Advantages and Challenges of Differential Immune Cell Count Determination in Blood and Milk for Monitoring the Health and Well-Being of Dairy Cows
Abstract
1. Introduction
2. Differential Cell Count as a Tool to Monitor Mammary Gland Immune Responses
2.1. Literature Research and Current Applications
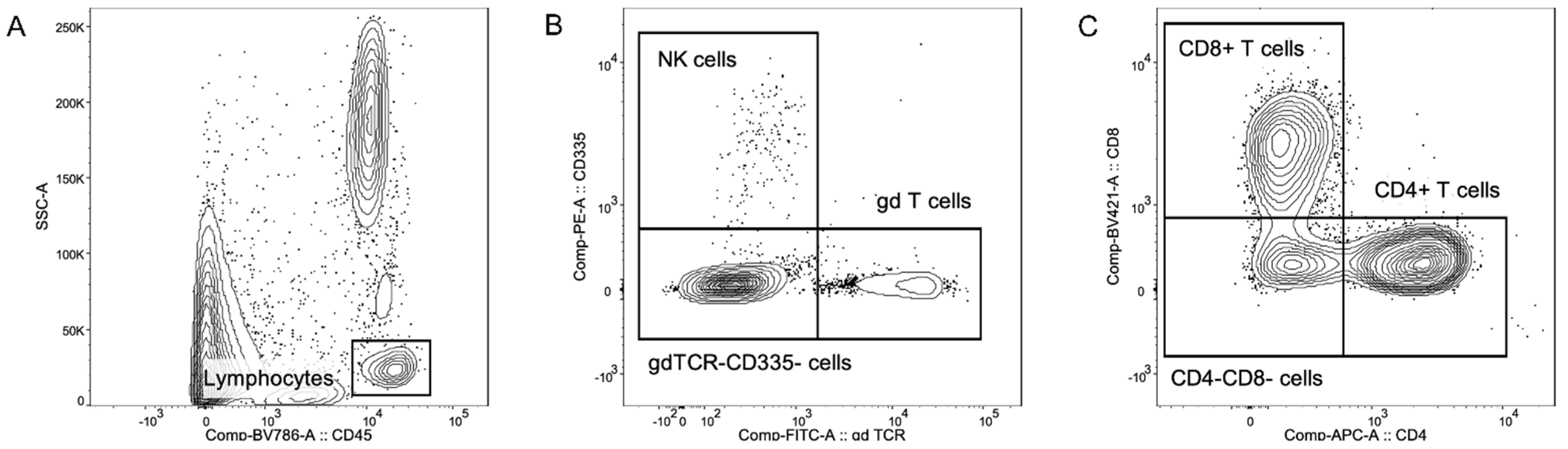
2.2. Multicolor Flow Cytometric Immunophenotyping
3. Extended Differential Cell Counts in Milk as a Tool to Monitor the General Health Status of Dairy Cattle
3.1. Background and Basic Assumptions
3.2. Similarities between Differential Cell Counts in Milk and Blood in Healthy Cattle
4. Differences between the Immune Systems of Humans and Dairy Cattle
4.1. Background and Basic Assumptions
4.2. Humoral Responses, Architecture, and Surface Marker Expression Profiles
5. The Special Role of γδ T Cells in Bovine Immune Responses
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koess, C.; Hamann, J. Detection of mastitis in the bovine mammary gland by flow cytometry at early stages. J. Dairy Res. 2008, 75, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Diesterbeck, U.S.; Konig, S.; Brugemann, K.; Schlez, K.; Zschock, M.; Wolter, W.; Czerny, C.P. Microscopic differential cell counts in milk for the evaluation of inflammatory reactions in clinically healthy and subclinically infected bovine mammary glands. J. Dairy Res. 2011, 78, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Boutinaud, M.; Jammes, H. Potential uses of milk epithelial cells: A review. Reprod. Nutr. Dev. 2002, 42, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Alhussien, M.; Kaur, M.; Manjari, P.; Kimothi, S.P.; Mohanty, A.K.; Dang, A.K. A comparative study on the blood and milk cell counts of healthy, subclinical, and clinical mastitis Karan Fries cows. Vet. World 2015, 8, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Rivas, A.L.; Quimby, F.A.; Blue, J.; Coksaygan, O. Longitudinal evaluation of bovine mammary gland health status by somatic cell counting, flow cytometry, and cytology. J. Vet. Diagn. Investig. 2001, 13, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Diesterbeck, U.S.; Konig, S.; Brugemann, K.; Schlez, K.; Zschock, M.; Wolter, W.; Czerny, C.P. Flow cytometric differential cell counts in milk for the evaluation of inflammatory reactions in clinically healthy and subclinically infected bovine mammary glands. J. Dairy Sci. 2011, 94, 5033–5044. [Google Scholar] [CrossRef]
- Adkins, P.R.F.; Middleton, J.R. Methods for Diagnosing Mastitis. Vet. Clin. N. Am. Food. Anim. Pract. 2018, 34, 479–491. [Google Scholar] [CrossRef]
- ICAR International Committee for Animal Recording. Yearly Survey on Milk Recording Systems (Years 2016, 2017 and 2018) for Cow, Sheep and Goats. Available online: https://www.icar.org/wp-content/uploads/2019/07/Survey-on-milk-recording-systems-in-cows-sheep-and-goats-2016-2017-and-2018.pdf (accessed on 23 March 2021).
- Bundesverband Rind und Schwein e.V. Aktuelle Ergebnisse der Milchkontrolle im Jahr 2020. Available online: https://www.rind-schwein.de/brs-news/aktuelle-ergebnisse-der-milchkontrolle-im-jahr-202.html (accessed on 23 March 2021).
- ICAR International Committee for Animal Recording. ICAR Guidelines. Section 07—Bovine Functional Traits. Available online: https://www.icar.org/Guidelines/07-Bovine-Functional-Traits.pdf (accessed on 23 March 2021).
