Immune Cell Degranulation in Fungal Host Defence
Abstract
1. Introduction
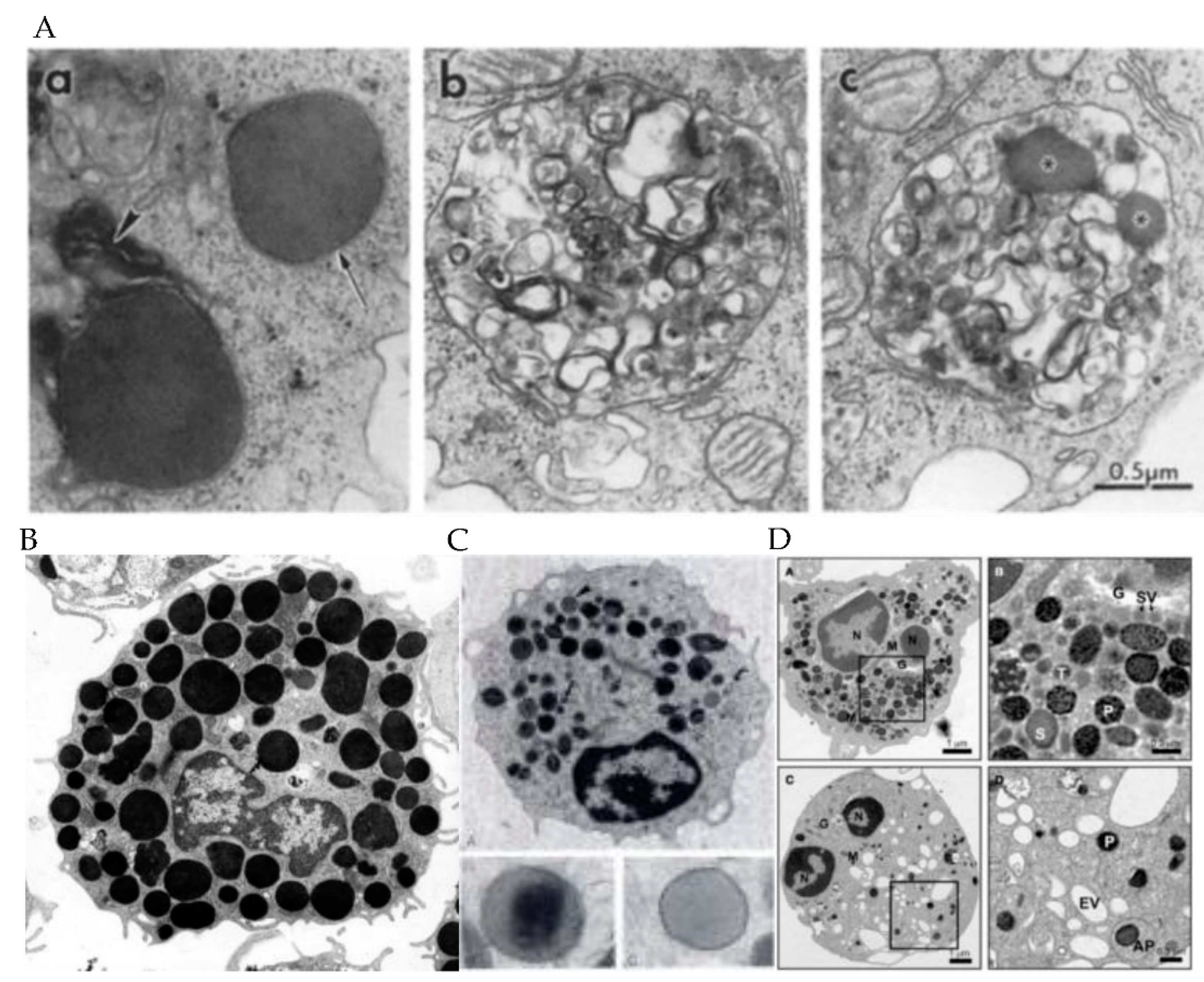
2. Granule Characteristics in Different Immune Cell Subsets
3. Cytolytic Contents of the Granules in Each Immune Cell Subtypes
4. Mechanisms of Fungal Recognition, Activation, and Cytotoxicity
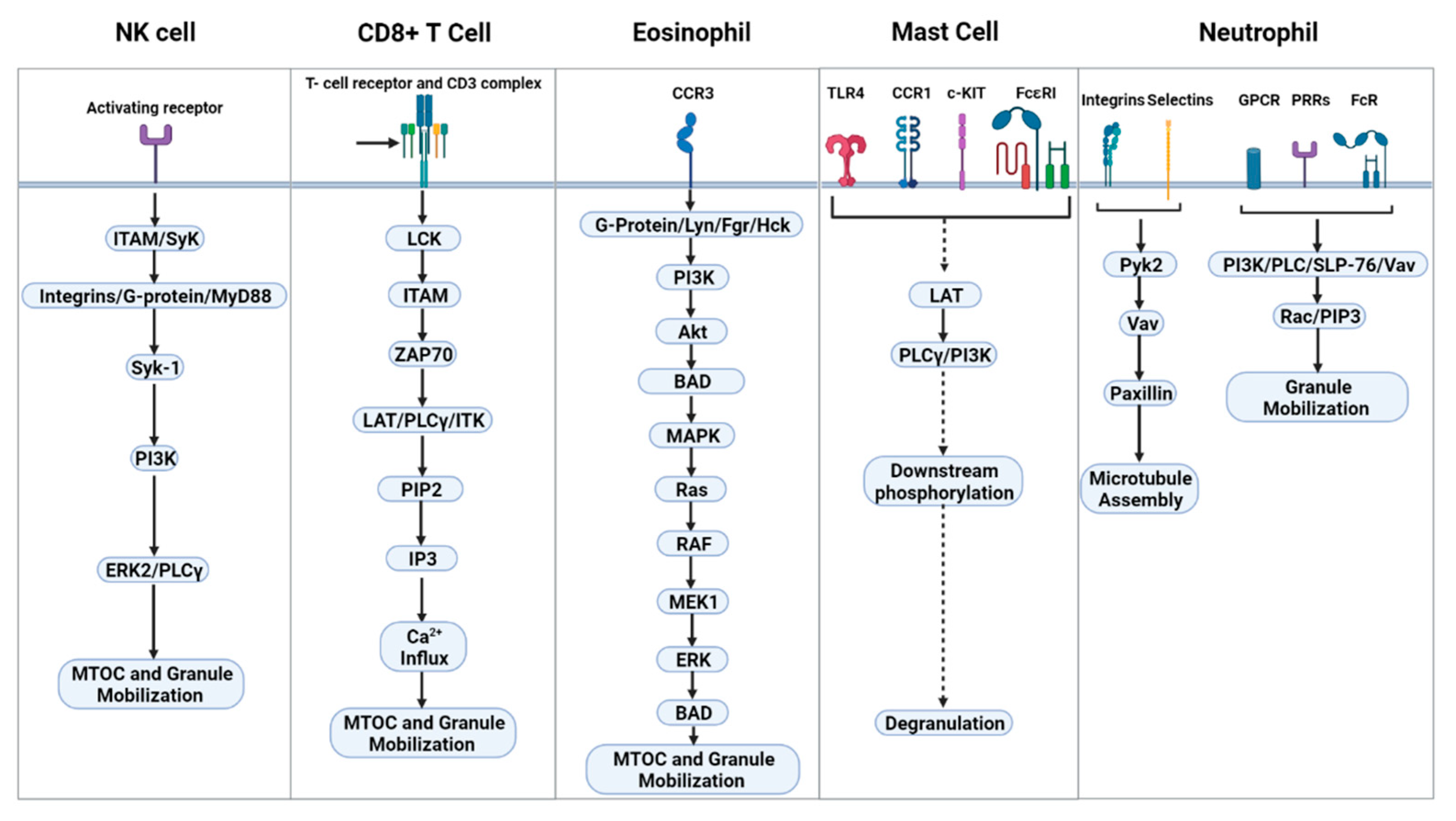
5. Signal Transduction Leading to Degranulation
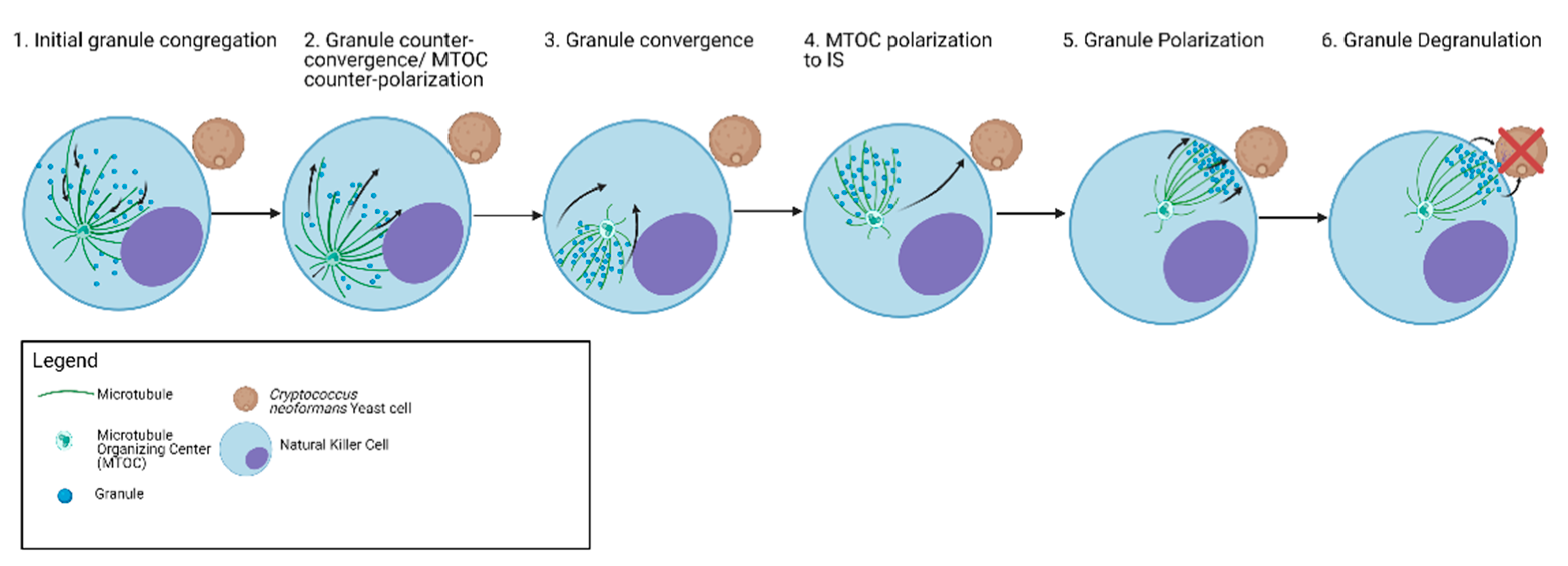
6. Granule Trafficking Leading to Degranulation
7. Function of Granule Proteins
7.1. Perforin
7.2. Granzymes
7.3. Other Proteins Causing Membrane Permeability
7.4. Proteases—Neutrophil Elastase (NE), Cathepsins, Proteinase 3, Matrix Metalloproteinases
7.5. Oxidative Agents: Myeloperoxidase (MPO) and Eosinophil Peroxidase (EPO)
7.6. Iron Scavengers
7.7. Alarmins: Azurocidin and Eosinophil-Derived Neurotoxin (EDN)
7.8. Iron scavengers
7.9. Major Basic Protein (MBP) and Eosinophil-Cationic Protein (ECP)
8. The Cells That Degranulate
8.1. NK Cells
8.2. CD8+ T Cells
8.3. Mast Cells
8.4. Eosinophils
8.5. Neutrophils
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lockhart, S.R.; Guarner, J. Emerging and Reemerging Fungal Infections. Semin. Diagn. Pathol. 2019, 36, 177–181. [Google Scholar] [CrossRef]
- Garbee, D.D.; Pierce, S.S.; Manning, J. Opportunistic Fungal Infections in Critical Care Units. Crit. Care Nurs. Clin. N. Am. 2017, 29, 67–79. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Bassetti, M.; Righi, E.; Ansaldi, F.; Merelli, M.; Trucchi, C.; Cecilia, T.; De Pascale, G.; Diaz-Martin, A.; Luzzati, R.; Rosin, C.; et al. A Multicenter Study of Septic Shock Due to Candidemia: Outcomes and Predictors of Mortality. Intensive Care Med. 2014, 40, 839–845. [Google Scholar] [CrossRef]
- Kontoyiannis, D.P. Antifungal Prophylaxis in Hematopoietic Stem Cell Transplant Recipients: The Unfinished Tale of Imperfect Success. Bone Marrow Transpl. 2011, 46, 165–173. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krzewski, K.; Coligan, J.E. Human NK Cell Lytic Granules and Regulation of Their Exocytosis. Front. Immunol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, J.K.; Hester, S.; Lapham, C.K.; Argon, Y. The Lytic Granules of Natural Killer Cells Are Dual-Function Organelles Combining Secretory and Pre-Lysosomal Compartments. J. Cell Biol. 1990, 111, 2327–2340. [Google Scholar] [CrossRef] [PubMed]
- Neighbour, P.A.; Huberman, H.S.; Kress, Y. Human Large Granular Lymphocytes and Natural Killing Ultrastructural Studies of Strontium-Induced Degranulation. Eur. J. Immunol. 1982, 12, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Ruiz, Y.; Valitutti, S.; Dupre, L. Stepwise Maturation of Lytic Granules during Differentiation and Activation of Human CD8+ T Lymphocytes. PLoS ONE 2011, 6, e27057. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Gelhaus, C.; Nebendahl, M.; Lettau, M.; Lucius, R.; Leippe, M.; Kabelitz, D.; Janssen, O. Effector Granules in Human T Lymphocytes: Proteomic Evidence for Two Distinct Species of Cytotoxic Effector Vesicles. J. Proteome. Res. 2011, 10, 1603–1620. [Google Scholar] [CrossRef]
- Siraganian, R.P. Mast Cells. In Encyclopedia of Immunology, 2nd ed.; Delves, P.J., Ed.; Elsevier: Oxford, UK, 1998; pp. 1667–1671. ISBN 978-0-12-226765-9. [Google Scholar]
- Dvorak, A.M.; Letourneau, L.; Login, G.R.; Weller, P.F.; Ackerman, S.J. Ultrastructural Localization of the Charcot-Leyden Crystal Protein (Lysophospholipase) to a Distinct Crystalloid-Free Granule Population in Mature Human Eosinophils. Blood 1988, 72, 150–158. [Google Scholar] [CrossRef]
- Sheshachalam, A.; Srivastava, N.; Mitchell, T.; Lacy, P.; Eitzen, G. Granule Protein Processing and Regulated Secretion in Neutrophils. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Moon, T.C.; Befus, A.D.; Kulka, M. Mast Cell Mediators: Their Differential Release and the Secretory Pathways Involved. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- McBrien, C.N.; Menzies-Gow, A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. 2017, 4. [Google Scholar] [CrossRef]
- Muniz, V.S.; Weller, P.F.; Neves, J.S. Eosinophil Crystalloid Granules: Structure, Function, and Beyond. J. Leukoc. Biol. 2012, 92, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Shamri, R.; Xenakis, J.J.; Spencer, L.A. Eosinophils in Innate Immunity: An Evolving Story. Cell Tissue Res. 2011, 343, 57–83. [Google Scholar] [CrossRef]
