stage and perhaps their abundance in plating assays is related to the preferential growth of lactobacilli on MRS over their lactococcal counterparts.
Microbiota analysis also revealed the presence of Pseudomonas, Enterococcus and Staphylococcus in some of the curd samples. The presence of Staphylococcus and Enterococcus in Pecorino Siciliano PDO cheese in the early stage of production is consistent with previous reports [61]. As some species of Enterococcus and Lactobacillus are associated with the production of biogenic amines, it is pertinent to establish the presence of microbes that may pose a risk to consumers [62]. The presence of enterococci, pseudomonads and staphylococci is likely due to the application of raw milk and the local farm hygiene and handling practices [63]. Microbiological analysis identified isolates of Enterococcus and Staphylococcus on LM17 and MRS agar and highlights the importance of the genotyping tools to establish the identity of the presumptive microbes in classical microbiological assays. The production of anti-microbial compounds by starter bacteria is an important attribute in assuring the overall microbiological safety of fermented dairy products. Several of the analyzed isolates exhibited anti-microbial activity against a sensitive indicator strain as well as S. thermophilus isolates emanating from this study. Cross-sensitivity in S. thermophilus has been reported for the BlpST bacteriocins while these compounds are also associated with broader bactericidal effects [64]. Future studies will focus on evaluating the exact nature, characteristics and functionality of these compounds and their role in eliminating pathogenic and spoilage microorganisms in fermented foods.
5. Conclusions
Farms producing Pecorino Siciliano PDO have distinct LAB communities that are believed to contribute to the fermentation process. While the abundance/dominance at the species level may not vary significantly in many cases, the strain-level diversity creates the signature of the produce from each farm. The present study identified the dominant starter lactic acid bacteria while future studies could apply the approaches applied here to a longitudinal analysis across the production and ripening periods to define the evolution of the microbial landscape of this regionally important cheese. Furthermore, the strain-level differentiation of isolates in this study provides significant insights into the diversity of lactococcal and streptococcal strains that may be derived from such foods to enhance global starter culture repertoires for the enhancement of defined and mixed starter culture systems.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9070620/s1, Table S1: List of primers used in this study; Table S2: Viable counts of presumptive LAB on LM17 and MRS agar at different temperatures.
Author Contributions
Conceptualization, J.M., L.S., M.T. and D.v.S.; methodology, M.M., S.R., G.A.L., B.M. and Z.K.; software, G.A.L., B.M. and M.V.; validation, M.M., S.R., G.A.L., B.M. and Z.K.; formal analysis, M.M., S.R., G.A.L., B.M. and Z.K.; investigation, M.M., S.R., M.T., G.A.L., B.M. and Z.K.; resources, J.M., D.v.S. and M.V.; data curation, M.M., S.R., G.A.L., B.M. and Z.K.; writing—original draft preparation, J.M. and S.R.; writing—review and editing, J.M., M.T., L.S., D.v.S. and M.V.; supervision, J.M., D.v.S., L.S. and M.V.; project administration, J.M. and S.R.; funding acquisition, J.M. and S.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Science Foundation Ireland, grant numbers 20/FFP-P/8664 and 12/RC/2273-P2. For the purpose of open access, we have applied a CC BY public copyright license to any author accepted manuscript version arising from this submission.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequences of S. thermophilus MM1, MM2, MM3, MM4, MM11, MM22, MM25, Lactococcus cremoris 32b and 71b and Lactococcus lactis 67b, 74b, 76b, 218, 463 and 464 have been deposited in the GenBank database (Table 5). The raw sequences of the 16S rRNA microbial profiling of each of the five curd samples are available through the Bioproject accession PRJNA983130.
