Abstract
The use of non-Saccharomyces yeast species generally involves sequential or co-inoculation of a Saccharomyces cerevisiae strain to complete fermentation. While most studies have focused on characterising the impact that S. cerevisiae has on the growth and metabolic activity of these non-Saccharomyces species, microbial interactions work reciprocally. Antagonism or competition of non-Saccharomyces species against S. cerevisiae has been shown to impact subsequent fermentation performance. To date, it remains unclear whether these negative interactions are strain specific. Hence, characterisation of strain-specific responses to co-inoculation would enable the identification of specific S. cerevisiae strain/non-Saccharomyces combinations that minimise the negative impacts of sequential fermentation on fermentation performance. The competitive fitness response of 93 S. cerevisiae strains to several non-Saccharomyces species was simultaneously investigated using a barcoded library to address this knowledge gap. Strain-specific fitness differences were observed across non-Saccharomyces treatments. Results obtained from experiments using selected S. cerevisiae strains sequentially inoculated after Metschnikowia pulcherrima and Torulaspora delbrueckii were consistent with the competitive barcoded library observations. The results presented in this study indicate that strain selection will influence fermentation performance when using non-Saccharomyces species, therefore, appropriate strain/yeast combinations are required to optimise fermentation.
1. Introduction
Inoculation with selected Saccharomyces cerevisiae strains is one way to initiate fermentation efficiently in compositionally and microbiologically diverse grape musts [1]. However, inoculated wines are often considered to lack the complexity of their uninoculated counterparts [2]. This lack of complexity may be attributed to a reduction in non-Saccharomyces metabolic activity. To address this perceived shortcoming, several non-Saccharomyces species have now been isolated and commercialised with the objective of enhancing the aroma and flavour attributes of wine [3]. Species such as Torulaspora delbrueckii and Metschnikowia pulcherrima are now commercially available as starter cultures and represent practical tools that can be used to reduce alcohol and volatile acidity concentrations in wine and to modulate the concentrations of varietal thiols and esters [4]. Due to the variable capacity of these yeasts to complete sugar consumption [5], strains of S. cerevisiae are sequentially inoculated to ensure the completion of alcoholic fermentation. However, inoculation with multiple species can introduce issues associated with microbial compatibility, especially when the purified strains are inoculated at high concentrations [6].
Alongside the characterisation of nutrient requirements, fermentation capabilities and metabolite production of non-Saccharomyces species [7,8,9,10,11,12,13], studies have investigated the impact of S. cerevisiae on growth to maximise non-Saccharomyces species’ persistence in grape juice. Several mechanisms of antagonism between S. cerevisiae and non-Saccharomyces species have been proposed, and these can differ depending on the non-Saccharomyces species, including cell-to-cell contact [14,15], the production of antimicrobial peptides [7,16] and toxic compounds [17].
Conversely, given the difference in inoculation times under a sequential inoculation scenario, initial uptake of nutrients such as amino acids and vitamins by non-Saccharomyces species has been suggested as an impediment for the subsequent growth of S. cerevisiae strains [18]. Non-Saccharomyces species such as T. delbrueckii, M. pulcherrima, Hanseniaspora vineae, H. guilliermondii and Kluyveromyces marxianus have also been shown to significantly decrease the nitrogen availability for S. cerevisiae [19,20,21]. Aside from nutrient competition, other antagonistic mechanisms of non-Saccharomyces species against S. cerevisiae have been reported, including cell-to-cell contact [20] and the production of killer toxins [22,23].
While the previous investigations have focused on characterising the features relating to competition between specific yeast species, recent research suggests that microbial interactions could be strain-specific [24,25]. Strain selection could therefore represent an important factor in maximising fermentation performance when inoculating with multiple yeast species. The fermentation fitness differences [26] and genetic diversity [27,28] of wine-associated S. cerevisiae strains have been well documented. However, it is unknown if these strain differences affect the ability to compete with other non-Saccharomyces species. This study simultaneously investigated the competitive fitness response of 93 S. cerevisiae wine strains to several non-Saccharomyces species using a genomically barcoded library. Experiments were also performed using single S. cerevisiae strains to further investigate specific strain–species interactions in a sequential inoculation regime.