- Pilla, R.; Malvisi, M.; Snel, G.G.; Schwarz, D.; Konig, S.; Czerny, C.P.; Piccinini, R. Differential cell count as an alternative method to diagnose dairy cow mastitis. J. Dairy Sci. 2013, 96, 1653–1660. [Google Scholar] [CrossRef]
- Schwarz, D.; Rivas, A.L.; Konig, S.; Diesterbeck, U.S.; Schlez, K.; Zschock, M.; Wolter, W.; Czerny, C.P. CD2/CD21 index: A new marker to evaluate udder health in dairy cows. J. Dairy Sci. 2013, 96, 5106–5119. [Google Scholar] [CrossRef]
- Soltys, J.; Quinn, M.T. Selective recruitment of T-cell subsets to the udder during staphylococcal and streptococcal mastitis: Analysis of lymphocyte subsets and adhesion molecule expression. Infect. Immun. 1999, 67, 6293–6302. [Google Scholar] [CrossRef]
- Fuenzalida, M.J.; Fricke, P.M.; Ruegg, P.L. The association between occurrence and severity of subclinical and clinical mastitis on pregnancies per artificial insemination at first service of Holstein cows. J. Dairy Sci. 2015, 98, 3791–3805. [Google Scholar] [CrossRef] [PubMed]
- Hudson, C.D.; Bradley, A.J.; Breen, J.E.; Green, M.J. Associations between udder health and reproductive performance in United Kingdom dairy cows. J. Dairy Sci. 2012, 95, 3683–3697. [Google Scholar] [CrossRef] [PubMed]
- Degen, S.; Knorr, N.; Paduch, J.H.; Klocke, D.; Zoche-Golob, V.; Hoedemaker, M.; Krömker, V. Cell differentiation assisting in evaluating mastitis treatment prognosis. Milchwissenschaft 2015, 68, 2–9. [Google Scholar]
- Barlow, J.W.; White, L.J.; Zadoks, R.N.; Schukken, Y.H. A mathematical model demonstrating indirect and overall effects of lactation therapy targeting subclinical mastitis in dairy herds. Prev. Vet. Med. 2009, 90, 31–42. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Critically important antimicrobials for human medicine, 6th revision. In Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019; pp. 26–40. [Google Scholar]
- Park, Y.H.; Joo, Y.S.; Park, J.Y.; Moon, J.S.; Kim, S.H.; Kwon, N.H.; Ahn, J.S.; Davis, W.C.; Davies, C.J. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 2004, 5, 29–39. [Google Scholar] [CrossRef]
- Mehrzad, J.; Duchateau, L.; Burvenich, C. Viability of Milk Neutrophils and Severity of Bovine Coliform Mastitis. J. Dairy Sci. 2004, 86, 4150–4162. [Google Scholar] [CrossRef]
- Bresolin, S.; Calderwood, D.A.; Heineck, T.M.; Newcomb, D.; Paul, C.; Pollard, J.N.; Rodriguez, R.R.; Young, D. Diagnostic Apparatus. U.S. Patent 9816982B2, 27 June 2013. [Google Scholar]
- Godden, S.M.; Royster, E.; Timmerman, J.; Rapnicki, P.; Green, H. Evaluation of an automated milk leukocyte differential test and the California Mastitis Test for detecting intramammary infection in early- and late-lactation quarters and cows. J. Dairy Sci. 2017, 100, 6527–6544. [Google Scholar] [CrossRef]
- Lozada-Soto, E.; Maltecca, C.; Anderson, K.; Tiezzi, F. Analysis of milk leukocyte differential measures for use in management practices for decreased mastitis incidence. J. Dairy Sci. 2020, 103, 572–582. [Google Scholar] [CrossRef]
- Holm, C. Method for Determining a Degree of Infection. EP Patent 2630487, 18 October 2012. [Google Scholar]
- Damm, M.; Holm, C.; Blaabjerg, M.; Bro, M.N.; Schwarz, D. Differential somatic cell count-A novel method for routine mastitis screening in the frame of Dairy Herd Improvement testing programs. J. Dairy Sci. 2017, 100, 4926–4940. [Google Scholar] [CrossRef]
- Schwarz, D.; Lipkens, Z.; Piepers, S.; De Vliegher, S. Investigation of differential somatic cell count as a potential new supplementary indicator to somatic cell count for identification of intramammary infection in dairy cows at the end of the lactation period. Prev. Vet. Med. 2019, 172, 104803. [Google Scholar] [CrossRef]
- Kirkeby, C.; Toft, N.; Schwarz, D.; Farre, M.; Nielsen, S.S.; Zervens, L.; Hechinger, S.; Halasa, T. Differential somatic cell count as an additional indicator for intramammary infections in dairy cows. J. Dairy Sci. 2020, 103, 1759–1775. [Google Scholar] [CrossRef] [PubMed]
- De Matteis, G.; Grandoni, F.; Scata, M.C.; Catillo, G.; Moioli, B.; Buttazzoni, L. Flow Cytometry-Detected Immunological Markers and on Farm Recorded Parameters in Composite Cow Milk as Related to Udder Health Status. Vet. Sci. 2020, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Clark, G.; Stockinger, H.; Balderas, R.; van Zelm, M.C.; Zola, H.; Hart, D.; Engel, P. Nomenclature of CD molecules from the Tenth Human Leucocyte Differentiation Antigen Workshop. Clin. Transl. Immunol. 2016, 5, e57. [Google Scholar] [CrossRef] [PubMed]
- Saalmuller, A.; Aasted, B. Summary of the animal homologue section of HLDA8. Vet. Immunol. Immunopathol. 2007, 119, 2–13. [Google Scholar] [CrossRef]
- Guzman, E.; Hope, J.; Taylor, G.; Smith, A.L.; Cubillos-Zapata, C.; Charleston, B. Bovine gammadelta T cells are a major regulatory T cell subset. J. Immunol. 2014, 193, 208–222. [Google Scholar] [CrossRef]
- Porwit, A.; Rajab, A. Flow cytometry immunophenotyping in integrated diagnostics of patients with newly diagnosed cytopenia: One tube 10-color 14-antibody screening panel and 3-tube extensive panel for detection of MDS-related features. Int. J. Lab. Hematol. 2015, 37 (Suppl. 1), 133–143. [Google Scholar] [CrossRef]
- Farschtschi, S.; Mattes, M.; Hildebrandt, A.; Chiang, D.; Kirchner, B.; Kliem, H.; Pfaffl, M.W. Development of an advanced flow cytometry based high-resolution immunophenotyping method to benchmark early immune response in dairy cows. Sci. Rep. 2021, 11, 22896. [Google Scholar] [CrossRef]