- Wickramasinghe, S.; Erber, W. CHAPTER 1—Normal blood cells. In Blood and Bone Marrow Pathology, 2nd ed.; Porwit, A., McCullough, J., Erber, W.N., Eds.; Churchill Livingstone: Edinburgh, Scotland, 2011; pp. 3–18. ISBN 978-0-7020-3147-2. [Google Scholar]
- Murav’ev, R.A.; Fomina, V.A.; Rogovin, V.V. Gelatinase Granules of Neutrophil Granulocytes. Biol. Bull. 2003, 30, 317–321. [Google Scholar] [CrossRef]
- Borregaard, N.; Sørensen, O.E.; Theilgaard-Mönch, K. Neutrophil Granules: A Library of Innate Immunity Proteins. Trends Immunol. 2007, 28, 340–345. [Google Scholar] [CrossRef]
- Melo, R.C.N.; Spencer, L.A.; Dvorak, A.M.; Weller, P.F. Mechanisms of Eosinophil Secretion: Large Vesiculotubular Carriers Mediate Transport and Release of Granule-Derived Cytokines and Other Proteins. J. Leukoc. Biol. 2008, 83, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Lundequist, A.; Tchougounova, E.; Åbrink, M.; Pejler, G. Cooperation between Mast Cell Carboxypeptidase A and the Chymase Mouse Mast Cell Protease 4 in the Formation and Degradation of Angiotensin II*. J. Biol. Chem. 2004, 279, 32339–32344. [Google Scholar] [CrossRef]
- Trevisan, E.; Vita, F.; Medic, N.; Soranzo, M.R.; Zabucchi, G.; Borelli, V. Mast Cells Kill Candida Albicans in the Extracellular Environment but Spare Ingested Fungi from Death. Inflammation 2014, 37, 2174–2189. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Kyei, S.K.; Timm-McCann, M.; Ogbomo, H.; Jones, G.J.; Shi, M.; Xiang, R.F.; Oykhman, P.; Huston, S.M.; Islam, A.; et al. The NK Receptor NKp30 Mediates Direct Fungal Recognition and Killing and Is Diminished in NK Cells from HIV-Infected Patients. Cell Host Microbe 2013, 14, 387–397. [Google Scholar] [CrossRef]
- Vitenshtein, A.; Charpak-Amikam, Y.; Yamin, R.; Bauman, Y.; Isaacson, B.; Stein, N.; Berhani, O.; Dassa, L.; Gamliel, M.; Gur, C.; et al. NK Cell Recognition of Candida Glabrata through Binding of NKp46 and NCR1 to Fungal Ligands Epa1, Epa6, and Epa7. Cell Host Microbe 2016, 20, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.; Weiss, E.; Schmitt, A.-L.; Schlegel, J.; Burgert, A.; Terpitz, U.; Sauer, M.; Moretta, L.; Sivori, S.; Leonhardt, I.; et al. CD56 Is a Pathogen Recognition Receptor on Human Natural Killer Cells. Sci. Rep. 2017, 7, 6138. [Google Scholar] [CrossRef]
- Salazar, F.; Brown, G.D. Antifungal Innate Immunity: A Perspective from the Last 10 Years. JIN 2018, 10, 373–397. [Google Scholar] [CrossRef]
- Backer, R.; van Leeuwen, F.; Kraal, G.; den Haan, J.M.M. CD8- Dendritic Cells Preferentially Cross-Present Saccharomyces Cerevisiae Antigens. Eur. J. Immunol. 2008, 38, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shmuel, A.; Sabag, B.; Biber, G.; Barda-Saad, M. The Role of the Cytoskeleton in Regulating the Natural Killer Cell Immune Response in Health and Disease: From Signaling Dynamics to Function. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Oykhman, P.; Mody, C.H. Direct Microbicidal Activity of Cytotoxic T-Lymphocytes. J. Biomed. Biotechnol. 2010, 2010, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Trivedi, P.P.; Ge, B.; Krzewski, K.; Strominger, J.L. Many NK Cell Receptors Activate ERK2 and JNK1 to Trigger Microtubule Organizing Center and Granule Polarization and Cytotoxicity. Proc. Natl. Acad. Sci. USA 2007, 104, 6329–6334. [Google Scholar] [CrossRef]
- Wiseman, J.C.D.; Ma, L.L.; Marr, K.J.; Jones, G.J.; Mody, C.H. Perforin-Dependent Cryptococcal Microbicidal Activity in NK Cells Requires PI3K-Dependent ERK1/2 Signaling. J. Immunol. 2007, 178, 6456–6464. [Google Scholar] [CrossRef]
- Bhat, R.; Watzl, C. Serial Killing of Tumor Cells by Human Natural Killer Cells – Enhancement by Therapeutic Antibodies. PLoS ONE 2007, 2, e326. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.L.; Wang, C.L.C.; Neely, G.G.; Epelman, S.; Krensky, A.M.; Mody, C.H. NK Cells Use Perforin Rather than Granulysin for Anticryptococcal Activity. J. Immunol. 2004, 173, 3357–3365. [Google Scholar] [CrossRef] [PubMed]
