Probiotic Potential of Lactic Acid Bacteria Strains Isolated from Artisanal Cheeses: Impact on Listeria monocytogenes Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and LAB Isolation
2.2. Strains and Culture Conditions
2.3. Bacterial Resistance to the Simulated Gastrointestinal (SGI) Tract and Bile Salt Hydrolase (BSH) Activity
2.3.1. Survival of LAB to SGI Conditions
2.3.2. Bile Salt Tolerance
2.4. Antimicrobial Activity Against L. Monocytogenes
2.5. Hydrophobicity and Auto-Aggregation Assay
2.6. Genetic Identification of the Selected Strains
2.6.1. Sequencing the 16S rRNA Gene
2.6.2. Multiplex PCR Assay
2.7. Adhesion Assays
2.8. Invasion Assays
2.9. Safety Evaluation
2.9.1. Antibiotic Sensitivity
2.9.2. Hemolytic, Gelatinase, and Lecithinase Activity
2.10. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hernandez-Milian, A.; Payeras-Cifre, A. What is new in listeriosis? BioMed Res. Int. 2014, 2014, 358051. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food. Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, M.M.; Brouwer, M.C.; Vázquez-Boland, J.A.; van de Beek, D. Human Listeriosis. Clin. Microbiol. Rev. 2023, 36, e0006019. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, M. Listeria monocytogenes, a model in infection biology. Cell. Microbiol. 2020, 22, e13186. [Google Scholar] [CrossRef]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef]
- Matera, M. Bifidobacteria, Lactobacilli… when, how and why to use them. Glob. Pediatr. 2024, 8, 100139. [Google Scholar] [CrossRef]
- Sarita, B.; Samadhan, D.; Hassan, M.Z.; Kovaleva, E.G. A comprehensive review of probiotics and human health-current prospective and applications. Front. Microbiol. 2025, 15, 1487641. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Yan, F.; Polk, D.B. Probiotics and probiotic-derived functional factors-mechanistic insights into applications for intestinal homeostasis. Front. Immunol. 2020, 11, 1428. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations/World Health Organization (FAO/WHO). Guidelines for the Evaluation of Probiotics in Food. Report of a Joint FAO/WHO working group in London Ontario, Canada, April 2002. Available online: https://www.mhlw.go.jp/file/05-Shingikai-11121000-Iyakushokuhinkyoku-Soumuka/0000197343.pdf (accessed on 20 December 2024).
- Mani-López, E.; Arrioja-Bretón, D.; López-Malo, A. The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Compr. Rev. Food Sci. Food Saf. 2022, 21, 604–641. [Google Scholar] [CrossRef]