- Sopp, P.; Werling, D.; Baldwin, C. Cross-reactivity of mAbs to human CD antigens with cells from cattle. Vet. Immunol. Immunopathol. 2007, 119, 106–114. [Google Scholar] [CrossRef]
- Dosogne, H.; Vangroenweghe, F.; Mehrzad, J.; Massart-Lee, A.M.; Burvenich, C. Differential Leukocyte Count Method for Bovine Low Somatic Cell Count Milk. J. Dairy Sci. 2003, 86, 828–834. [Google Scholar] [CrossRef]
- Pillai, S.R.; Kunze, E.; Sordillo, L.M.; Jayarao, B.M. Application of differential inflammatory cell count as a tool to monitor udder health. J. Dairy Sci. 2001, 84, 1413–1420. [Google Scholar] [CrossRef]
- Faucher, J.L.; Lacronique-Gazaille, C.; Frebet, E.; Trimoreau, F.; Donnard, M.; Bordessoule, D.; Lacombe, F.; Feuillard, J. “6 markers/5 colors” extended white blood cell differential by flow cytometry. Cytom. A 2007, 71, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Sarikaya, H.; Prgomet, C.; Pfaffl, M.W.; Bruckmaier, R.M. Differentiation of leukocytes in bovine milk. Milchwissenschaft 2004, 59, 586–589. [Google Scholar]
- Karim, M.R.; Parvin, M.S.; Hossain, M.Z.; Islam, M.T.; Hussan, M.T. A Report on Clinical Prevalence of Diseases and Disorders in Cattle and Goats at The Upazilla Veterinary Hospital, Mohammadpur, Magura. Bangl. J. Vet. Med. 2014, 12, 47–53. [Google Scholar] [CrossRef]
- Halasa, T.; Huijps, K.; Osteras, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Thompson-Crispi, K.; Atalla, H.; Miglior, F.; Mallard, B.A. Bovine mastitis: Frontiers in immunogenetics. Front. Immunol. 2014, 5, 493. [Google Scholar] [CrossRef]
- Van Soest, F.J.; Santman-Berends, I.M.; Lam, T.J.; Hogeveen, H. Failure and preventive costs of mastitis on Dutch dairy farms. J. Dairy Sci. 2016, 99, 8365–8374. [Google Scholar] [CrossRef]
- Bennett, R.M.; Christiansen, K.; Clifton-Hadley, R.S. Estimating the costs associated with endemic diseases of dairy cattle. J. Dairy Sci. 1999, 66, 455–459. [Google Scholar] [CrossRef]
- Sariozkan, S.; Yalcin, C. Estimating the total cost of bovine fasciolosis in Turkey. Ann. Trop. Med. Parasitol. 2011, 105, 439–444. [Google Scholar] [CrossRef]
- Dolecheck, K.; Bewley, J. Animal board invited review: Dairy cow lameness expenditures, losses and total cost. Animal 2018, 12, 1462–1474. [Google Scholar] [CrossRef]
- Bruijnis, M.R.; Beerda, B.; Hogeveen, H.; Stassen, E.N. Assessing the welfare impact of foot disorders in dairy cattle by a modeling approach. Animal 2012, 6, 962–970. [Google Scholar] [CrossRef]
- Leslie, K.E.; Petersson-Wolfe, C.S. Assessment and management of pain in dairy cows with clinical mastitis. Vet. Clin. N. Am. Food. Anim. Pract. 2012, 28, 289–305. [Google Scholar] [CrossRef]
- Weiss, D.J.; Wardrop, K.J. Schalm’s Veterinary Hematology, 6th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Saito, A. On the essential nature of hematopoietic function of bone marrow. 5. On the leucocyte reaction in infectious diseases with special reference to Schilling’s biological leucocyte curve. Tohoku J. Exp. Med. 1961, 74, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Ravin, K.A.; Loy, M. The Eosinophil in Infection. Clin. Rev. Allergy Immunol. 2016, 50, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Paape, M.J.; Bannerman, D.D.; Zhao, X.; Lee, J.W. The bovine neutrophil: Structure and function in blood and milk. Vet. Res. 2003, 34, 597–627. [Google Scholar] [CrossRef]
- Sladek, Z.; Ryznarova, H.; Rysanek, D. Macrophages of the bovine heifer mammary gland: Morphological features during initiation and resolution of the inflammatory response. Anat. Histol. Embryol. 2006, 35, 116–124. [Google Scholar] [CrossRef]
- Li, N.; Richoux, R.; Perruchot, M.H.; Boutinaud, M.; Mayol, J.F.; Gagnaire, V. Flow Cytometry Approach to Quantify the Viability of Milk Somatic Cell Counts after Various Physico-Chemical Treatments. PLoS ONE 2015, 10, e0146071. [Google Scholar] [CrossRef]
- Baumert, A.; Bruckmaier, R.M.; Wellnitz, O. Cell population, viability, and some key immunomodulatory molecules in different milk somatic cell samples in dairy cows. J. Dairy Res. 2009, 76, 356–364. [Google Scholar] [CrossRef]
- von Engelhardt, W.; Breves, G.; Diener, M.; Gäbel, G. Physiologie der Haustiere, 5th ed.; Enke: Stuttgart, Germany, 2015. [Google Scholar]
- Macedo, A.A.; Marciano, A.P.; Rocha, L.M.; Alves-Junior, J.R.; Faria, A.M.; Bittar, J.F.; Araujo, M.S.; Santos, R.L.; Martins-Filho, O.A. Comparative phenotypic profile of subpopulations of peripheral blood leukocytes in European (Bos taurus taurus) and Zebu cattle (Bos taurus indicus). Genet. Mol. Res. 2013, 12, 6838–6849. [Google Scholar] [CrossRef]
- Pelan-Mattocks, L.S.; Pesch, B.A.; Kehrli, M.E. Flow cytometric analysis of intracellular complexity and CD45 expression for use in rapid differentiation of leukocytes in bovine blood samples. Am. J. Vet. Res. 2001, 62, 1740–1744. [Google Scholar] [CrossRef]
- Souza, F.N.; Blagitz, M.G.; Batista, C.F.; Takano, P.V.; Gargano, R.G.; Diniz, S.A.; Silva, M.X.; Ferronatto, J.A.; Santos, K.R.; Heinemann, M.B.; et al. Immune response in nonspecific mastitis: What can it tell us? J. Dairy Sci. 2020, 103, 5376–5386. [Google Scholar] [CrossRef]
- Ezzat Alnakip, M.; Quintela-Baluja, M.; Bohme, K.; Fernandez-No, I.; Caamano-Antelo, S.; Calo-Mata, P.; Barros-Velazquez, J. The Immunology of Mammary Gland of Dairy Ruminants between Healthy and Inflammatory Conditions. J. Vet. Med. 2014, 2014, 659801. [Google Scholar] [CrossRef]
- Harp, J.A.; Waters, T.E.; Goff, J.P. Lymphocyte subsets and adhesion molecule expression in milk and blood of periparturient dairy cattle. Vet. Immunol. Immunopathol. 2004, 102, 9–17. [Google Scholar] [CrossRef]