- Ogbomo, H.; Timm-McCann, M.; Barnes, T.; Xiang, R.F.; Jamil, K.; Ganguly, A.; Stack, D.; Huston, S.M.; Li, S.S.; Colarusso, P.; et al. Granule-Dependent NK Cell Killing of Cryptococcus Requires Kinesin to Reposition the Cytolytic Machinery for Directed Cytotoxicity. Cell Rep. 2018, 24, 3017–3032. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.F.; Stack, D.; Huston, S.M.; Li, S.S.; Ogbomo, H.; Kyei, S.K.; Mody, C.H. Ras-Related C3 Botulinum Toxin Substrate (Rac) and Src Family Kinases (SFK) Are Proximal and Essential for Phosphatidylinositol 3-Kinase (PI3K) Activation in Natural Killer (NK) Cell-Mediated Direct Cytotoxicity against Cryptococcus Neoformans. J. Biol. Chem. 2016, 291, 6912–6922. [Google Scholar] [CrossRef] [PubMed]
- Kabanova, A.; Zurli, V.; Baldari, C.T. Signals Controlling Lytic Granule Polarization at the Cytotoxic Immune Synapse. Front. Immunol. 2018, 9, 307. [Google Scholar] [CrossRef]
- Chang, H.W.; Kanegasaki, S.; Jin, F.; Deng, Y.; You, Z.; Chang, J.-H.; Kim, D.Y.; Timilshina, M.; Kim, J.-R.; Lee, Y.J.; et al. A Common Signaling Pathway Leading to Degranulation in Mast Cells and Its Regulation by CCR1-Ligand. Allergy 2020, 75, 1371–1381. [Google Scholar] [CrossRef]
- Gorska, M.M.; Alam, R. The Signaling Mechanism of Eosinophil Activation. Expert Rev. Clin. Immunol. 2005, 1, 247–256. [Google Scholar] [CrossRef]
- Yoon, J.; Ponikau, J.U.; Lawrence, C.B.; Kita, H. Innate Antifungal Immunity of Human Eosinophils Mediated by a Β2 Integrin, CD11b. J. Immunol. 2008, 181, 2907–2915. [Google Scholar] [CrossRef]
- Kapitein, L.C.; Peterman, E.J.G.; Kwok, B.H.; Kim, J.H.; Kapoor, T.M.; Schmidt, C.F. The Bipolar Mitotic Kinesin Eg5 Moves on Both Microtubules That It Crosslinks. Nature 2005, 435, 114–118. [Google Scholar] [CrossRef]
- Kurowska, M.; Goudin, N.; Nehme, N.T.; Court, M.; Garin, J.; Fischer, A.; de Saint Basile, G.; Ménasché, G. Terminal Transport of Lytic Granules to the Immune Synapse Is Mediated by the Kinesin-1/Slp3/Rab27a Complex. Blood 2012, 119, 3879–3889. [Google Scholar] [CrossRef] [PubMed]
- Tuli, A.; Thiery, J.; James, A.M.; Michelet, X.; Sharma, M.; Garg, S.; Sanborn, K.B.; Orange, J.S.; Lieberman, J.; Brenner, M.B. Arf-like GTPase Arl8b Regulates Lytic Granule Polarization and Natural Killer Cell-Mediated Cytotoxicity. Mol. Biol. Cell 2013, 24, 3721–3735. [Google Scholar] [CrossRef] [PubMed]
- Munoz, I.; Danelli, L.; Claver, J.; Goudin, N.; Kurowska, M.; Madera-Salcedo, I.K.; Huang, J.-D.; Fischer, A.; González-Espinosa, C.; de Saint Basile, G.; et al. Kinesin-1 Controls Mast Cell Degranulation and Anaphylaxis through PI3K-Dependent Recruitment to the Granular Slp3/Rab27b Complex. J. Cell Biol. 2016, 215, 203–216. [Google Scholar] [CrossRef]
- Tang, B.L. A Unique SNARE Machinery for Exocytosis of Cytotoxic Granules and Platelets Granules. Mol. Membr. Biol. 2015, 32, 120–126. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and Granzymes: Function, Dysfunction and Human Pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Wagner, C.; Iking-Konert, C.; Denefleh, B.; Stegmaier, S.; Hug, F.; Hänsch, G.M. Granzyme B and Perforin: Constitutive Expression in Human Polymorphonuclear Neutrophils. Blood 2004, 103, 1099–1104. [Google Scholar] [CrossRef]
- Osińska, I.; Popko, K.; Demkow, U. Perforin: An Important Player in Immune Response. Cent. Eur J. Immunol. 2014, 39, 109–115. [Google Scholar] [CrossRef]
- Ma, L.L.; Spurrell, J.C.L.; Wang, J.F.; Neely, G.G.; Epelman, S.; Krensky, A.M.; Mody, C.H. CD8 T Cell-Mediated Killing of Cryptococcus Neoformans Requires Granulysin and Is Dependent on CD4 T Cells and IL-15. J. Immunol. 2002, 169, 5787–5795. [Google Scholar] [CrossRef]
- Ewen, C.L.; Kane, K.P.; Bleackley, R.C. A Quarter Century of Granzymes. Cell Death Differ. 2012, 19, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Gamen, S.; Hanson, D.A.; Kaspar, A.; Naval, J.; Krensky, A.M.; Anel, A. Granulysin-Induced Apoptosis. I. Involvement of at Least Two Distinct Pathways. J. Immunol. 1998, 161, 1758–1764. [Google Scholar] [PubMed]
- Caligiuri, M.A. Human Natural Killer Cells. Blood 2008, 112, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Dotiwala, F.; Santara, S.S.; Binker-Cosen, A.A.; Li, B.; Chandrasekaran, S.; Lieberman, J. Granzyme B Disrupts Central Metabolism and Protein Synthesis in Bacteria to Promote an Immune Cell Death Program. Cell 2017, 171, 1125–1137.e11. [Google Scholar] [CrossRef] [PubMed]