Acknowledgments
The authors wish to thank the farmers for kindly providing the samples for analysis in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bintsis, T. Lactic Acid Bacteria as Starter Cultures: An Update in Their Metabolism and Genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Anastasiou, R.; Georgalaki, M.; Bounenni, R.; Paximadaki, A.; Charmpi, C.; Alexandraki, V.; Kazou, M.; Tsakalidou, E. Comparison of the Microbiome of Artisanal Homemade and Industrial Feta Cheese through Amplicon Sequencing and Shotgun Metagenomics. Microorganisms 2022, 10, 1073. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, I.; Di Cagno, R.; Buchin, S.; De Angelis, M.; Gobbetti, M. Spatial Distribution of the Metabolically Active Microbiota within Italian PDO Ewes’ Milk Cheeses. PLoS ONE 2016, 11, e0153213. [Google Scholar] [CrossRef]
- Erhardt, M.M.; Oliveira, W.D.C.; Fröder, H.; Marques, P.H.; Oliveira, M.B.P.P.; Richards, N.S.P.D.S. Lactic Bacteria in Artisanal Cheese: Characterization through Metagenomics. Fermentation 2023, 9, 41. [Google Scholar] [CrossRef]
- EU Commission Implementing Regulation (EU). Commission Implementing Regulation (EU) 2020/1338 of 21 September 2020 Approving Non-Minor Amendments to the Specification for a Name Entered in the Register of Protected Designations of Origin and Protected Geographical Indications (‘Pecorino Siciliano’ (PDO)). 2020. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32020R1338 (accessed on 8 June 2023).
- Schimmenti, E.; Viola, E.; Funsten, C.; Borsellino, V. The Contribution of Geographical Certification Programs to Farm Income and Rural Economies: The Case of Pecorino Siciliano PDO. Sustainability 2021, 13, 1977. [Google Scholar] [CrossRef]
- European Union Regulation (EU). No. 1151/2012 of the European Parliament and of the Council of 21 November 2012 on Quality Schemes for Agricultural Products and Foodstuffs. Off. J. Eur. Union 2012, 343, 1–29. [Google Scholar]
- Todaro, M.; Francesca, N.; Reale, S.; Moschetti, G.; Vitale, F.; Settanni, L. Effect of Different Salting Technologies on the Chemical and Microbiological Characteristics of PDO Pecorino Siciliano Cheese. Eur. Food Res. Technol. 2011, 233, 931–940. [Google Scholar] [CrossRef]
- Todaro, M.; Lo Presti, V.; Macaluso, A.; Alleri, M.; Licitra, G.; Chiofalo, V. Alkaline Phosphatase Survey in Pecorino Siciliano PDO Cheese. Foods 2021, 10, 1648. [Google Scholar] [CrossRef]
- Gaglio, R.; Todaro, M.; Settanni, L. Improvement of Raw Milk Cheese Hygiene through the Selection of Starter and Non-Starter Lactic Acid Bacteria: The Successful Case of PDO Pecorino Siciliano Cheese. Int. J. Environ. Res. Public Health 2021, 18, 1834. [Google Scholar] [CrossRef]
- Settanni, L.; Busetta, G.; Puccio, V.; Licitra, G.; Franciosi, E.; Botta, L.; Di Gerlando, R.; Todaro, M.; Gaglio, R. In-Depth Investigation of the Safety of Wooden Shelves Used for Traditional Cheese Ripening. Appl. Environ. Microbiol. 2021, 87, e01524-21. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Torriani, S.; Akkermans, A.D.L.; De Vos, W.M.; Vaughan, E.E. Diversity, Dynamics, and Activity of Bacterial Communities during Production of an Artisanal Sicilian Cheese as Evaluated by 16S RRNA Analysis. Appl. Environ. Microbiol. 2002, 68, 1882–1892. [Google Scholar] [CrossRef]