2. Materials and Methods
2.1. Non-Saccharomyces Strains and Barcoded Library
The strains used in this study are available in The Australian Wine Research Institute (AWRI) culture collection. Five non-Saccharomyces species (Table 1) were used for the competitive fitness experiments and were selected based on their use as commercial starter cultures and their reported natural abundance in grape juice. The S. cerevisiae barcode library is comprised of 93 wine strains (Table S1) that represent several of the phylogenetic clades within this species [26,28]. Each strain contains a unique pair of barcodes inserted in the HO ORF region, allowing the quantification of each strain under mixed culture competitive conditions. Details on the construction of the wine yeast barcoded library (WYBC) and mixed pooled inoculum have been described previously [26].

Table 1.
Yeast strains used in this study.
2.2. Barcoded Library Competitive Fitness Experiments
Competitive fitness experiments were performed in a chemically defined medium (CDM) [26]. The overall approach has been described in detail previously [26]. Briefly, each non-Saccharomyces species was inoculated at a concentration of 106 CFU/mL into 100 mL sealed Schott flasks fitted with gas release valves and stirred at 250 rpm for 24 h at 18 °C, after which 1 mL of freshly thawed WYBC pool was added into each flask. A control treatment without the presence of a non-Saccharomyces species was also included. After 3 days (T1) of growth, samples were taken for DNA extraction, and 1 mL of each flask was used to inoculate another set of treatments and control flasks using a serial batch approach. Sequential inoculations were performed twice for a total of 3 sampling time points (T1–T3). All treatments were performed in triplicate. DNA was also extracted directly from the WYBC pool to use as an intercept (control) in the statistical analyses of the barcode counts’ data.
2.3. Amplicon Sequencing, Bioinformatic and Data Analyses
Genomic DNA from each time point was extracted using a Gentra Puregene Yeast/Bact kit (Qiagen, Doncaster, Victoria, Australia) following the manufacturer’s instructions with minor modifications. A total of 3 uL of Zymolyase (10 mg/mL) (MP Biomedicals, NSW, Australia) was used instead of lytic enzyme. Amplicons for Illumina sequencing were generated using primers Illum_P5_S50(1-6) and Illum_P7_N70(1-12), and PCR conditions as previously described [26]. A total of 300 ng of each PCR reaction was pooled and then cleaned using a NucleoSpin PCR cleanup kit (Machery-Nagel GMBH, Düren, Germany) and sent for sequencing to the Ramaciotti Centre for Genomics (University of New South Wales, Sydney, Australia) on an Illumina NextSeq500 instrument using 2 × 36 bp run format. Forward and reverse reads were fused using BBMap v.39.90 and converted into fasta files. Unique dual barcodes corresponding to each strain were demultiplexed from fasta files using the Python packages demultiplex and TSSV [29], searching for the unique pair of barcodes in each read allowing no nucleotide mismatches. Demultiplexed reads corresponding to each strain and treatment were counted and imported into R [30] (File S1). Statistical analysis of strain counts was performed using EdgeR [31], as previously described by Schmidt et al. [26], using a pairwise design with the WYBC pool strain counts data as an intercept. Strain fitness was evaluated as the contrast of each time point in each non-Saccharomyces treatment against the control treatment (e.g., T3 MP = Timepoint 3 S. cerevisiae strain counts (M. pulcherrima treatment) − Timepoint 3 S. cerevisiae strain counts (WYBC control treatment)) (Tables S2–S5).
2.4. Single Strain Competitive Experiments
Experiments using single S. cerevisiae strains selected according to their competitive fitness performance (high and low fitness performance) were performed in the presence of non-Saccharomyces species. A blue fluorescent protein (BFP) gene was introduced in the HO ORF of the parental non-barcoded S. cerevisiae strains to allow cell counting of S. cerevisiae by flow cytometry in the presence of a different yeast species. Transformations were carried out using the lithium acetate-polyethylene glycol method [32]. DNA cassettes for transformation were obtained by PCR from plasmid pCV2_HO-BFP, a modified version of plasmid pCV2_BB-BFP [33] containing 312 bp of homology to the HO ORF. Positive transformants were screened using geneticin and confirmed by PCR.