- Mattila, J.T.; Maiello, P.; Sun, T.; Via, L.E.; Flynn, J.L. Granzyme B-Expressing Neutrophils Correlate with Bacterial Load in Granulomas from Mycobacterium Tuberculosis-Infected Cynomolgus Macaques. Cell Microbiol. 2015, 17, 1085–1097. [Google Scholar] [CrossRef]
- Legrand, F.; Driss, V.; Delbeke, M.; Loiseau, S.; Hermann, E.; Dombrowicz, D.; Capron, M. Human Eosinophils Exert TNF-α and Granzyme A-Mediated Tumoricidal Activity toward Colon Carcinoma Cells. J. Immunol. 2010, 185, 7443–7451. [Google Scholar] [CrossRef]
- Strik, M.C.M.; de Koning, P.J.A.; Kleijmeer, M.J.; Bladergroen, B.A.; Wolbink, A.M.; Griffith, J.M.; Wouters, D.; Fukuoka, Y.; Schwartz, L.B.; Hack, C.E.; et al. Human Mast Cells Produce and Release the Cytotoxic Lymphocyte Associated Protease Granzyme B upon Activation. Mol. Immunol. 2007, 44, 3462–3472. [Google Scholar] [CrossRef]
- Tomalka, J.; Azodi, E.; Narra, H.P.; Patel, K.; O’Neill, S.; Cardwell, C.; Hall, B.A.; Wilson, J.M.; Hise, A.G. Beta-Defensin 1 Plays a Role in Acute Mucosal Defence to Candida Albicans. J. Immunol. 2015, 194, 1788–1795. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.R.; Ottolini, B. An Evolutionary History of Defensins: A Role for Copy Number Variation in Maximizing Host Innate and Adaptive Immune Responses. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, E.; Li, C.; Zeng, P.; Li, C.; Diepeveen-de Buin, M.; Lu, W.-Y.; Breukink, E.; Lu, W. Functional Interaction of Human Neutrophil Peptide-1 with the Cell Wall Precursor Lipid II. FEBS Lett. 2010, 584, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, W.; Hong, M. Membrane-Bound Structure and Topology of a Human Alpha Defensin Indicates A Dimer Pore Mechanism for Membrane Disruption. Biochemistry 2010, 49, 9770–9782. [Google Scholar] [CrossRef] [PubMed]
- Kurschus, F.C.; Fellows, E.; Stegmann, E.; Jenne, D.E. Granzyme B Delivery via Perforin Is Restricted by Size, but Not by Heparan Sulfate-Dependent Endocytosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13799–13804. [Google Scholar] [CrossRef]
- Tsai, P.-W.; Cheng, Y.-L.; Hsieh, W.-P.; Lan, C.-Y. Responses of Candida Albicans to the Human Antimicrobial Peptide LL-37. J. Microbiol. 2014, 52, 581–589. [Google Scholar] [CrossRef]
- Levy, O. A Neutrophil-Derived Anti-Infective Molecule: Bactericidal/Permeability-Increasing Protein. Antimicrob. Agents Chemother. 2000, 44, 2925–2931. [Google Scholar] [CrossRef] [PubMed]
- Tkalcevic, J.; Novelli, M.; Phylactides, M.; Iredale, J.P.; Segal, A.W.; Roes, J. Impaired Immunity and Enhanced Resistance to Endotoxin in the Absence of Neutrophil Elastase and Cathepsin G. Immunity 2000, 12, 201–210. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Weng, C.-L.; Jheng, M.-J.; Kan, H.-W.; Hsieh, S.-T.; Liu, F.-T.; Wu-Hsieh, B.A. Candida Albicans Triggers NADPH Oxidase-Independent Neutrophil Extracellular Traps through Dectin-2. PLoS Pathog. 2019, 15, e1008096. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, B.; Horwitz, M.S.; Jenne, D.E.; Gauthier, F. Neutrophil Elastase, Proteinase 3, and Cathepsin G as Therapeutic Targets in Human Diseases. Pharm. Rev. 2010, 62, 726–759. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, O.E.; Follin, P.; Johnsen, A.H.; Calafat, J.; Tjabringa, G.S.; Hiemstra, P.S.; Borregaard, N. Human Cathelicidin, HCAP-18, Is Processed to the Antimicrobial Peptide LL-37 by Extracellular Cleavage with Proteinase 3. Blood 2001, 97, 3951–3959. [Google Scholar] [CrossRef]
- Cassatella, M.A.; Östberg, N.K.; Tamassia, N.; Soehnlein, O. Biological Roles of Neutrophil-Derived Granule Proteins and Cytokines. Trends Immunol. 2019, 40, 648–664. [Google Scholar] [CrossRef]
- Tomee, J.F.C.; Hiemstra, P.S.; Heinzel-Wieland, R.; Kauffman, H.F. Antileukoprotease: An Endogenous Protein in the Innate Mucosal Defence against Fungi. J. Infect. Dis. 1997, 176, 740–747. [Google Scholar] [CrossRef]
- Nauseef, W.M.; Metcalf, J.A.; Root, R.K. Role of Myeloperoxidase in the Respiratory Burst of Human Neutrophils. Blood 1983, 61, 483–492. [Google Scholar] [CrossRef]
- Borelli, V.; Vita, F.; Shankar, S.; Soranzo, M.R.; Banfi, E.; Scialino, G.; Brochetta, C.; Zabucchi, G. Human Eosinophil Peroxidase Induces Surface Alteration, Killing, and Lysis of Mycobacterium Tuberculosis. Infect. Immun. 2003, 71, 605–613. [Google Scholar] [CrossRef]