- Dolci, P.; Alessandria, V.; Zeppa, G.; Rantsiou, K.; Cocolin, L. Microbiological Characterization of Artisanal Raschera PDO Cheese: Analysis of Its Indigenous Lactic Acid Bacteria. Food Microbiol. 2008, 25, 392–399. [Google Scholar] [CrossRef]
- Cardamone, C.; Cirlincione, F.; Gaglio, R.; Puccio, V.; Daidone, F.; Sciortino, S.; Mancuso, I.; Scatassa, M.L. Behavior of Four Main Dairy Pathogenic Bacteria during Manufacturing and Ripening of Pecorino Siciliano Cheese. J. Food Qual. Hazards Control 2020, 7, 27–35. [Google Scholar] [CrossRef]
- Mahony, J.; Kot, W.; Murphy, J.; Ainsworth, S.; Neve, H.; Hansen, L.H.; Heller, K.J.; Sørensen, S.J.; Hammer, K.; Cambillau, C.; et al. Investigation of the Relationship between Lactococcal Host Cell Wall Polysaccharide Genotype and 936 Phage Receptor Binding Protein Phylogeny. Appl. Environ. Microbiol. 2013, 79, 4385–4392. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, K.; Sadovskaya, I.; Vinogradov, E.; Kelleher, P.; Lugli, G.A.; Ventura, M.; van Sinderen, D.; Mahony, J. Bipartite Rgp Locus Diversity in Streptococcus Thermophilus Corresponds to Backbone and Side Chain Differences of Its Rhamnose-Containing Cell Wall Polysaccharide. Appl. Environ. Microbiol. 2022, 88, e0150422. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.-P.; van Sinderen, D. Differences in Lactococcal Cell Wall Polysaccharide Structure Are Major Determining Factors in Bacteriophage Sensitivity. mBio 2014, 5, e00880-14. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Frantzen, C.; Vinogradov, E.; Sadovskaya, I.; Theodorou, I.; Kelleher, P.; Chapot-Chartier, M.-P.; Cambillau, C.; Holo, H.; van Sinderen, D. The CWPS Rubik’s Cube: Linking Diversity of Cell Wall Polysaccharide Structures with the Encoded Biosynthetic Machinery of Selected Lactococcus Lactis Strains. Mol. Microbiol. 2020, 114, 582–596. [Google Scholar] [CrossRef]
- Chopin, A.; Bolotin, A.; Sorokin, A.; Ehrlich, S.D.; Chopin, M. Analysis of Six Prophages in Lactococcus Lactis IL1403: Different Genetic Structure of Temperate and Virulent Phage Populations. Nucleic Acids Res. 2001, 29, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Zomer, A.; Canchaya, C.; O’Connell-Motherway, M.; Kuipers, O.; Turroni, F.; Ribbera, A.; Foroni, E.; Buist, G.; Wegmann, U.; et al. Comparative Analyses of Prophage-like Elements Present in Two Lactococcus Lactis Strains. Appl. Environ. Microbiol. 2007, 73, 7771–7780. [Google Scholar] [CrossRef]
- Alexandraki, V.; Kazou, M.; Blom, J.; Pot, B.; Papadimitriou, K.; Tsakalidou, E. Comparative Genomics of Streptococcus Thermophilus Support Important Traits Concerning the Evolution, Biology and Technological Properties of the Species. Front. Microbiol. 2019, 10, 2916. [Google Scholar] [CrossRef]
- Da Silva Duarte, V.; Giaretta, S.; Campanaro, S.; Treu, L.; Armani, A.; Tarrah, A.; Oliveira De Paula, S.; Giacomini, A.; Corich, V. A Cryptic Non-Inducible Prophage Confers Phage-Immunity on the Streptococcus Thermophilus M17PTZA496. Viruses 2018, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Cruz, S.; Parlindungan, E.; Erazo Garzon, A.; Alqarni, M.; Lugli, G.A.; Ventura, M.; van Sinderen, D.; Mahony, J. Lysogenization of a Lactococcal Host with Three Distinct Temperate Phages Provides Homologous and Heterologous Phage Resistance. Microorganisms 2020, 8, 1685. [Google Scholar] [CrossRef] [PubMed]