- Darbandi, A.; Asadi, A.; Mahdizade Ari, M.; Ohadi, E.; Talebi, M.; Halaj Zadeh, M.; Darb Emamie, A.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and potential use as antimicrobials. J. Clin. Lab. Anal. 2022, 36, e24093. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.; Cocconcelli, P.; Vignolo, G.; Saavedra, L. Occurrence of antilisterial structural bacteriocins genes in meat borne lactic acid bacteria. Food Control 2015, 47, 53–59. [Google Scholar] [CrossRef]
- Chua, J.C.L.; Hale, J.D.F.; Silcock, P.; Bremer, P.J. Bacterial survival and adhesion for formulating new oral probiotic foods. Crit. Rev. Food Sci. Nutr. 2020, 60, 2926–2937. [Google Scholar] [CrossRef]
- Castellano, P.; Vignolo, G. Inhibition of Listeria innocua and Brochothrix thermosphacta in vacuum-packaged meat by addition of bacteriocinogenic Lactobacillus curvatus CRL705 and its bacteriocins. Lett. Appl. Microbiol. 2006, 43, 194–199. [Google Scholar] [CrossRef]
- Marchesi, A.; Silva, J.A.; Wiese, B.; Nader-Macías, M.E.F. Survival of beneficial vaginal Lactobacilli (BVL) to different gastrointestinal tract conditions. Curr. Pharm. Des. 2020, 26, 3608–3618. [Google Scholar] [CrossRef]
- Franz, C.M.; Specht, I.; Haberer, P.; Holzapfel, W.H. Bile salt hydrolase activity of Enterococci isolated from food: Screening and quantitative determination. J. Food Prot. 2001, 64, 725–729. [Google Scholar] [CrossRef]
- Pérez Ibarreche, M.; Castellano, P.; Vignolo, G. Evaluation of anti-Listeria meat borne Lactobacillus for biofilm formation on selected abiotic surfaces. Meat Sci. 2014, 96, 295–303. [Google Scholar] [CrossRef]
- Maldonado, N.C.; de Ruiz, C.S.; Otero, M.C.; Sesma, F.; Nader-Macías, M.E. Lactic acid bacteria isolated from young calves-characterization and potential as probiotics. Res. Vet. Sci. 2012, 92, 342–349. [Google Scholar] [CrossRef]
- Pospiech, A.; Neumann, B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995, 11, 217–218. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; Volume 4, pp. 115–175. [Google Scholar]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- Castellano, P.; Pérez Ibarreche, M.; Longo Borges, L.; Niño Arias, F.C.; Ross, G.R.; De Martinis, E.C.P. Lactobacillus spp. impair the ability of Listeria monocytogenes FBUNT to adhere to and invade Caco-2 cells. Biotechnol. Lett. 2018, 40, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- ISO 10932:2010/IDF 223:2010 standard; Milk and Milk Products—Determination of the Minimal Inhibitory Concentration (Mic) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB). International Organization for Standardization: Geneva, Switzerland, 2010.