- Lehrer, R.I. Antifungal Effects of Peroxidase Systems. J. Bacteriol. 1969, 99, 361–365. [Google Scholar] [CrossRef]
- Al-Sheikh, H. Effect of Lactoferrin and Iron on the Growth of Human Pathogenic Candida Species. Pak. J. Biol. Sci. 2009, 12, 91–94. [Google Scholar] [CrossRef]
- Lai, Y.-W.; Campbell, L.T.; Wilkins, M.R.; Pang, C.N.I.; Chen, S.; Carter, D.A. Synergy and Antagonism between Iron Chelators and Antifungal Drugs in Cryptococcus. Int. J. Antimicrob. Agents 2016, 48, 388–394. [Google Scholar] [CrossRef]
- Ballard, E.; Yucel, R.; Melchers, W.J.G.; Brown, A.J.P.; Verweij, P.E.; Warris, A. Antifungal Activity of Antimicrobial Peptides and Proteins against Aspergillus Fumigatus. J. Fungi 2020, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- McCabe, D.; Cukierman, T.; Gabay, J.E. Basic Residues in Azurocidin/HBP Contribute to Both Heparin Binding and Antimicrobial Activity*. J. Biol. Chem. 2002, 277, 27477–27488. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Matsuwaki, Y.; Shin, S.-H.; Ponikau, J.U.; Kita, H. Nonpathogenic, Environmental Fungi Induce Activation and Degranulation of Human Eosinophils. J. Immunol. 2005, 175, 5439–5447. [Google Scholar] [CrossRef] [PubMed]
- Hamann, K.J.; Barker, R.L.; Loegering, D.A.; Gleich, G.J. Comparative Toxicity of Purified Human Eosinophil Granule Proteins for Newborn Larvae of Trichinella Spiralis. J. Parasitol. 1987, 73, 523–529. [Google Scholar] [CrossRef]
- Rosenberg, H.F.; Domachowske, J.B. Eosinophils, Eosinophil Ribonucleases, and Their Role in Host Defence against Respiratory Virus Pathogens. J. Leukoc. Biol. 2001, 70, 691–698. [Google Scholar] [CrossRef]
- Skerrett, S.J. Lysozyme in Pulmonary Host Defence. Am. J. Respir Crit. Care Med. 2004, 169, 435–436. [Google Scholar] [CrossRef]
- Gleich, G.J.; Adolphson, C.R.; Leiferman, K.M. The Biology of the Eosinophilic Leukocyte. Annu. Rev. Med. 1993, 44, 85–101. [Google Scholar] [CrossRef]
- Bystrom, J.; Amin, K.; Bishop-Bailey, D. Analysing the Eosinophil Cationic Protein - a Clue to the Function of the Eosinophil Granulocyte. Respir. Res. 2011, 12, 10. [Google Scholar] [CrossRef]
- Schmidt, S.; Tramsen, L.; Lehrnbecher, T. Natural Killer Cells in Antifungal Immunity. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Mentlik, A.N.; Sanborn, K.B.; Holzbaur, E.L.; Orange, J.S. Rapid Lytic Granule Convergence to the MTOC in Natural Killer Cells Is Dependent on Dynein but Not Cytolytic Commitment. Mol. Biol. Cell 2010, 21, 2241–2256. [Google Scholar] [CrossRef] [PubMed]
- Marr, K.J.; Jones, G.J.; Zheng, C.; Huston, S.M.; Timm-McCann, M.; Islam, A.; Berenger, B.M.; Ma, L.L.; Wiseman, J.C.D.; Mody, C.H. Cryptococcus Neoformans Directly Stimulates Perforin Production and Rearms NK Cells for Enhanced Anticryptococcal Microbicidal Activity. Infect. Immun. 2009, 77, 2436–2446. [Google Scholar] [CrossRef] [PubMed]
- Arneson, L.N.; Brickshawana, A.; Segovis, C.M.; Schoon, R.A.; Dick, C.J.; Leibson, P.J. Cutting Edge: Syntaxin 11 Regulates Lymphocyte-Mediated Secretion and Cytotoxicity. J. Immunol. 2007, 179, 3397–3401. [Google Scholar] [CrossRef]
- Syme, R.M.; Wood, C.J.; Wong, H.; Mody, C.H. Both CD4+ and CD8+ Human Lymphocytes Are Activated and Proliferate in Response to Cryptococcus Neoformans. Immunology 1997, 92, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Breinig, T.; Scheller, N.; Glombitza, B.; Breinig, F.; Meyerhans, A. Human Yeast-Specific CD8 T Lymphocytes Show a Nonclassical Effector Molecule Profile. Med. Microbiol. Immunol. 2012, 201, 127–136. [Google Scholar] [CrossRef]
- Kumaresan, P.R.; da Silva, T.A.; Kontoyiannis, D.P. Methods of Controlling Invasive Fungal Infections Using CD8+ T Cells. Front. Immunol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.; De Luca, A.; Bozza, S.; Cunha, C.; D’Angelo, C.; Moretti, S.; Perruccio, K.; Iannitti, R.G.; Fallarino, F.; Pierini, A.; et al. TLR3 Essentially Promotes Protective Class I-Restricted Memory CD8+ T-Cell Responses to Aspergillus Fumigatus in Hematopoietic Transplanted Patients. Blood 2012, 119, 967–977. [Google Scholar] [CrossRef]
- Firinu, D.; Pisanu, M.; Piras, B.; Meleddu, R.; Lorrai, M.M.; Manconi, P.E.; Giacco, S.D. Genetic Susceptibility to Candida Infection: A New Look at an Old Entity. Chin. Med. J. 2013, 126, 378–381. [Google Scholar] [CrossRef]