Competitive experiments were performed in CDM using two non-Saccharomyces species (M. pulcherrima and T. delbrueckii) and eight S. cerevisiae strains (Table 1). All treatments were undertaken in triplicate. Non-Saccharomyces species were inoculated (106 CFU/mL) into 100 mL sealed Schott flasks fitted with gas release valves and stirred at 250 rpm for 24 h after which a S. cerevisiae strain was inoculated at a final OD600 of 0.02 (approx. 106 CFU/mL). All fermentations were conducted at 18 °C. Samples were taken daily to determine cell concentration using flow cytometry and sugar concentration by enzymatic assay.
2.5. Analytical Methods
Glucose and fructose concentrations were determined enzymatically as previously described by [34] with adaptations described by [35] to allow the measurement in a 96-well microplate format.
Cells were counted by flow cytometry using a Guava® easyCyte 12HT instrument (Merck Millipore, Burlington, MA, USA). Prior to cell counting, cells were diluted in PBS (NaCl 8 g/L, KCl 0.2 g/L, Na2HPO4 1.44 g/L, KH2PO4 0.24 g/L pH 7.4) containing propidium iodide (PI) (1% v/v) to a concentration lower than 5 × 106 cells/mL. Forward and side scatter detectors were used to determine particle size and estimate cell numbers. A minimum of 5000 events with a throughput lower than 500 event/μL was measured in all samples. The viable cell concentration of S. cerevisiae was determined by measuring the number of positive blue fluorescent (405 nm excitation and 448/50 nm detection filter) and PI-negative events per mL.
3. Results and Discussion
3.1. S. cerevisiae Competitive Fitness Response to the Presence of a Non-Saccharomyces Species
The strain-specific effect resulting from preculturing of medium with different non-Saccharomyces species (A. pullulans, H. uvarum, L. thermotolerans, M. pulcherrima and T. delbrueckii) on the growth of S. cerevisiae was investigated. A sequential-batch experiment estimated competitive fitness responses for each S. cerevisiae strain using a pooled-inoculum approach (93 barcoded S. cerevisiae strains). At each time point, the strain-wise barcode counts for samples taken from ferments containing non-Saccharomyces species were determined by sequencing and compared to the strain-wise barcode counts obtained from ferments without non-Saccharomyces species.
Sugar concentrations at the time of passage revealed a differential effect of the non-Saccharomyces species on the general fermentation performance of the mixed cultures (Table 2). Ferments containing A. pullulans and M. pulcherrima displayed the slowest sugar consumption rates (Table 2). Relative to the control condition, all mixed cultures had reduced sugar consumption rates in T2 and T3, suggesting a direct competition/antagonism of these non-Saccharomyces species with S. cerevisiae. While the majority of these non-Saccharomyces species are associated with beneficial organoleptic effects in wine [12,13,36,37], several studies have reported slower fermentation rates when these species were used in co-culture [11,20,37].

Table 2.
Sugar concentration (g/L) at each time point (3 days after S. cerevisiae inoculation). All data points correspond to the mean of three replicates ± sd.
S. cerevisiae strain-specific fitness responses to different non-Saccharomyces treatments varied depending on the non-Saccharomyces species present (Figure 1). Clustering of treatments resolved three groups which included T. delbrueckii and H. uvarum in one group, M. pulcherrima and L. thermotolerans in the second and the third containing A. pullulans as the only member (Figure 1). These observations indicate that S. cerevisiae strain fitness variability is species-dependent.
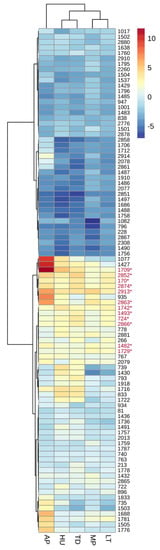
Figure 1.