- EFSA-FEEDAP. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740–2749. [Google Scholar] [CrossRef]
- De Vuyst, L.; Foulquie Moreno, M.R.; Revets, H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 2003, 84, 299–318. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.H.; Aristimuño Ficoseco, C.; Nader-Macías, M.E.F. Safety, environmental and technological characterization of beneficial autochthonous lactic bacteria, and their vaginal administration to pregnant cows for the design of homologous multi-strain probiotic formulas. Braz. J. Microbiol. 2021, 52, 2455–2473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lv, J.; Pan, L.; Zhang, Y. Roles and applications of probiotic Lactobacillus strains. Appl. Microbiol. Biotechnol. 2018, 102, 8135–8143. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Aljasir, S.F.; D’Amico, D.J. Probiotic potential of commercial dairy-associated protective cultures: In vitro and in vivo protection against Listeria monocytogenes infection. Food Res. Int. 2021, 149, 110699. [Google Scholar] [CrossRef]
- Choi, Y.; Park, E.; Kim, S.; Ha, J.; Oh, H.; Kim, Y.; Lee, Y.; Seo, Y.; Kang, J.; Lee, S.; et al. Fermented milk with Lactobacillus curvatus SMFM2016-NK alleviates periodontal and gut inflammation, and alters oral and gut microbiota. J. Dairy Sci. 2021, 104, 5197–5207. [Google Scholar] [CrossRef]
- Zommiti, M.; Connil, N.; Hamida, J.B.; Ferchichi, M. Probiotic characteristics of Lactobacillus curvatus DN317, a strain isolated from chicken ceca. Probiotics Antimicrob. Proteins 2017, 9, 415–424. [Google Scholar] [CrossRef]
- Jo, S.G.; Noh, E.J.; Lee, J.Y.; Kim, G.; Choi, J.H.; Lee, M.E.; Song, J.H.; Chang, J.Y.; Park, J.H. Lactobacillus curvatus WiKim38 isolated from kimchi induces IL-10 production in dendritic cells and alleviates DSS-induced colitis in mice. J. Microbiol. 2016, 54, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Rubayet Ul Alam, A.S.M.; Jahid, I.K. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.D.; Kim, S.H.; Jeong, J.W.; Lee, D.E.; Huh, C.S.; Hong, S.S.; Sim, J.H.; Ahn, Y.T. Triglyceride-lowering effects of two probiotics, Lactobacillus plantarum KY1032 and Lactobacillus curvatus HY7601, in a rat model of high-fat diet-induced hypertriglyceridemia. J. Microbiol. Biotechnol. 2016, 26, 483–487. [Google Scholar] [CrossRef]