- Riollet, C.; Rainard, P.; Poutrel, B. Cell subpopulations and cytokine expression in cow milk in response to chronic Staphylococcus aureus infection. J. Dairy Sci. 2001, 84, 1077–1084. [Google Scholar] [CrossRef]
- Mackay, C.R.; Hein, W.R. A large proportion of bovine T cells express the gamma delta T cell receptor and show a distinct tissue distribution and surface phenotype. Int. Immunol. 1989, 1, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Davis, W.C.; Brown, W.C.; Hamilton, M.J.; Wyatt, C.R.; Orden, J.A.; Khalid, A.M.; Naessens, J. Analysis of monoclonal antibodies specific for the gamma delta TcR. Vet. Immunol. Immunopathol. 1996, 52, 275–283. [Google Scholar] [CrossRef]
- Faldyna, M.; Leva, L.; Sladek, Z.; Rysanek, D.; Toman, M. γδ-TCR+ CD2– lymphocytes are recruited into bovine mammary gland after stimulation. Vet. Med. 2006, 51, 258–264. [Google Scholar] [CrossRef]
- Boysen, P.; Storset, A.K. Bovine natural killer cells. Vet. Immunol. Immunopathol. 2009, 130, 163–177. [Google Scholar] [CrossRef]
- Scata, M.C.; Grandoni, F.; de Matteis, G.; Roncoroni, C.; Barile, V.L. Flow cytometric differential cell count (DCC) in buffalo milk. In Proceedings of the 8th Asian Buffalo Congress, Istanbul, Turkey, 21–25 April 2015. [Google Scholar]
- Sarikaya, H.; Schlamberger, G.; Meyer, H.H.; Bruckmaier, R.M. Leukocyte populations and mRNA expression of inflammatory factors in quarter milk fractions at different somatic cell score levels in dairy cows. J. Dairy Sci. 2006, 89, 2479–2486. [Google Scholar] [CrossRef]
- Roland, L.; Drillich, M.; Iwersen, M. Hematology as a diagnostic tool in bovine medicine. J. Vet. Diagn. Investig. 2014, 26, 592–598. [Google Scholar] [CrossRef]
- Kakinuma, S.; Maeda, Y.; Ohtsuka, H.; Ohmae, K.; Ayabe, K.; Konnai, S.; Oikawa, M. The Leukocyte Population in the Peripheral Blood and the Colostrum of Cows Infected with Bovine Leukemia Virus is Skewed towards Humoral Immunity. Intern. J. Appl. Res. Vet. Med. 2012, 10, 323–327. [Google Scholar]
- Badi, F.A.; Haroon, A.I.; Alluwaimi, A.M. The gammadelta cells as marker of non-seroconverted cattle naturally infected with Mycobacterium avium subspecies paratuberculosis. Res. Vet. Sci. 2010, 88, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Seong, S.Y.; Matzinger, P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004, 4, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, D.L.; Spencer, J. Mouse and human intestinal immunity: Same ballpark, different players; different rules, same score. Mucosal Immunol. 2011, 4, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Mestas, J.; Hughes, C.C.W. Of Mice and Not Men: Differences between Mouse and Human Immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef]
- Zschaler, J.; Schlorke, D.; Arnhold, J. Differences in innate immune response between man and mouse. Crit. Rev. Immunol. 2014, 34, 433–454. [Google Scholar] [CrossRef]
- Bailey, M.; Christoforidou, Z.; Lewis, M.C. The evolutionary basis for differences between the immune systems of man, mouse, pig and ruminants. Vet. Immunol. Immunopathol. 2013, 152, 13–19. [Google Scholar] [CrossRef]
- Entrican, G.; Wattegedera, S.R.; Griffiths, D.J. Exploiting ovine immunology to improve the relevance of biomedical models. Mol. Immunol. 2015, 66, 68–77. [Google Scholar] [CrossRef]
- Murphy, W.J.; Pevzner, P.A.; O’Brien, S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004, 20, 631–639. [Google Scholar] [CrossRef]
- Elsik, C.G.; Tellam, R.L.; Worley, K.C.; Gibbs, R.A.; Muzny, D.M.; Weinstock, G.M.; Adelson, D.L.; Eichler, E.E.; Elnitski, L.; Guigo, R.; et al. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 2009, 324, 522–528. [Google Scholar] [CrossRef]
- Kondrashov, F.A. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 2012, 279, 5048–5057. [Google Scholar] [CrossRef]
- Tadepalli, S.; Narayanan, S.K.; Stewart, G.C.; Chengappa, M.M.; Nagaraja, T.G. Fusobacterium necrophorum: A ruminal bacterium that invades liver to cause abscesses in cattle. Anaerobe 2009, 15, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Troussard, X.; Vol, S.; Cornet, E.; Bardet, V.; Couaillac, J.P.; Fossat, C.; Luce, J.C.; Maldonado, E.; Siguret, V.; Tichet, J.; et al. Full blood count normal reference values for adults in France. J. Clin. Pathol. 2014, 67, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Green, R.; Wachsmann-Hogiu, S. Development, history, and future of automated cell counters. Clin. Lab. Med. 2015, 35, 1–10. [Google Scholar] [CrossRef]
- Kim, D.J.; Noh, J.H.; Lee, B.W.; Choi, Y.H.; Chung, J.H.; Min, Y.K.; Lee, M.S.; Lee, M.K.; Kim, K.W. The associations of total and differential white blood cell counts with obesity, hypertension, dyslipidemia and glucose intolerance in a Korean population. J. Korean Med. Sci. 2008, 23, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Negewo, N.A.; McDonald, V.M.; Baines, K.J.; Wark, P.A.; Simpson, J.L.; Jones, P.W.; Gibson, P.G. Peripheral blood eosinophils: A surrogate marker for airway eosinophilia in stable COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 2016, 11, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Trend, S.; de Jong, E.; Lloyd, M.L.; Kok, C.H.; Richmond, P.; Doherty, D.A.; Simmer, K.; Kakulas, F.; Strunk, T.; Currie, A. Leukocyte Populations in Human Preterm and Term Breast Milk Identified by Multicolour Flow Cytometry. PLoS ONE 2015, 10, e0135580. [Google Scholar] [CrossRef]