- Nahum, A.; Dadi, H.; Bates, A.; Roifman, C.M. The Biological Significance of TLR3 Variant, L412F, in Conferring Susceptibility to Cutaneous Candidiasis, CMV and Autoimmunity. Autoimmun. Rev. 2012, 11, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Bochud, P.-Y.; Chien, J.W.; Marr, K.A.; Leisenring, W.M.; Upton, A.; Janer, M.; Rodrigues, S.D.; Li, S.; Hansen, J.A.; Zhao, L.P.; et al. Toll-like Receptor 4 Polymorphisms and Aspergillosis in Stem-Cell Transplantation. N. Engl. J. Med. 2008, 359, 1766–1777. [Google Scholar] [CrossRef] [PubMed]
- Ritter, A.T.; Angus, K.L.; Griffiths, G.M. The Role of the Cytoskeleton at the Immunological Synapse. Immunol. Rev. 2013, 256, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, J.K.; McIlvain, J.M.; Sheetz, M.P.; Argon, Y. Lytic Granules from Cytotoxic T Cells Exhibit Kinesin-Dependent Motility on Microtubules in Vitro. J. Cell Sci. 1993, 104, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Hooikaas, P.J.; Damstra, H.G.; Gros, O.J.; van Riel, W.E.; Martin, M.; Smits, Y.T.; van Loosdregt, J.; Kapitein, L.C.; Berger, F.; Akhmanova, A. Kinesin-4 KIF21B Limits Microtubule Growth to Allow Rapid Centrosome Polarization in T Cells. Elife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- De Zuani, M.; Paolicelli, G.; Zelante, T.; Renga, G.; Romani, L.; Arzese, A.; Pucillo, C.E.M.; Frossi, B. Mast Cells Respond to Candida Albicans Infections and Modulate Macrophages Phagocytosis of the Fungus. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Horiguchi, K.; Yoshikawa, S.; Saito, A.; Haddad, S.; Ohta, T.; Miyake, K.; Yamanishi, Y.; Karasuyama, H. Real-Time Imaging of Mast Cell Degranulation in Vitro and in Vivo. Biochem. Biophys. Res. Commun. 2016, 479, 517–522. [Google Scholar] [CrossRef]
- Joulia, R.; Gaudenzio, N.; Rodrigues, M.; Lopez, J.; Blanchard, N.; Valitutti, S.; Espinosa, E. Mast Cells Form Antibody-Dependent Degranulatory Synapse for Dedicated Secretion and Defence. Nat. Commun. 2015, 6, 6174. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Yamasaki, S.; Ito, Y.; Kabu, K.; Hattori, K.; Tezuka, T.; Nishizumi, H.; Kitamura, D.; Goitsuka, R.; Geha, R.S.; et al. FcεRI-Mediated Mast Cell Degranulation Requires Calcium-Independent Microtubule-Dependent Translocation of Granules to the Plasma Membrane. J. Cell Biol. 2005, 170, 115–126. [Google Scholar] [CrossRef]
- Efergan, A.; Azouz, N.P.; Klein, O.; Noguchi, K.; Rothenberg, M.E.; Fukuda, M.; Sagi-Eisenberg, R. Rab12 Regulates Retrograde Transport of Mast Cell Secretory Granules by Interacting with the RILP-Dynein Complex. J. Immunol. 2016, 196, 1091–1101. [Google Scholar] [CrossRef]
- Woska, J.R.; Gillespie, M.E. SNARE Complex-Mediated Degranulation in Mast Cells. J. Cell Mol. Med. 2012, 16, 649–656. [Google Scholar] [CrossRef]
- Figueiredo, R.T.; Neves, J.S. Eosinophils in Fungal Diseases: An Overview. J. Leukoc. Biol. 2018, 104, 49–60. [Google Scholar] [CrossRef]
- Carmo, L.A.S.; Dias, F.F.; Malta, K.K.; Amaral, K.B.; Shamri, R.; Weller, P.F.; Melo, R.C.N. Expression and Subcellular Localization of the Qa-SNARE Syntaxin17 in Human Eosinophils. Exp. Cell Res. 2015, 337, 129–135. [Google Scholar] [CrossRef][Green Version]
- Melo, R.C.N.; Weller, P.F. Piecemeal Degranulation in Human Eosinophils: A Distinct Secretion Mechanism Underlying Inflammatory Responses. Histol. Histopathol. 2010, 25, 1341–1354. [Google Scholar] [CrossRef]
- Rocha, J.D.B.; Nascimento, M.T.C.; Decote-Ricardo, D.; Côrte-Real, S.; Morrot, A.; Heise, N.; Nunes, M.P.; Previato, J.O.; Mendonça-Previato, L.; DosReis, G.A.; et al. Capsular Polysaccharides from Cryptococcus Neoformans Modulate Production of Neutrophil Extracellular Traps (NETs) by Human Neutrophils. Sci. Rep. 2015, 5, 8008. [Google Scholar] [CrossRef] [PubMed]
- Springer, D.J.; Ren, P.; Raina, R.; Dong, Y.; Behr, M.J.; McEwen, B.F.; Bowser, S.S.; Samsonoff, W.A.; Chaturvedi, S.; Chaturvedi, V. Extracellular Fibrils of Pathogenic Yeast Cryptococcus Gattii Are Important for Ecological Niche, Murine Virulence and Human Neutrophil Interactions. PLoS ONE 2010, 5, e10978. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.F.; Reichard, U.; Brinkmann, V.; Zychlinsky, A. Neutrophil Extracellular Traps Capture and Kill Candida Albicans Yeast and Hyphal Forms. Cell Microbiol. 2006, 8, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.A.; Grinstein, S. Phagocytosis: Receptors, Signal Integration, and the Cytoskeleton. Immunol. Rev. 2014, 262, 193–215. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Heit, B. Armed for Destruction: Formation, Function and Trafficking of Neutrophil Granules. Cell Tissue Res. 2018, 371, 455–471. [Google Scholar] [CrossRef]