- Jeung, W.H.; Shim, J.-J.; Woo, S.-W.; Sim, J.-H.; Lee, J.-L. Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 cell extracts inhibit adipogenesis in 3T3-L1 and HepG2 cells. J. Med. Food 2018, 21, 876–886. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef]
- Aazami, N.; Salehi Jouzani, G.; Khodaei, Z.; Meimandipour, A.; Safari, M.; Goudarzvand, M. Characterization of some potentially probiotic Lactobacillus strains isolated from Iranian native chickens. J. Gen. Appl. Microbiol. 2014, 60, 215–221. [Google Scholar] [CrossRef]
- Mukherjee, S.; Ramesh, A. Dual-label flow cytometry-based host cell adhesion assay to ascertain the prospect of probiotic Lactobacillus plantarum in niche-specific antibacterial therapy. Microbiology 2017, 163, 1822–1834. [Google Scholar] [CrossRef]
- Garriga, M.; Rubio, R.; Aymerich, T.; Ruas-Madiedo, P. Potentially probiotic and bioprotective lactic acid bacteria starter cultures antagonise the Listeria monocytogenes adhesion to HT29 colonocyte-like cells. Benef. Microbes 2015, 6, 337–343. [Google Scholar] [CrossRef]
- Kotzamanidis, C.; Kourelis, A.; Litopoulou-Tzanetaki, E.; Tzanetakis, N.; Yiangou, M. Evaluation of adhesion capacity, cell surface traits and immunomodulatory activity of presumptive probiotic Lactobacillus strains. Int. J. Food Microbiol. 2010, 140, 154–163. [Google Scholar] [CrossRef]
- Nandha, M.C.; Shukla, R.M. Exploration of probiotic attributes in lactic acid bacteria isolated from fermented Theobroma cacao L. fruit using in vitro techniques. Front. Microbiol. 2023, 14, 1274636. [Google Scholar] [CrossRef]
- Liu, C.; Xue, W.J.; Ding, H.; An, C.; Ma, S.J.; Liu, Y. Probiotic potential of Lactobacillus strains isolated from fermented vegetables in shaanxi, China. Front. Microbiol. 2022, 12, 774903. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.C.; Rodrigues, M.R.; Winkelströter, L.K.; Nomizo, A.; de Martinis, E.C. In vitro evaluation of the probiotic potential of bacteriocin producer Lactobacillus sakei 1. J. Food Prot. 2012, 75, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Zhang, X.; Huang, Y.; Liu, J.; Liu, M.; Zhou, G. Bacteriocinogenic Enterococcus faecium inhibits the virulence property of Listeria monocytogenes. LWT-Food Sci. Technol. 2018, 89, 87–92. [Google Scholar] [CrossRef]