Fitness (log2 FC relative to control conditions) of Saccharomyces cerevisiae strains in the presence of a non-Saccharomyces species. Data are clustered across fitness and treatments. Strains with an asterisk (*) showed a log2 FC ≥ 1 across all treatments. AP: Aureobasidium pullulans, HU: Hanseniaspora uvarum, TD: Torulaspora delbrueckii, MP: Metschnikowia pulcherrima and LT: Lachancea thermotolerans.
Inspection of the fitness response of specific S. cerevisiae strains revealed 12 strains with a log2 fold change (FC) ≥ 1 (Figure 1, highlighted in red) and 30 strains with a log2 FC ≤ −1 across all treatments. The largest increase in relative abundance was observed in strain AWRI1709 (log2 FC of 11.8) in the presence of A. pullulans (Figure 1) (Table S4). The increased relative abundance of AWRI1709 suggests that this specific strain might have the ability to overcome the nutrient competition previously associated with A. pullulans [37]. Mapping onto pre-existing wine yeast phylogenies of the 12 strains that showed a log2 FC ≥ 1 revealed that none of these strains corresponded to the previously reported Prize de Mousse (PdM) clade [28], which is represented by 17 strains in the WYBC. Furthermore, from these 17 strains, 10 showed a log2 FC ≤ −1, and 4 showed a negative log2 FC in all treatments. Strains belonging to the PdM clade are considered highly domesticated to the grape juice/wine environment; however, the data suggests that these strains might not be the fittest in a highly competitive scenario.
All treatments showed a higher number of strains with reduced representation in the community relative to the control condition (MP: 45 strains down, 23 strains up | TD: 44 down, 29 up | HU: 41 down, 30 up | LT: 50 down, 22 up | AP: 37 down, 28 up, Figure 1). Although it is not possible to determine if the strains showing a relative increase in abundance are indeed growing at faster rates when a non-Saccharomyces species is present, the sugar consumption data suggest the contrary, where strains with a positive log2 FC are likely less affected by the antagonism/competition of these non-Saccharomyces species.
3.2. Single Inoculum Competition Experiments
To confirm the barcoded competitive growth assay results, eight S. cerevisiae strains that showed either a strongly positive or negative fitness response to the presence of two non-Saccharomyces species (Table 1, Figure 1) were individually assessed in dual-species fermentation experiments.
In agreement with the WYBC library experiments, sugar consumption kinetics (Figure 2A,C) were delayed in all treatments. Although slower sugar consumption rates were explicitly observed in all treatments containing M. pulcherrima (Figure 2A), ferments inoculated with S. cerevisiae strains AWRI2867 and AWRI1490 contained high residual sugar concentrations after 40 days of fermentation (10.7 ± 1.7 and 7.3 ± 4.5 g/L). Despite the observed delay in sugar consumption compared to the control condition, all treatments containing T. delbrueckii finished fermentation after 14 days (Figure 2C). In both non-Saccharomyces treatments, S. cerevisiae strains that showed a positive and negative log2 FC in competitive experiments correlated with faster and slower sugar consumption rates, respectively (Figure 2A,C). The correlation between the two experimental outcomes confirms that there is S. cerevisiae strain-dependent variation in response to a non-Saccharomyces species. These results suggest that the previously reported detrimental effects on fermentation performance caused by non-Saccharomyces species [9,11,20] could be ameliorated by inoculation with specific S. cerevisiae strains. Furthermore, in the case of M. pulcherrima, the differences observed between strains describe the possibility of a stuck fermentation if specific S. cerevisiae strains are selected for sequential inoculation (Figure 2A).
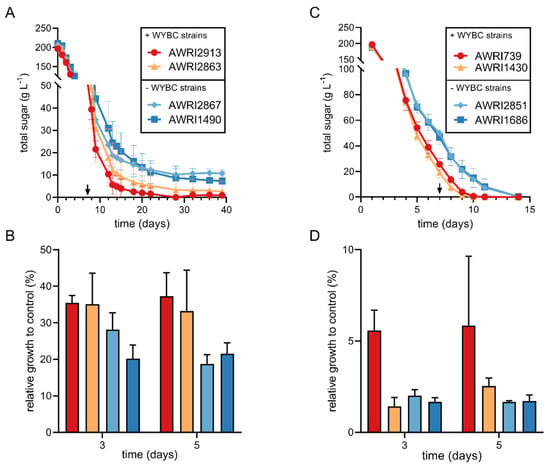
Figure 2.