- Hassiotou, F.; Hepworth, A.R.; Metzger, P.; Tat Lai, C.; Trengove, N.; Hartmann, P.E.; Filgueira, L. Maternal and infant infections stimulate a rapid leukocyte response in breastmilk. Clin. Transl. Immunol. 2013, 2, e3. [Google Scholar] [CrossRef]
- Passlick, B.; Flieger, D.; Ziegler-Heitbrock, H.W. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 1989, 74, 2527–2534. [Google Scholar] [CrossRef]
- Hussen, J.; Duvel, A.; Sandra, O.; Smith, D.; Sheldon, I.M.; Zieger, P.; Schuberth, H.J. Phenotypic and functional heterogeneity of bovine blood monocytes. PLoS ONE 2013, 8, e71502. [Google Scholar] [CrossRef]
- Storset, A.K.; Kulberg, S.; Berg, I.; Boysen, P.; Hope, J.C.; Dissen, E. NKp46 defines a subset of bovine leukocytes with natural killer cell characteristics. Eur. J. Immunol. 2004, 34, 669–676. [Google Scholar] [CrossRef]
- Widdison, S.; Coffey, T.J. Cattle and chemokines: Evidence for species-specific evolution of the bovine chemokine system. Anim. Genet. 2011, 42, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E. Immunoglobulin gene organization and the mechanism of repertoire development. Scan. J. Immunol. 1997, 45, 455–462. [Google Scholar] [CrossRef]
- Butler, J.E.; Kehrli, M.E.J. Immunoglobulins and immunocytes in the mammary gland and its secretion. In Mucosal Immunology, 3rd ed.; Mestecky, J.F., Bienenstock, J., Lamm, M.E., Mayer, L.R.M.J., Strober, W., Eds.; Elsevier Academic Press: Burlington, MA, USA, 2005; Volume 2, pp. 1763–1793. [Google Scholar]
- Liljavirta, J.; Ekman, A.; Knight, J.S.; Pernthaner, A.; Iivanainen, A.; Niku, M. Activation-induced cytidine deaminase (AID) is strongly expressed in the fetal bovine ileal Peyer’s patch and spleen and is associated with expansion of the primary antibody repertoire in the absence of exogenous antigens. Mucosal Immunol. 2013, 6, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Jenne, C.N.; Kennedy, L.J.; Reynolds, J.D. The sheep and cattle Peyer’s patch as a site of B-cell development. Vet. Res. 2006, 37, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Wood, P.R.; Seow, H.F. T cell cytokines and disease prevention. Vet. Immunol. Immunopathol. 1996, 54, 33–44. [Google Scholar] [CrossRef]
- Brown, W.C.; Rice-Ficht, A.C.; Estes, D.M. Bovine type 1 and type 2 responses. Vet. Immunol. Immunopathol. 1998, 63, 45–55. [Google Scholar] [CrossRef]
- Magombedze, G.; Eda, S.; Ganusov, V.V. Competition for antigen between Th1 and Th2 responses determines the timing of the immune response switch during Mycobaterium avium subspecies paratuberulosis infection in ruminants. PLoS Comput. Biol. 2014, 10, e1003414. [Google Scholar] [CrossRef]
- Washington State University. Monoclonal Antibody Center. Available online: https://vmp.vetmed.wsu.edu/resources/monoclonal-antibody-center (accessed on 20 April 2022).
- Murphy, K.; Weaver, C. Janeway’s Immunobiology, 9th ed.; Garland Science: New York, NY, USA, 2016. [Google Scholar]
- Bensaid, A.; Hadam, M. Individual antigens of cattle. Bovine CD4 (BoCD4). Vet. Immunol. Immunopathol. 1991, 27, 51–54. [Google Scholar] [CrossRef]
- Elhmouzi-Younes, J.; Boysen, P.; Pende, D.; Storset, A.K.; Le Vern, Y.; Laurent, F.; Drouet, F. Ovine CD16+/CD14− blood lymphocytes present all the major characteristics of natural killer cells. Vet. Res. 2010, 41, 4. [Google Scholar] [CrossRef]
- MacHugh, N.D.; Sopp, P. Individual antigens of cattle. Bovine CD8 (BoCD8). Vet. Immunol. Immunopathol. 1991, 27, 65–69. [Google Scholar] [CrossRef]
- Wilson, E.; Hedges, J.F.; Butcher, E.C.; Briskin, M.; Jutila, M.A. Bovine gamma delta T cell subsets express distinct patterns of chemokine responsiveness and adhesion molecules: A mechanism for tissue-specific gamma delta T cell subset accumulation. J. Immunol. 2002, 169, 4970–4975. [Google Scholar] [CrossRef] [PubMed]
- Toka, F.N.; Kenney, M.A.; Golde, W.T. Rapid and transient activation of gammadelta T cells to IFN-gamma production, NK cell-like killing, and antigen processing during acute virus infection. J. Immunol. 2011, 186, 4853–4861. [Google Scholar] [CrossRef]
- Russano, A.M.; Bassotti, G.; Agea, E.; Bistoni, O.; Mazzocchi, A.; Morelli, A.; Porcelli, S.A.; Spinozzi, F. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. J. Immunol. 2007, 178, 3620–3626. [Google Scholar] [CrossRef] [PubMed]
- Pang, D.J.; Neves, J.F.; Sumaria, N.; Pennington, D.J. Understanding the complexity of gammadelta T-cell subsets in mouse and human. Immunology 2012, 136, 283–290. [Google Scholar] [CrossRef]
- Mossad, A.A.; Elbagoury, A.R.; Khalid, A.M.; Waters, W.R.; Tibary, A.; Hamilton, M.J.; Davis, W.C. Identification of Monoclonal Antibody Reagents for Use in the Study of Immune Response in Camel and Water Buffalo. J. Camel Pract. Res. 2006, 13, 91–101. [Google Scholar]
- Wang, F.; Herzig, C.; Ozer, D.; Baldwin, C.L.; Telfer, J.C. Tyrosine phosphorylation of scavenger receptor cysteine-rich WC1 is required for the WC1-mediated potentiation of TCR-induced T-cell proliferation. Eur. J. Immunol. 2009, 39, 254–266. [Google Scholar] [CrossRef]
- Naessens, J.; Newson, J.; McHugh, N.; Howard, C.J.; Parsons, K.; Jones, B. Characterization of a bovine leucocyte differentiation antigen of 145,000 MW restricted to B lymphocytes. Immunology 1990, 69, 525–530. [Google Scholar] [PubMed]
- Bembridge, G.P.; Howard, C.J.; Parsons, K.R.; Sopp, P. Identification of monoclonal antibodies specific for bovine leukocyte common antigen (CD45) together with a novel broadly expressed leukocyte differentiation antigen, BoWC11. Vet. Immunol. Immunopathol. 1993, 39, 115–120. [Google Scholar] [CrossRef]