- Montiel, R.; Quesille-Villalobos, A.; Alessandria, V.; Medina, M.; Cocolin, L.S.; Rantsiou, K. Antilisterial effect and influence on Listeria monocytogenes gene expression of enterocin or Enterococcus faecalis in sliced dry-cured ham stored at 7 °C. J. Food Prot. 2019, 82, 1598–1606. [Google Scholar] [CrossRef]
- Melian, C.; Bentencourt, E.; Castellano, P.; Ploper, D.; Vignolo, G.; Mendoza, L.M. Biofilm genes expression of Listeria monocytogenes exposed to Latilactobacillus curvatus bacteriocins at 10 °C. Int. J. Food Microbiol. 2022, 370, 109648. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards. Scientific opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed. EFSA J. 2013, 11, 3449. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.A.; Tsakalidou, E.; Nychas, G.J.; Panagou, E.Z.; Tassou, C.C.l. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, L.; Qiao, N.; Xiao, Y.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Latilactobacillus curvatus: A candidate probiotic with excellent fermentation properties and health benefits. Foods 2020, 9, 1366. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, S.; Zhao, M.D.; Yang, X.; Choi, S.H.; Li, G.Y. Screening and identification of Latilactobacillus curvatus Z12 from rumen fluid of an adult female sika deer as a potential probiotic for feed additives. Front. Vet. Sci. 2021, 8, 753527. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Olufemi, F.O.; Chijioke, O.M.; Popoola, S.T.; Oluwafemi, O.F. In vitro study of potential probiotic lactic acid bacteria isolated from the gut of chickens in Abeokuta, Nigeria. Alex. J. Vet. Sci. 2018, 58, 73–84. [Google Scholar]
- Jawan, R.; Abbasiliasi, S.; Mustafa, S.; Kapri, M.R.; Halim, M.; Ariff, A.B. In vitro evaluation of potential probiotic strain Lactococcus lactis Gh1 and its bacteriocin-like inhibitory substances for potential use in the food industry. Probiotics Antimicrob. Proteins 2021, 13, 422–440. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.A.; Marchesi, A.; Aristimuño Ficosecco, M.C.; Nader-Macías, M.E.F. Functional and safety characterization of beneficial vaginal lactic acid bacteria for the design of vaginal hygiene products. J. Appl. Microbiol. 2022, 133, 3041–3058. [Google Scholar] [CrossRef] [PubMed]

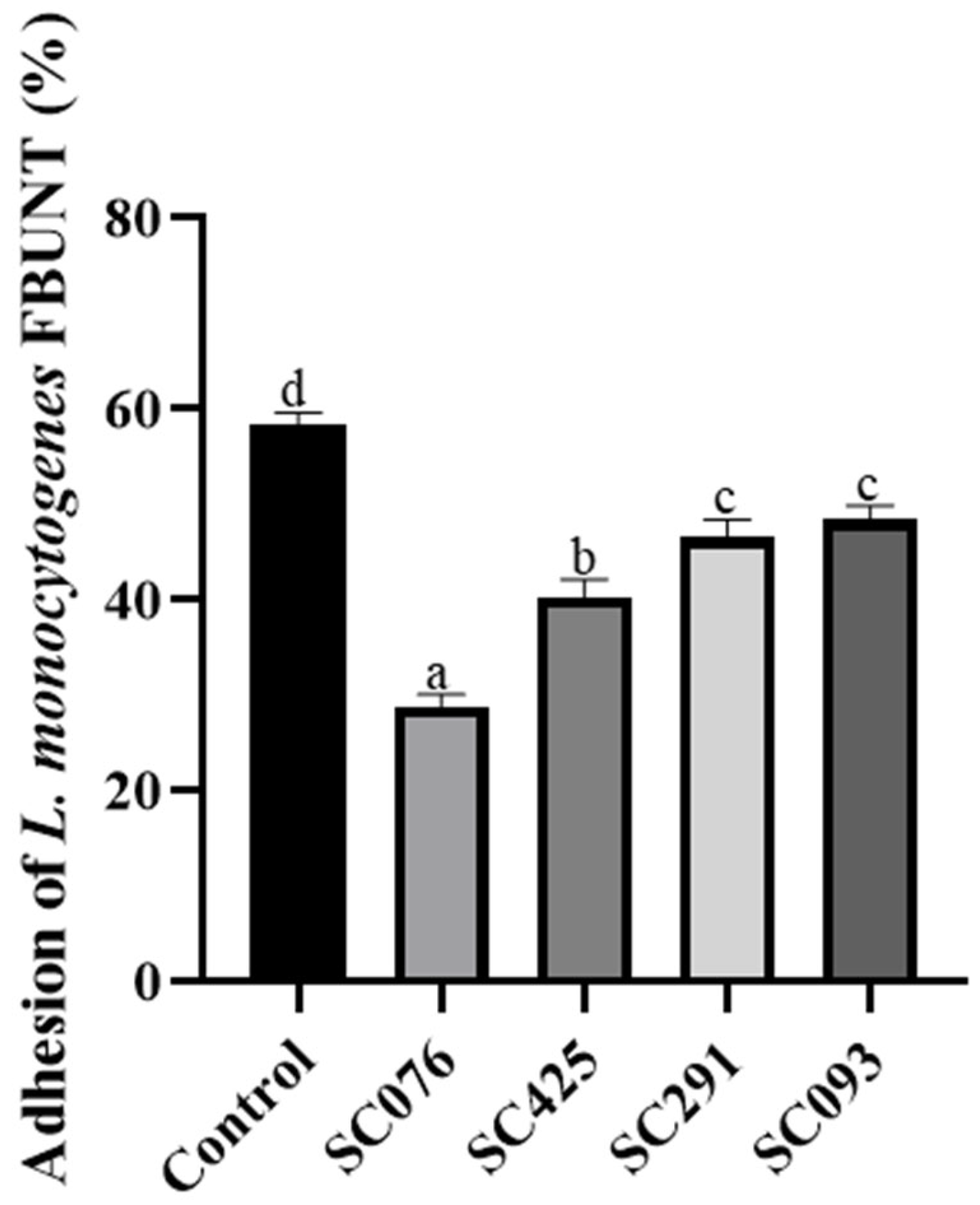
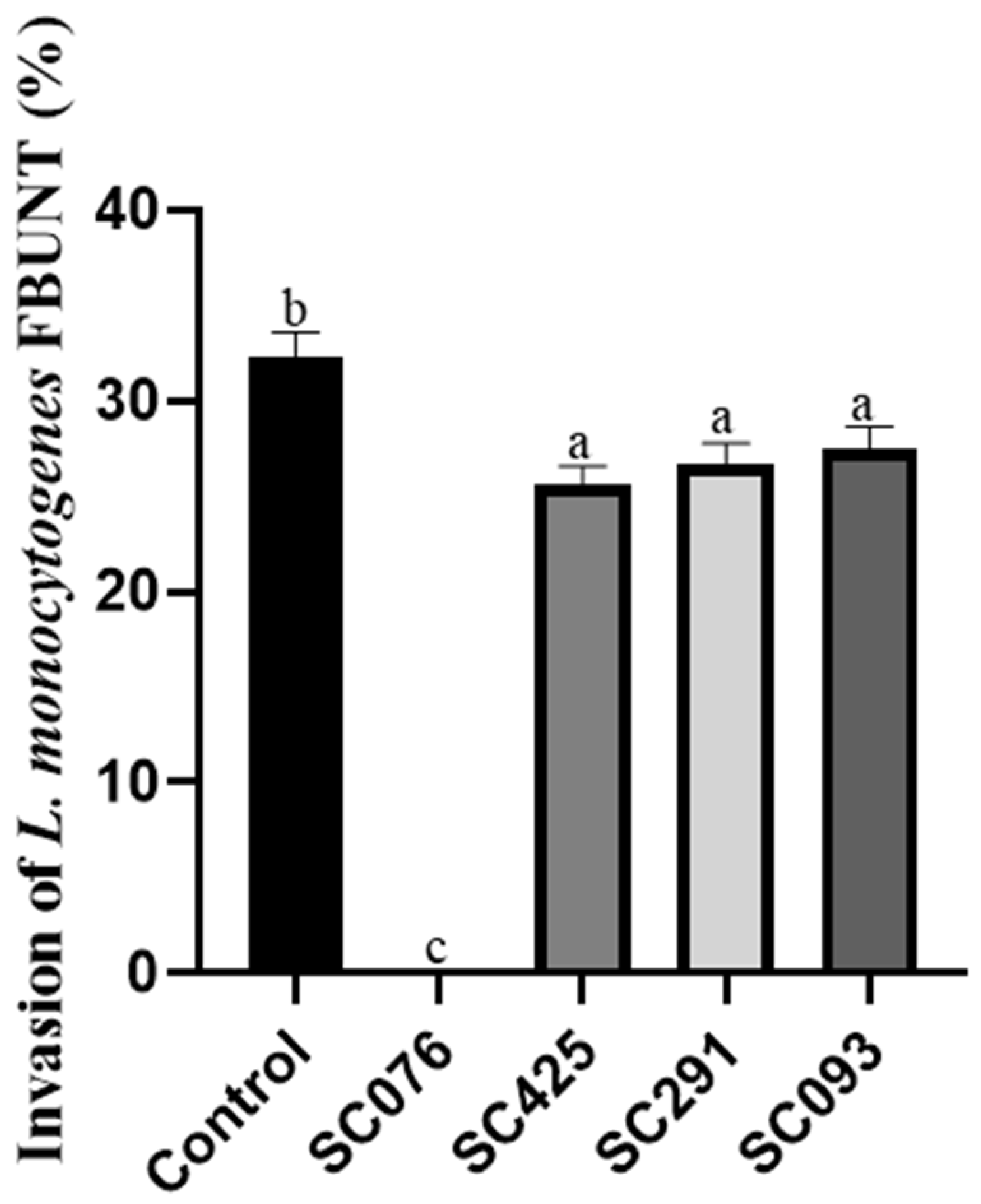