- Ali, Y.; Koberg, S.; Heßner, S.; Sun, X.; Rabe, B.; Back, A.; Neve, H.; Heller, K.J. Temperate Streptococcus Thermophilus Phages Expressing Superinfection Exclusion Proteins of the Ltp Type. Front. Microbiol. 2014, 5, 98. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage Resistance Mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, T.; Sberro, H.; Weinstock, E.; Cohen, O.; Doron, S.; Charpak-Amikam, Y.; Afik, S.; Ofir, G.; Sorek, R. BREX Is a Novel Phage Resistance System Widespread in Microbial Genomes. EMBO J. 2015, 34, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Sattler, L.; Shafqat, S.; Graumann, P.L.; Bramkamp, M. A Bacterial Dynamin-Like Protein Confers a Novel Phage Resistance Strategy on the Population Level in Bacillus Subtilis. mBio 2022, 13, e03753-21. [Google Scholar] [CrossRef]
- Vassallo, C.N.; Doering, C.R.; Littlehale, M.L.; Teodoro, G.I.C.; Laub, M.T. A Functional Selection Reveals Previously Undetected Anti-Phage Defence Systems in the E. Coli Pangenome. Nat. Microbiol. 2022, 7, 1568–1579. [Google Scholar] [CrossRef]
- Rousset, F.; Depardieu, F.; Miele, S.; Dowding, J.; Laval, A.-L.; Lieberman, E.; Garry, D.; Rocha, E.P.C.; Bernheim, A.; Bikard, D. Phages and Their Satellites Encode Hotspots of Antiviral Systems. Cell Host Microbe 2022, 30, 740–753.e5. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Payne, L.J.; Meaden, S.; Mestre, M.R.; Palmer, C.; Toro, N.; Fineran, P.C.; Jackson, S.A. PADLOC: A Web Server for the Identification of Antiviral Defence Systems in Microbial Genomes. Nucleic Acids Res. 2022, 50, W541–W550. [Google Scholar] [CrossRef]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; d’Humières, C.; Cury, J.; Bernheim, A. Systematic and Quantitative View of the Antiviral Arsenal of Prokaryotes. Nat. Commun. 2022, 13, 2561. [Google Scholar] [CrossRef]
- Niccum, B.A.; Kastman, E.K.; Kfoury, N.; Robbat, A.; Wolfe, B.E. Strain-Level Diversity Impacts Cheese Rind Microbiome Assembly and Function. mSystems 2020, 5, e00149-20. [Google Scholar] [CrossRef]
- Somerville, V.; Berthoud, H.; Schmidt, R.S.; Bachmann, H.-P.; Meng, Y.H.; Fuchsmann, P.; Von Ah, U.; Engel, P. Functional Strain Redundancy and Persistent Phage Infection in Swiss Hard Cheese Starter Cultures. ISME J. 2022, 16, 388–399. [Google Scholar] [CrossRef]
- Hayes, S.; Mahony, J.; Vincentelli, R.; Ramond, L.; Nauta, A.; van Sinderen, D.; Cambillau, C. Ubiquitous Carbohydrate Binding Modules Decorate 936 Lactococcal Siphophage Virions. Viruses 2019, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Erkus, O.; De Jager, V.C.; Spus, M.; Van Alen-Boerrigter, I.J.; Van Rijswijck, I.M.; Hazelwood, L.; Janssen, P.W.; Van Hijum, S.A.; Kleerebezem, M.; Smid, E.J. Multifactorial Diversity Sustains Microbial Community Stability. ISME J. 2013, 7, 2126–2136. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Hevia, A.; Foroni, E.; Duranti, S.; Turroni, F.; Lugli, G.A.; Sanchez, B.; Martín, R.; Gueimonde, M.; van Sinderen, D.; et al. Assessing the Fecal Microbiota: An Optimized Ion Torrent 16S RRNA Gene-Based Analysis Protocol. PLoS ONE 2013, 8, e68739. [Google Scholar] [CrossRef] [PubMed]