Fermentation kinetics and relative growth in CDM of Saccharomyces cerevisiae strains in the presence of Metschnikowia pulcherrima (A,B) and Torulaspora delbrueckii (C,D). + and - WBYC strains refer to strains that showed a positive or negative log2 FC in the WYBC library competitive fitness experiments for the specific non-Saccharomyces treatment (Figure 1). The arrows in panels (A,C) indicate the time point when all S. cerevisiae controls finished fermentation. The relative growth in panels (B,D) are expressed as a percentage of cells in the specified timepoint relative to control conditions without the presence of a non-Saccharomyces species. All data points correspond to the mean of three replicates ± sd.
Determination of S. cerevisiae cell concentration by flow cytometry during the initial stages of fermentation revealed detrimental effects on cell growth that differed between strains and non-Saccharomyces treatments (Figure 2B,D). In the presence of M. pulcherrima (Figure 2B), S. cerevisiae strains differed in the cell concentration they were able to achieve relative to the control condition. These differences in cell concentration correlated with results obtained in the competitive fitness experiments (Figure 1). In treatments including T. delbrueckii, differences between positive and negative WYBC were only apparent in day 5 and an overall strong inhibition of cell growth was observed (Figure 2D). It can be speculated that larger differences in cell growth between positive and negative WYBC occurred later in fermentation, reflecting the differences observed in sugar consumption rates (Figure 2C). Other studies have reported similar effects on the growth of S. cerevisiae when using T. delbrueckii in a sequential inoculation regime; however, the mechanisms of inhibition have not yet been determined [20,38,39]. Despite this, consumption of more than 50% of the available sugar was already observed by day 5, indicating that T. delbrueckii was a significant contributor to sugar utilisation in this treatment. The overall cell counts confirm that S. cerevisiae strains are affected differently by the antagonism/competition caused by these non-Saccharomyces species. Comparison between treatments also highlights different mechanisms of antagonism/competition between M. pulcherrima and T. delbrueckii that should be further investigated.
4. Conclusions
The competitive fitness response to the presence of five non-Saccharomyces species was investigated in 93 S. cerevisiae wine strains using a genomically barcoded library of wine yeast. Analysis of unique barcode counts revealed considerable differences in the response of S. cerevisiae strains to the different non-Saccharomyces treatments. Dual species experiments that evaluated individual S. cerevisiae strain responses confirmed the presence of strain-specific fitness variation in response to a non-Saccharomyces species with consequences to fermentation timeframes. The information presented in this study will serve as an initial tool for the screen of yeast species interaction and selection of compatible strains that enhance fermentation performance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/fermentation7030165/s1, Table S1: Strain names of the barcoded library, Table S2: Log2 FC of barcoded strains at timepoint 1, Table S3: Log2 FC of barcoded strains at timepoint 2, Table S4: Log2 FC of barcoded strains at timepoint 3, Table S5: Metadata of raw barcode count files, File S1: Raw barcode count files.