- Fujimoto, H.; Sakata, T.; Hamaguchi, Y.; Shiga, S.; Tohyama, K.; Ichiyama, S.; Wang, F.S.; Houwen, B. Flow cytometric method for enumeration and classification of reactive immature granulocyte populations. Cytometry 2000, 42, 371–378. [Google Scholar] [CrossRef]
- Pillay, J.; Kamp, V.M.; van Hoffen, E.; Visser, T.; Tak, T.; Lammers, J.W.; Ulfman, L.H.; Leenen, L.P.; Pickkers, P.; Koenderman, L. A subset of neutrophils in human systemic inflammation inhibits T cell responses through Mac-1. J. Clin. Investig. 2012, 122, 327–336. [Google Scholar] [CrossRef]
- Lahmers, K.K.; Hedges, J.F.; Jutila, M.A.; Deng, M.; Abrahamsen, M.S.; Brown, W.C. Comparative gene expression by WC1+ gammadelta and CD4+ alphabeta T lymphocytes, which respond to Anaplasma marginale, demonstrates higher expression of chemokines and other myeloid cell-associated genes by WC1+ gammadelta T cells. J. Leukoc. Biol. 2006, 80, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Connelley, T.; Storset, A.K.; Pemberton, A.; MacHugh, N.; Brown, J.; Lund, H.; Morrison, I.W. NKp46 defines ovine cells that have characteristics corresponding to NK cells. Vet. Res. 2011, 42, 37. [Google Scholar] [CrossRef] [PubMed]
- Van Merris, V.; Meyer, E.; Burvenich, C. Functional Maturation during Bovine Granulopoiesis. J. Dairy Sci. 2002, 85, 2859–2868. [Google Scholar] [CrossRef]
- Elnaggar, M.M.; Abdellrazeq, G.S.; Mack, V.; Fry, L.M.; Davis, W.C.; Park, K.T. Characterization and use of new monoclonal antibodies to CD11c, CD14, and CD163 to analyze the phenotypic complexity of ruminant monocyte subsets. Vet. Immunol. Immunopathol. 2016, 178, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ziegler-Heitbrock, L. Monocyte subsets in man and other species. Cell. Immunol. 2014, 289, 135–139. [Google Scholar] [CrossRef]
- Corripio-Miyar, Y.; Hope, J.; McInnes, C.J.; Wattegedera, S.R.; Jensen, K.; Pang, Y.; Entrican, G.; Glass, E.J. Phenotypic and functional analysis of monocyte populations in cattle peripheral blood identifies a subset with high endocytic and allogeneic T-cell stimulatory capacity. Vet. Res. 2015, 46, 112. [Google Scholar] [CrossRef]
- Noronha, L.E.; Harman, R.M.; Wagner, B.; Antczak, D.F. Generation and characterization of monoclonal antibodies to equine CD16. Vet. Immunol. Immunopathol. 2012, 146, 135–142. [Google Scholar] [CrossRef]
- Dato, M.E.; Wierda, W.G.; Kim, Y.B. A triggering structure recognized by G7 monoclonal antibody on porcine lymphocytes and granulocytes. Cell. Immunol. 1992, 140, 468–477. [Google Scholar] [CrossRef]
- Boysen, P.; Gunnes, G.; Pende, D.; Valheim, M.; Storset, A.K. Natural killer cells in lymph nodes of healthy calves express CD16 and show both cytotoxic and cytokine-producing properties. Dev. Comp. Immunol. 2008, 32, 773–783. [Google Scholar] [CrossRef]
- Girardi, M. Immunosurveillance and immunoregulation by gammadelta T cells. J. Investig. Dermatol. 2006, 126, 25–31. [Google Scholar] [CrossRef]
- Herzig, C.T.; Lefranc, M.P.; Baldwin, C.L. Annotation and classification of the bovine T cell receptor delta genes. BMC Genom. 2010, 11, 100. [Google Scholar] [CrossRef]
- Van Rhijn, I.; Spiering, R.; Smits, M.; van Blokland, M.T.; de Weger, R.; van Eden, W.; Rutten, V.P.; Koets, A.P. Highly diverse TCR delta chain repertoire in bovine tissues due to the use of up to four D segments per delta chain. Mol. Immunol. 2007, 44, 3155–3161. [Google Scholar] [CrossRef] [PubMed]
- Hayday, A.C. γδ cells: A right time and a right place for a conserved third way of protection. Annu. Rev. Immunol. 2000, 18, 975–1026. [Google Scholar] [CrossRef] [PubMed]
- Guzman, E.; Price, S.; Poulsom, H.; Hope, J. Bovine gammadelta T cells: Cells with multiple functions and important roles in immunity. Vet. Immunol. Immunopathol. 2012, 148, 161–167. [Google Scholar] [CrossRef] [PubMed]
- MacHugh, N.D.; Mburu, J.K.; Carol, M.J.; Wyatt, C.R.; Orden, J.A.; Davis, W.C. Identification of two distinct subsets of bovine gamma delta T cells with unique cell surface phenotype and tissue distribution. Immunology 1997, 92, 340–345. [Google Scholar] [CrossRef]
- Wijngaard, P.L.; MacHugh, N.D.; Metzelaar, M.J.; Romberg, S.; Bensaid, A.; Pepin, L.; Davis, W.C.; Clevers, H.C. Members of the novel WC1 gene family are differentially expressed on subsets of bovine CD4−CD8− gamma delta T lymphocytes. J. Immunol. 1994, 152, 3476–3482. [Google Scholar]
- Chen, C.; Herzig, C.T.; Telfer, J.C.; Baldwin, C.L. Antigenic basis of diversity in the gammadelta T cell co-receptor WC1 family. Mol. Immunol. 2009, 46, 2565–2575. [Google Scholar] [CrossRef]
- Hoek, A.; Rutten, V.P.; Kool, J.; Arkesteijn, G.J.; Bouwstra, R.J.; Van Rhijn, I.; Koets, A.P. Subpopulations of bovine WC1+ gammadelta T cells rather than CD4+CD25high Foxp3+ T cells act as immune regulatory cells ex vivo. Vet. Res. 2009, 40, 6. [Google Scholar] [CrossRef]
- Hedges, J.F.; Cockrell, D.; Jackiw, L.; Meissner, N.; Jutila, M.A. Differential mRNA expression in circulating gammadelta T lymphocyte subsets defines unique tissue-specific functions. J. Leukoc. Biol. 2003, 73, 306–314. [Google Scholar] [CrossRef]
- Wesch, D.; Glatzel, A.; Kabelitz, D. Differentiation of resting human peripheral blood gamma delta T cells toward Th1- or Th2-phenotype. Cell. Immunol. 2001, 212, 110–117. [Google Scholar] [CrossRef]