- Alagna, L.; Mancabelli, L.; Magni, F.; Chatenoud, L.; Bassi, G.; Del Bianco, S.; Fumagalli, R.; Turroni, F.; Mangioni, D.; Migliorino, G.M.; et al. Changes in Upper Airways Microbiota in Ventilator-Associated Pneumonia. Intensive Care Med. Exp. 2023, 11, 17. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Lugli, G.A.; Milani, C.; Mancabelli, L.; Van Sinderen, D.; Ventura, M. MEGAnnotator: A User-Friendly Pipeline for Microbial Genomes Assembly and Annotation. FEMS Microbiol. Lett. 2016, 363, fnw049. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J. Heuristic Approach to Deriving Models for Gene Finding. Nucleic Acids Res. 1999, 27, 3911–3920. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Parlindungan, E.; Lugli, G.A.; Ventura, M.; Van Sinderen, D.; Mahony, J. Lactic Acid Bacteria Diversity and Characterization of Probiotic Candidates in Fermented Meats. Foods 2021, 10, 1519. [Google Scholar] [CrossRef] [PubMed]
- Gaglio, R.; Franciosi, E.; Todaro, A.; Guarcello, R.; Alfeo, V.; Randazzo, C.L.; Settanni, L.; Todaro, M. Addition of Selected Starter/Non-Starter Lactic Acid Bacterial Inoculums to Stabilise PDO Pecorino Siciliano Cheese Production. Food Res. Int. 2020, 136, 109335. [Google Scholar] [CrossRef] [PubMed]
- Busetta, G.; Gaglio, R.; Mangione, G.; Garofalo, G.; Franciosi, E.; Gannuscio, R.; Caccamo, M.; Todaro, M.; Di Gerlando, R.; Settanni, L.; et al. Effect of Commission Implementing Regulation (EU) 2020/1319 on the Bacterial Composition of PDO Provola Dei Nebrodi Cheese. Int. J. Food Microbiol. 2023, 394, 110188. [Google Scholar] [CrossRef] [PubMed]
- Cruciata, M.; Gaglio, R.; Scatassa, M.L.; Sala, G.; Cardamone, C.; Palmeri, M.; Moschetti, G.; La Mantia, T.; Settanni, L. Formation and Characterization of Early Bacterial Biofilms on Different Wood Typologies Applied in Dairy Production. Appl. Environ. Microbiol. 2018, 84, e02107-17. [Google Scholar] [CrossRef]
- Didienne, R.; Defargues, C.; Callon, C.; Meylheuc, T.; Hulin, S.; Montel, M.-C. Characteristics of Microbial Biofilm on Wooden Vats (‘Gerles’) in PDO Salers Cheese. Int. J. Food Microbiol. 2012, 156, 91–101. [Google Scholar] [CrossRef]
- Gaglio, R.; Cruciata, M.; Di Gerlando, R.; Scatassa, M.L.; Cardamone, C.; Mancuso, I.; Sardina, M.T.; Moschetti, G.; Portolano, B.; Settanni, L. Microbial Activation of Wooden Vats Used for Traditional Cheese Production and Evolution of Neoformed Biofilms. Appl. Environ. Microbiol. 2016, 82, 585–595. [Google Scholar] [CrossRef]
- Busetta, G.; Garofalo, G.; Barbera, M.; Di Trana, A.; Claps, S.; Lovallo, C.; Franciosi, E.; Gaglio, R.; Settanni, L. Metagenomic, Microbiological, Chemical and Sensory Profiling of Caciocavallo Podolico Lucano Cheese. Food Res. Int. 2023, 169, 112926. [Google Scholar] [CrossRef]
- Licitra, G.; Caccamo, M.; Valence, F.; Lortal, S. Traditional Wooden Equipment Used for Cheesemaking and Their Effect on Quality. In Global Cheesemaking Technology; Papademas, P., Bintsis, T., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2017; pp. 157–172. ISBN 978-1-119-04616-5. [Google Scholar]