Author Contributions
Conceptualization, C.A.O., A.R.B., S.A.S.; methodology, C.A.O. and S.A.S.; formal analysis, C.A.O.; investigation, C.A.O. and S.A.S.; data curation, C.A.O.; writing—original draft preparation, C.A.O.; writing—review and editing, C.A.O., A.R.B., S.A.S.; supervision, S.A.S. and A.R.B.; project administration, S.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Australian grapegrowers and winemakers through their investment body Wine Australia, with matching funds from the Australian Government. The AWRI is a member of the Wine Innovation Cluster (WIC) in Adelaide.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Acknowledgments
The authors would like to thank Jane McCarthy and Radka Kolouchova for their assistance in laboratory procedures.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. Handbook of Enology, Volume 1: The Microbiology of Wine and Vinifications; John Wiley & Sons: Hoboken, NJ, USA, 2006; Volume 1. [Google Scholar]
- Medina, K.; Boido, E.; Fariña, L.; Gioia, O.; Gomez, M.E.; Barquet, M.; Gaggero, C.; Dellacassa, E.; Carrau, F. Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem. 2013, 141, 2513–2521. [Google Scholar] [CrossRef]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benito, Á.; Calderón, F.; Benito, S. The influence of non-Saccharomyces species on wine fermentation quality parameters. Fermentation 2019, 5, 54. [Google Scholar] [CrossRef] [Green Version]
- Roca-Mesa, H.; Sendra, S.; Mas, A.; Beltran, G.; Torija, M.J. Nitrogen preferences during alcoholic fermentation of different non-Saccharomyces yeasts of oenological interest. Microorganisms 2020, 8, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pretorius, I.S. Tasting the terroir of wine yeast innovation. FEMS Yeast Res. 2019, 20, foz084. [Google Scholar] [CrossRef] [Green Version]
- Kemsawasd, V.; Viana, T.; Ardö, Y.; Arneborg, N. Influence of nitrogen sources on growth and fermentation performance of different wine yeast species during alcoholic fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 10191–10207. [Google Scholar] [CrossRef] [PubMed]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: Role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef]
- Benito, S. The impact of Torulaspora delbrueckii yeast in winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 3081–3094. [Google Scholar] [CrossRef] [PubMed]
- Rollero, S.; Bloem, A.; Ortiz-Julien, A.; Camarasa, C.; Divol, B. Fermentation performances and aroma production of non-conventional wine yeasts are influenced by nitrogen preferences. FEMS Yeast Res. 2018, 18, foy055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prior, K.J.; Bauer, F.F.; Divol, B. The utilization of nitrogenous compounds by commercial non-Saccharomyces yeasts associated with wine. Food Microbiol. 2019, 79, 75–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varela, C.; Bartel, C.; Espinase Nandorfy, D.; Bilogrevic, E.; Tran, T.; Heinrich, A.; Balzan, T.; Bindon, K.; Borneman, A. Volatile aroma composition and sensory profile of Shiraz and Cabernet Sauvignon wines produced with novel Metschnikowia pulcherrima yeast starter cultures. Aust. J. Grape Wine Res. 2021, 27, 406–418. [Google Scholar] [CrossRef]
- Hranilovic, A.; Albertin, W.; Capone, D.L.; Gallo, A.; Grbin, P.R.; Danner, L.; Bastian, S.E.P.; Masneuf-Pomarede, I.; Coulon, J.; Bely, M.; et al. Impact of Lachancea thermotolerans on chemical composition and sensory profiles of Merlot wines. Food Chem. 2021, 349, 129015. [Google Scholar] [CrossRef] [PubMed]
- Nissen, P.; Nielsen, D.; Arneborg, N. Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell–cell contact-mediated mechanism. Yeast 2003, 20, 331–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renault, P.E.; Albertin, W.; Bely, M. An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl. Microbiol. Biotechnol. 2013, 97, 4105–4119. [Google Scholar] [CrossRef] [PubMed]
- Albergaria, H.; Francisco, D.; Gori, K.; Arneborg, N.; Gírio, F. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 2010, 86, 965–972. [Google Scholar] [CrossRef]