- Casetti, R.; Agrati, C.; Wallace, M.; Sacchi, A.; Martini, F.; Martino, A.; Rinaldi, A.; Malkovsky, M. Cutting edge: TGF-beta1 and IL-15 Induce FOXP3+ gammadelta regulatory T cells in the presence of antigen stimulation. J. Immunol. 2009, 183, 3574–3577. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, N.; La Mendola, C.; Orlando, V.; Meraviglia, S.; Todaro, M.; Stassi, G.; Sireci, G.; Fournie, J.J.; Dieli, F. Differentiation, phenotype, and function of interleukin-17-producing human Vgamma9Vdelta2 T cells. Blood 2011, 118, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.R.; Mackay, C.R.; Moser, B.; Eberl, M. IL-21 enhances the potential of human gammadelta T cells to provide B-cell help. Eur. J. Immunol. 2012, 42, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Brandes, M.; Willimann, K.; Moser, B. Professional antigen-presentation function by human gammadelta T cells. Science 2005, 309, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Correia, D.V.; Fogli, M.; Hudspeth, K.; da Silva, M.G.; Mavilio, D.; Silva-Santos, B. Differentiation of human peripheral blood Vdelta1+ T cells expressing the natural cytotoxicity receptor NKp30 for recognition of lymphoid leukemia cells. Blood 2011, 118, 992–1001. [Google Scholar] [CrossRef]
- Toulon, A.; Breton, L.; Taylor, K.R.; Tenenhaus, M.; Bhavsar, D.; Lanigan, C.; Rudolph, R.; Jameson, J.; Havran, W.L. A role for human skin-resident T cells in wound healing. J. Exp. Med. 2009, 206, 743–750. [Google Scholar] [CrossRef]
- Holderness, J.; Hedges, J.F.; Ramstead, A.; Jutila, M.A. Comparative biology of gammadelta T cell function in humans, mice, and domestic animals. Annu. Rev. Anim. Biosci. 2013, 1, 99–124. [Google Scholar] [CrossRef]
- Hedges, J.F.; Buckner, D.L.; Rask, K.M.; Kerns, H.M.; Jackiw, L.O.; Trunkle, T.C.; Pascual, D.W.; Jutila, M.A. Mucosal lymphatic-derived gammadelta T cells respond early to experimental Salmonella enterocolitis by increasing expression of IL-2R alpha. Cell. Immunol. 2007, 246, 8–16. [Google Scholar] [CrossRef]
- Lundberg, P.; Splitter, G.A. gd T-lymphocyte cytotoxicity against envelope-expressing target cells is unique to the alymphocytic state of bovine leukemia virus infection in the natural host. J. Virol. 2000, 74, 8299–8306. [Google Scholar] [CrossRef]
- McKenna, S.L.; Keefe, G.P.; Tiwari, A.; Van Leeuwen, J.; Barkema, H.W. Johne’s disease in Canada Part II: Disease impacts, risk factors, and control programs for dairy producers. Can. Vet. J. 2006, 47, 1089–1099. [Google Scholar]
- Albarrak, S.M.; Waters, W.R.; Stabel, J.R.; Hostetter, J.M. Evaluating the cytokine profile of the WC1+ gammadelta T cell subset in the ileum of cattle with the subclinical and clinical forms of MAP infection. Vet. Immunol. Immunopathol. 2018, 201, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Thompson-Crispi, K.A.; Miglior, F.; Mallard, B.A. Incidence rates of clinical mastitis among Canadian Holsteins classified as high, average, or low immune responders. Clin. Vaccine Immunol. 2013, 20, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Vance, R.E.; Isberg, R.R.; Portnoy, D.A. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 2009, 6, 10–21. [Google Scholar] [CrossRef]
- Rojas, R.E.; Torres, M.; Fournie, J.J.; Harding, C.V.; Boom, W.H. Phosphoantigen presentation by macrophages to mycobacterium tuberculosis--reactive Vgamma9Vdelta2+ T cells: Modulation by chloroquine. Infect. Immun. 2002, 70, 4019–4027. [Google Scholar] [CrossRef]
- Heuston, S.; Begley, M.; Gahan, C.G.; Hill, C. Isoprenoid biosynthesis in bacterial pathogens. Microbiology 2012, 158, 1389–1401. [Google Scholar] [CrossRef]
- Lindberg, B.G.; Merritt, E.A.; Rayl, M.; Liu, C.; Parmryd, I.; Olofsson, B.; Faye, I. Immunogenic and antioxidant effects of a pathogen-associated prenyl pyrophosphate in Anopheles gambiae. PLoS ONE 2013, 8, e73868. [Google Scholar] [CrossRef] [PubMed]
- Uchida, R.; Ashihara, E.; Sato, K.; Kimura, S.; Kuroda, J.; Takeuchi, M.; Kawata, E.; Taniguchi, K.; Okamoto, M.; Shimura, K.; et al. Gamma delta T cells kill myeloma cells by sensing mevalonate metabolites and ICAM-1 molecules on cell surface. Biochem. Biophys. Res. Commun. 2007, 354, 613–618. [Google Scholar] [CrossRef]
- Meissner, N.; Radke, J.; Hedges, J.F.; White, M.; Behnke, M.; Bertolino, S.; Abrahamsen, M.; Jutila, M.A. Serial analysis of gene expression in circulating gamma delta T cell subsets defines distinct immunoregulatory phenotypes and unexpected gene expression profiles. J. Immunol. 2003, 170, 356–364. [Google Scholar] [CrossRef]
- Liang, D.; Arnold, L.M.; Stowe, C.J.; Harmon, R.J.; Bewley, J.M. Estimating US dairy clinical disease costs with a stochastic simulation model. J. Dairy Sci. 2017, 100, 1472–1486. [Google Scholar] [CrossRef]
- Roche, J.R.; Friggens, N.C.; Kay, J.K.; Fisher, M.W.; Stafford, K.J.; Berry, D.P. Invited review: Body condition score and its association with dairy cow productivity, health, and welfare. J. Dairy Sci. 2009, 92, 5769–5801. [Google Scholar] [CrossRef]
| Biomarker in Bovine Raw Milk | Limit | Information about Udder Health | Reference |
|---|---|---|---|
| T cells per B cell → CD2/CD21 index | <10 | Subclinical mastitis | [12] |
| Log (neutrophils per lymphocyte) → Log PMN/lymphocyte ratio | >0.495 | [11] | |
| CD18 expression level on neutrophils and lymphocytes | “High” | [13] | |
| Granulocytes per macrophage → PMN/M ratio | <2.39 | Mastitis in resolution phase | [5] |
| Percentages of macrophages AND granulated cells AND non-vital cells | >4.5%, >25.5%, >22% | Chronic mastitis | [16] |
| T helper cells per cytotoxic T cell → CD4/CD8 ratio | <1 | Low mastitis resistance | [19] |
| Viability of neutrophils | “Low” | [20] | |
| CD11b expression level on leukocytes | “High” | Udder inflammation | [28] |
| Cell Type | Percentage of all Leukocytes in Bovine Peripheral Blood | Percentage of All Somatic Cells in Bovine Raw Milk |