- Settanni, L.; Di Grigoli, A.; Tornambé, G.; Bellina, V.; Francesca, N.; Moschetti, G.; Bonanno, A. Persistence of Wild Streptococcus Thermophilus Strains on Wooden Vat and during the Manufacture of a Traditional Caciocavallo Type Cheese. Int. J. Food Microbiol. 2012, 155, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Di Grigoli, A.; Francesca, N.; Gaglio, R.; Guarrasi, V.; Moschetti, M.; Scatassa, M.L.; Settanni, L.; Bonanno, A. The Influence of the Wooden Equipment Employed for Cheese Manufacture on the Characteristics of a Traditional Stretched Cheese during Ripening. Food Microbiol. 2015, 46, 81–91. [Google Scholar] [CrossRef]
- Bernheim, A.; Millman, A.; Ofir, G.; Meitav, G.; Avraham, C.; Shomar, H.; Rosenberg, M.M.; Tal, N.; Melamed, S.; Amitai, G.; et al. Prokaryotic Viperins Produce Diverse Antiviral Molecules. Nature 2021, 589, 120–124. [Google Scholar] [CrossRef]
- Cheng, R.; Huang, F.; Wu, H.; Lu, X.; Yan, Y.; Yu, B.; Wang, X.; Zhu, B. A Nucleotide-Sensing Endonuclease from the Gabija Bacterial Defense System. Nucleic Acids Res. 2021, 49, 5216–5229. [Google Scholar] [CrossRef]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Lowey, B.; Whiteley, A.T.; Keszei, A.F.A.; Morehouse, B.R.; Mathews, I.T.; Antine, S.P.; Cabrera, V.J.; Kashin, D.; Niemann, P.; Jain, M.; et al. CBASS Immunity Uses CARF-Related Effectors to Sense 3′–5′- and 2′–5′-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection. Cell 2020, 182, 38–49.e17. [Google Scholar] [CrossRef]
- Gao, L.; Altae-Tran, H.; Böhning, F.; Makarova, K.S.; Segel, M.; Schmid-Burgk, J.L.; Koob, J.; Wolf, Y.I.; Koonin, E.V.; Zhang, F. Diverse Enzymatic Activities Mediate Antiviral Immunity in Prokaryotes. Science 2020, 369, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Afshari, R.; Pillidge, C.J.; Read, E.; Rochfort, S.; Dias, D.A.; Osborn, A.M.; Gill, H. New Insights into Cheddar Cheese Microbiota-Metabolome Relationships Revealed by Integrative Analysis of Multi-Omics Data. Sci. Rep. 2020, 10, 3164. [Google Scholar] [CrossRef]
- Vernile, A.; Giammanco, G.; Spano, G.; Beresford, T.P.; Fox, P.F.; Massa, S. Genotypic Characterization of Lactic Acid Bacteria Isolated from Traditional Pecorino Siciliano Cheese. Dairy Sci. Technol. 2008, 88, 619–629. [Google Scholar] [CrossRef]
- Yeluri Jonnala, B.R.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D. Sequencing of the Cheese Microbiome and Its Relevance to Industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef]
- Ercolini, D.; De Filippis, F.; La Storia, A.; Iacono, M. “Remake” by High-Throughput Sequencing of the Microbiota Involved in the Production of Water Buffalo Mozzarella Cheese. Appl. Environ. Microbiol. 2012, 78, 8142–8145. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, L.; Hols, P. The Inhibitory Spectrum of Thermophilin 9 from Streptococcus thermophilus LMD-9 Depends on the Production of Multiple Peptides and the Activity of BlpGSt, a Thiol-Disulfide Oxidase. Appl. Environ. Microbiol. 2008, 74, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).