- Pérez-Nevado, F.; Albergaria, H.; Hogg, T.; Girio, F. Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 2006, 108, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Growth of non-Saccharomyces yeasts affects nutrient availability for Saccharomyces cerevisiae during wine fermentation. Int. J. Food Microbiol. 2012, 157, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Taillandier, P.; Lai, Q.P.; Julien-Ortiz, A.; Brandam, C. Interactions between Torulaspora delbrueckii and Saccharomyces cerevisiae in wine fermentation: Influence of inoculation and nitrogen content. World J. Microbiol. Biotechnol. 2014, 30, 1959–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rollero, S.; Bloem, A.; Ortiz-Julien, A.; Camarasa, C.; Divol, B. Altered fermentation performances, growth, and metabolic footprints reveal competition for nutrients between yeast species inoculated in synthetic grape juice-like medium. Front. Microbiol. 2018, 9, 196. [Google Scholar] [CrossRef]
- Santos, A.; San Mauro, M.; Bravo, E.; Marquina, D. PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 2009, 155, 624–634. [Google Scholar] [CrossRef] [Green Version]
- Fernández de Ullivarri, M.; Mendoza, L.M.; Raya, R.R. Characterization of the killer toxin KTCf20 from Wickerhamomyces anomalus, a potential biocontrol agent against wine spoilage yeasts. Biol. Control. 2018, 121, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Oro, L.; Ciani, M.; Comitini, F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 2014, 116, 1209–1217. [Google Scholar] [CrossRef]
- Wang, C.; Mas, A.; Esteve-Zarzoso, B. Interaction between Hanseniaspora uvarum and Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 2015, 206, 67–74. [Google Scholar] [CrossRef]
- Schmidt, S.A.; Kolouchova, R.; Forgan, A.H.; Borneman, A.R. Evaluation of Saccharomyces cerevisiae wine yeast competitive fitness in enologically relevant environments by barcode sequencing. G3 Genes|Genomes|Genet. 2020, 10, 591–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borneman, A.R.; Desany, B.A.; Riches, D.; Affourtit, J.P.; Forgan, A.H.; Pretorius, I.S.; Egholm, M.; Chambers, P.J. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLOS Genet. 2011, 7, e1001287. [Google Scholar] [CrossRef] [Green Version]
- Borneman, A.R.; Forgan, A.H.; Kolouchova, R.; Fraser, J.A.; Schmidt, S.A. Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3: Genes|Genomes|Genet. 2016, 6, 957–971. [Google Scholar] [CrossRef] [Green Version]
- Anvar, S.Y.; van der Gaag, K.J.; van der Heijden, J.W.; Veltrop, M.H.; Vossen, R.H.; de Leeuw, R.H.; Breukel, C.; Buermans, H.P.; Verbeek, J.S.; de Knijff, P. TSSV: A tool for characterization of complex allelic variants in pure and mixed genomes. Bioinformatics 2014, 30, 1651–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. 2013. Available online: https://www.yumpu.com/en/document/view/6853895/r-a-language-and-environment-for-statistical-computing (accessed on 24 August 2021).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef] [PubMed]
- Varela, C.; Lleixà, J.; Curtin, C.; Borneman, A. Development of a genetic transformation toolkit for Brettanomyces bruxellensis. FEMS Yeast Res. 2018, 18, foy070. [Google Scholar] [CrossRef]
- Hohorst, H.J. d-Glucose-6-phosphate and d-Fructose-6-phosphate: Determination with glucose-6-phosphate dehydrogenase and phosphoglucose isomerase. In Methods of Enzymatic Analysis; Bergmeyer, H.U., Ed.; Academic Press: Cambridge, MA, USA, 1965; pp. 134–138. [Google Scholar]
- Vermeir, S.; Nicolaï, B.M.; Jans, K.; Maes, G.; Lammertyn, J. High-throughput microplate enzymatic assays for fast sugar and acid quantification in apple and tomato. J. Agric. Food Chem. 2007, 55, 3240–3248. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Beisert, B.; Navascués, E.; Marquina, D.; Calderón, F.; Rauhut, D.; Benito, S.; Santos, A. Influence of Torulaspora delbrueckii in varietal thiol (3-SH and 4-MSP) release in wine sequential fermentations. Int. J. Food Microbiol. 2017, 257, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Onetto, C.A.; Borneman, A.R.; Schmidt, S.A. Investigating the effects of Aureobasidium pullulans on grape juice composition and fermentation. Food Microbiol. 2020, 90, 103451. [Google Scholar] [CrossRef] [PubMed]
- Renault, P.; Coulon, J.; de Revel, G.; Barbe, J.-C.; Bely, M. Increase of fruity aroma during mixed T. delbrueckii/S. cerevisiae wine fermentation is linked to specific esters enhancement. Int. J. Food Microbiol. 2015, 207, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Renault, P.; Coulon, J.; Moine, V.; Thibon, C.; Bely, M. Enhanced 3-sulfanylhexan-1-ol production in sequential mixed fermentation with Torulaspora delbrueckii/Saccharomyces cerevisiae reveals a situation of synergistic interaction between two industrial strains. Front. Microbiol. 2016, 7, 293. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).