|---|---|---|
| PMNs | 20–50 [55] 22 [56] 36 [57] | 34 [38] 31–50 [1] 9 [58] 41 [28] |
| Eosinophils | 2–6 [55] 8 [56] 5 [57] | <1 [38] <1 [1] |
| Basophils | 0–2 [55] | <1 [38] |
| Monocytes | 2–6 [55] 10 [56] | |
| Macrophages | 35–79 [59] 46 [38] 35–43 [1] 52 [58] 13 [28] | |
| Lymphocytes | 45–65 [55] 55 [56] | 10–28 [59] 21 [38] 14–26 [1] 46 [28] |
| Mammary epithelial cells | 1–3 [1] <1 [38] |
| Lymphocyte Subset | Percentage of All Lymphocytes in Bovine Peripheral Blood | Percentage of All Lymphocytes in Bovine Raw Milk |
|---|---|---|
| T cells (αβ and γδ) | 62 * [60] | 40–50 [59] 84 * [60] 90 [13] |
| αβ T cells | 44 * [60] 50 [56] | 64 * [60] 54 [61] 80 [13] |
| CD8+ cells | 20 * [60] 13 * [19] 20 [56] | 40 * [60] 21 [61] 10 * [19] 50 [13] |
| CD4+ cells | 24 * [60] 31 * [19] 30 [56] | 24 * [60] 33 [61] 31 * [19] 30 [13] |
| γδ T cells | 18 * [60] 15–30 [62] <60 in calves [31] 7–20 [63] 28 [64] | 20 * [60] 9 # [64] 10 [13] |
| WC1+ cells | 13 * [19] 5–15 [63] 26 [64] | 5 [61] 22 * [19] 1 # [64] |
| WC1− cells | 2–5 [63] 2 [64] | 8 # [64] |
| B cells | 38 * [60] 44 * [19] 16 [56] | 20–25 [59] 8 * [60] 1 [61] 36 * [19] |
| NK cells | 2–10 [65] | 2–4 in buffalo raw milk [66] |
| CD Marker | Major Functions | Expression on Human Leukocytes | Expression on Bovine Leukocytes |
|---|---|---|---|
| CD4 | Co-receptor with MHC class-II-restricted TCRs in antigen recognition. | On T cells that recognize antigens associated with MHC class II molecules (T helper cells and regulatory T cells), monocytes, macrophages. | Only on T cells [100]. |
| CD8 | Co-receptor with MHC class I-restricted TCRs in antigen recognition. | On T cells that recognize antigens associated with MHC class I molecules (cytotoxic T cells), subsets of γδ T cells, NK cells and monocytes. | Similar [101,102,103]. |
| CD335 | Major cytotoxicity-activating receptor (induces the lysis of virus-infected cells and tumor cells). | On NK cells. |
|
| gdTCR | Antigen receptor, e.g., to antigens presented by antigen presenting cells (APCs) via nonclassical MHC-molecules [105]. | Only on γδ T cells [106]. | Only on γδ T cells [63,107], but differences with regard to the co-expression of Workshop Cluster 1 (WC1), a possible costimulatory molecule for the gdTCR [108] that is exclusively expressed on ruminant γδ T-cell subsets. |
| CD21 | Complement receptor that binds to the breakdown products of Complement component 3 (C3). Associated with CD19 and CD81 (B cell coreceptor complex). | On mature B cells, follicular dendritic cells. | Only on mature B cells [109]. |
| CD Marker | Major Functions | Expression on Human Leukocytes | Expression on Bovine Leukocytes |
|---|---|---|---|
| CD45 | Signaling molecule (protein tyrosine phosphatase) that regulates a variety of cellular processes including cell growth, differentiation. Critical requirement for antigen receptor-mediated activation of T cells and B cells. | On all leukocytes. | Similar [57,110]. |
| CD11b |
|
|
|
| CD14 | Receptor for complex of LPS and soluble LBP (lipopolysaccharide-binding protein). | High expression level on monocytes and macrophages, weak expression level on granulocytes. | Only on monocytes and macrophages [6,57], in contrast to other ruminant species (sheep and goats) which also show a high CD14 expression on granulocytes [116]. |
| CD16a | Low affinity Fc receptor for IgG2 and IgG3. Binds to IgG on opsonized antigens and mediates phagocytosis or antibody-dependent cellular cytotoxicity (ADCC) plus cytokine production. | On NK cells (in blood), macrophages, γδ T cells and monocyte subsets (nonclassical monocytes (ncM) and intermediate monocytes (intM), not classical monocytes (cM)). |
|
| CD16b | Low affinity Fc receptor for IgG1 and IgG3, similar to CD16a. | On neutrophils, absent in eosinophils (but inducible by interferon gamma, IFN-γ). On all mature neutrophils (band cells and segmented cells), with no elevated expression level upon LPS stimulation [112]. Not on early and late immature neutrophils (promyelocytes, myelocytes, metamyelocytes) and thus a possible marker for early inflammatory responses, especially when CD11b expression is also taken into account [111]. | With regard to CD16b expression, bovine neutrophils are similar to equine neutrophils [119] but dissimilar to the largely (>90%) CD16b+ neutrophils in pigs [120], goats, and sheep [116]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farschtschi, S.; Mattes, M.; Pfaffl, M.W. Advantages and Challenges of Differential Immune Cell Count Determination in Blood and Milk for Monitoring the Health and Well-Being of Dairy Cows. Vet. Sci. 2022, 9, 255. https://doi.org/10.3390/vetsci9060255
Farschtschi S, Mattes M, Pfaffl MW. Advantages and Challenges of Differential Immune Cell Count Determination in Blood and Milk for Monitoring the Health and Well-Being of Dairy Cows. Veterinary Sciences. 2022; 9(6):255. https://doi.org/10.3390/vetsci9060255
Chicago/Turabian StyleFarschtschi, Sabine, Martin Mattes, and Michael W. Pfaffl. 2022. "Advantages and Challenges of Differential Immune Cell Count Determination in Blood and Milk for Monitoring the Health and Well-Being of Dairy Cows" Veterinary Sciences 9, no. 6: 255. https://doi.org/10.3390/vetsci9060255
APA StyleFarschtschi, S., Mattes, M., & Pfaffl, M. W. (2022). Advantages and Challenges of Differential Immune Cell Count Determination in Blood and Milk for Monitoring the Health and Well-Being of Dairy Cows. Veterinary Sciences, 9(6), 255. https://doi.org/10.3390/vetsci9060255