- Mollinedo, F.; Calafat, J.; Janssen, H.; Martín-Martín, B.; Canchado, J.; Nabokina, S.M.; Gajate, C. Combinatorial SNARE Complexes Modulate the Secretion of Cytoplasmic Granules in Human Neutrophils. J. Immunol. 2006, 177, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Lacy, P.; Eitzen, G. Control of Granule Exocytosis in Neutrophils. Front. Biosci. 2008, 13, 5559–5570. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, S.W.; Deal, C.C.; Pinto, J.; Wright, D.G. Affinity Purification and Subcellular Localization of Kinesin in Human Neutrophils. J. Leukoc. Biol. 1993, 53, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, S.W.; Nath, J.; Wright, D.G. Interactions of Cytoplasmic Granules with Microtubules in Human Neutrophils. J. Cell Biol. 1989, 108, 2313–2326. [Google Scholar] [CrossRef] [PubMed]
- Metzler, K.D.; Goosmann, C.; Lubojemska, A.; Zychlinsky, A.; Papayannopoulos, V. A Myeloperoxidase-Containing Complex Regulates Neutrophil Elastase Release and Actin Dynamics during NETosis. Cell Rep. 2014, 8, 883–896. [Google Scholar] [CrossRef] [PubMed]
| NK Cells | CD8+ T Cells | Mast Cells | Eosinophil | Neutrophil | |
|---|---|---|---|---|---|
| Types | Type 1 Granule (fully Formed) 50–700 nm Contains a dense core surrounded by thin layer of vesicles Type 2 Granule 200–1000 nm Contains multiple vesicles and membrane whorls Intermediate Granule Contains dense cores and multiple vesicles, less abundant than type 2 granules | CytotoxicGranule 100–1300 nm Exists in tiny droplets, dark-core bodies surrounded by a thin membrane, or large granules containing small internal vesicles | Type 1 Granule MHC class II, β-hexosaminidase, lysosomal membrane protein (LAMP)-1/2, Mannose- 6-phosphatereceptors (M6PR) Type 2 Granule MHC class II, β- hexosaminidase, LAMP-1/2, M6PR, Serotonin Type 3 Granule β-hexosaminidase, serotonin | Primary Granule: 500–1000 nm Lack crystalline core Secondary (Specific) Granule: 500–1000 nm Contain distinctive dense crystalline core that is surrounded by a less dense matrix and enclosed by a trilaminar membrane | Primary Azurophilic Granule electron dense 500–1000 nm Secondary SpecificGranule 200–500 nm Tertiary (gelatinase)Granule Mean size of 187 nm |
| Content | In all granule types: Perforin Granzymes Defensins 1–3 LL-37 Granulysin FasL and TRAIL | In all granule types: Perforin Granzymes Defensins 1–3 LL-37 Granulysin FasL and TRAIL May be separated by granule density | No distinct difference in content between granule types but are: chymase, tryptase, mast cell carboxypeptidase A3 (CPA3), β-hexosaminidase, histamine, granzyme | Primary Granule: Charcot–Leyden crystal protein (galactin-10) Secondary Granule: eosinophil peroxidase (EPO) major basic protein (MBP) eosinophil cationic protein (ECP) eosinophil-derived neurotoxin (EDN) | Primary Granule: neutrophil elastase, myeloperoxidase (MPO), defensins, cathepsin G, proteinase 3 Secondary Granule: lactoferrins, defensins, BPI, MPO, lysozyme, LL-37 Tertiary Granule: matrix metalloproteinases, azurocidin, lysozyme |
| NK Cells | CD8+ T Cells | Mast Cells | Eosinophil | Neutrophil | |
|---|---|---|---|---|---|
| Pathway | ERK2 ➔ JNK1 ➔ MTOC, granule polarization and cytotoxicity ITAM dependent and independent signaling ➔ MAPK cascade ➔ NK cell effector functions | TCR ➔ LCK/ZAP70 ➔ LAT/PLCγ/ITK ➔ PIP2 ➔ IP3 ➔ Ca2+ influx ➔ degranulation | Surface receptors (CCR1, TLR4, KIT, or FcεRI). G-protein, MyD88, Jak/STAT, ➔ Lck-phos ➔ LAT-phos ➔ PLCγ ➔ degranulation | CCR3 ➔ G-protein/Lyn, Fgr, Hck ➔ PI3K ➔ Akt ➔ BAD ➔ MAPK ➔ Ras ➔ RAF ➔ MEK1 ➔ ERK ➔ BAD | Microtubule assembly: selectins/integrins ➔ Pyk2 ➔ Vav ➔ paxillin granule mobilization: surface receptors (GPCR, Fc-R, PPRs) ➔ PI3K/PLC/SLP-76/Vav complex ➔ Rac and PIP3 |