| Strains | Antimicrobial Activity Against L. monocytogenes FBUNT | Bile Salt Hydrolysis | |||
|---|---|---|---|---|---|
| CFS* | CFS Neutralized (N) | CFS N+Catalase | CFS N+Proteinase K | ||
| SC076 | +++ | +++ | +++ | - | + |
| SC425 | + | - | - | - | + |
| SC291 | + | - | - | - | + |
| SC093 | + | - | - | - | + |
| CRL705† | +++ | +++ | +++ | - | + |
| Strains | Auto-Aggregation (%) | Hydrophobicity (%) | Adhesion (%) | |
|---|---|---|---|---|
| Xilene | Toluene | |||
| SC076 | 33.62a ± 1.13 | 46.00a ± 6.60 | 49.96a ± 5.13 | 43.17a ± 3.52 |
| SC425 | 38.64a ± 2.12 | 53.37a ± 0.79 | 45.74a ± 5.13 | 48.29a ± 1.17 |
| SC291 | 37.39a ± 2.13 | 55.09a ± 2.48 | 48.89a ± 12.57 | 46.24a ± 1.90 |
| SC093 | 35.82a ± 0.10 | 59.09a ± 4.99 | 55.35a ± 5.99 | 43.18a ±1.52 |
| Strains | ATB | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | GEN | KAN | ERY | TET | CHL | STR | |||
| Cut-Off Value | 2 | 16 | 64 | 1 | 32 | 8 | n.r | ||
| Lact. paraplantarum | SC093 | MIC | 4 | 16 | 128 | 0.125 | 64 | 8 | |
| SC291 | MIC | 4 | 8 | 64 | 0.125 | 64 | 8 | ||
| SC425 | MIC | 4 | 8 | 128 | 0.125 | 32 | 4 | ||
| Lat. curvatus | SC076 | Cut-off value | 4 | 16 | 64 | 1 | 8 | 4 | 64 |
| MIC | 4 | 4 | 8 | 0 | 4 | 2 | 16 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burgos, C.; Melian, C.; Mendoza, L.M.; Salva, S.; Castellano, P. Probiotic Potential of Lactic Acid Bacteria Strains Isolated from Artisanal Cheeses: Impact on Listeria monocytogenes Infection. Fermentation 2025, 11, 343. https://doi.org/10.3390/fermentation11060343
Burgos C, Melian C, Mendoza LM, Salva S, Castellano P. Probiotic Potential of Lactic Acid Bacteria Strains Isolated from Artisanal Cheeses: Impact on Listeria monocytogenes Infection. Fermentation. 2025; 11(6):343. https://doi.org/10.3390/fermentation11060343
Chicago/Turabian StyleBurgos, Carla, Constanza Melian, Lucía M. Mendoza, Susana Salva, and Patricia Castellano. 2025. "Probiotic Potential of Lactic Acid Bacteria Strains Isolated from Artisanal Cheeses: Impact on Listeria monocytogenes Infection" Fermentation 11, no. 6: 343. https://doi.org/10.3390/fermentation11060343
APA StyleBurgos, C., Melian, C., Mendoza, L. M., Salva, S., & Castellano, P. (2025). Probiotic Potential of Lactic Acid Bacteria Strains Isolated from Artisanal Cheeses: Impact on Listeria monocytogenes Infection. Fermentation, 11(6), 343. https://doi.org/10.3390/fermentation11060343