| Mode | Cytotoxic degranulation through direct contact of target cells | Cytotoxic degranulation through direct contact of target cells | Anaphylactic/cytotoxic degranulation Phagosomal granule fusion and degranulation | Piecemeal degranulation Intact granule exocytosis and EETosis Phagosomal granule fusion and degranulation | Cytotoxic degranulation Phagosomal granule fusion NET formation and degranulation onto NETs |
| NK Cells | CD8+ T Cells | Mast Cells | Eosinophil | Neutrophil | |
|---|---|---|---|---|---|
| Microtubules | Microtubules facilitate the delivery of lytic granules to the synaptic cleft between NK cells and target cells through microtubule associated motor proteins | Microtubules facilitate secretory granule dynamics and degranulation by microtubule protrusion formation and reorganization | Microtubules serve as scaffold for Kinesin and Dynein | Microtubule Reorganization facilitates granule release | Granules are recruited and mobilized by microtubules |
| Dynein/Kinesin | Dynein mediates minus directed movement of granules to converge to MTOC Kinesin-1 mediates terminal granule movement and degranulation to IS Eg5 Kinesin is involved in NK cell granule trafficking during antifungal activity | Terminal transport of lytic granules is mediated by the kinesin-1/Slp3/Rab27a complex Kinesin-4 KIF21B limits microtubule growth to allow rapid centrosome polarization in T cells. Lytic granules have kinesin-dependent motility on microtubules in vitro | Kinesin-1 controls mast cell degranulation through PI3K-dependent recruitment to the granular Slp3/Rab27b complex Dynein is involved in retrograde transport of secretory vesicles | Role of motor proteins are unknown | Kinesins are involved in granule-microtubule interactions and movement |
| SNAPs/SNAREs | atypical Q-SNARE syntaxin 11/Sec/Munc (SM) family Mediate granule exocytosis by providing the tight complex that brings the granule to the cell membrane and enabling for granule fusion | atypical Q-SNARE syntaxin 11/Sec/Munc (SM) family Mediate granule exocytosis by providing the tight complex that brings the granule to the cell membrane and enabling for granule fusion | SNARE proteins function to mediate constitutive trafficking events through both endocytic and secretory pathways | Specific membrane docking of granules through interaction with plasma membrane t-SNARES, SNAP-23, and syntaxin-4 Qa SNARE (Syntaxin17) is involved in granule transport | Two SNARE complexes, made up of syntaxin 4/SNAP-23/VAMP-1 and syntaxin 4/SNAP-23/VAMP-2, are involved in the exocytosis of specific and tertiary granules Interactions between syntaxin 4 and VAMP-1/VAMP-7 are involved in the exocytosis of azurophilic granules. |
| Classification of Granule Proteins | Proteins |
|---|---|
| Cytolytic, Cell wall and Membrane disrupting/pore forming | Perforin, granulysin, defensins, LL-37, eosinophil cationic protein, major basic protein, bactericidal/permeability-increasing protein, azurocidin |
| Peptidoglycanases | lysozyme |
| Protease Inhibitors | Secretory leukocyte protease inhibitor (SLPI) |
| Immune modifying/cytokines | IFN-γ, TNF-α, GM-CSF, VEGF IL-1a, IL-10, IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, IL-12, IL-13, IL-16, IL-17A, IL-17F, IL-21, IL-22, IL-25, IL-27 |
| Oxidative agents | MPO, EPO |
| Pro-apoptotic agents (serine proteases including tryptases and chymases) | Mast cell tryptase and chymases, CPA3, granzymes A, B, H, K, M, |
| Chymotrypsin-like serine proteases | Neutrophil elastase, cathepsin G |
| Iron binding proteins | Lactoferrin |
| Extracellular matrix degrading Matrix Metalloproteinases/Gelatinases | MMP-8, MMP-9 |
| Ribonucleases including Cationic proteins | ECP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mok, A.C.; Mody, C.H.; Li, S.S. Immune Cell Degranulation in Fungal Host Defence. J. Fungi 2021, 7, 484. https://doi.org/10.3390/jof7060484
Mok AC, Mody CH, Li SS. Immune Cell Degranulation in Fungal Host Defence. Journal of Fungi. 2021; 7(6):484. https://doi.org/10.3390/jof7060484
Chicago/Turabian StyleMok, Adley CH., Christopher H. Mody, and Shu Shun Li. 2021. "Immune Cell Degranulation in Fungal Host Defence" Journal of Fungi 7, no. 6: 484. https://doi.org/10.3390/jof7060484
APA StyleMok, A. C., Mody, C. H., & Li, S. S. (2021). Immune Cell Degranulation in Fungal Host Defence. Journal of Fungi, 7(6), 484. https://doi.org/10.3390/jof7060484






