Influence of Colostrum and Vitamins A, D3, and E on Early Intestinal Colonization of Neonatal Holstein Calves Infected with Mycobacterium avium subsp. paratuberculosis
Abstract
1. Introduction
2. Materials and Methods
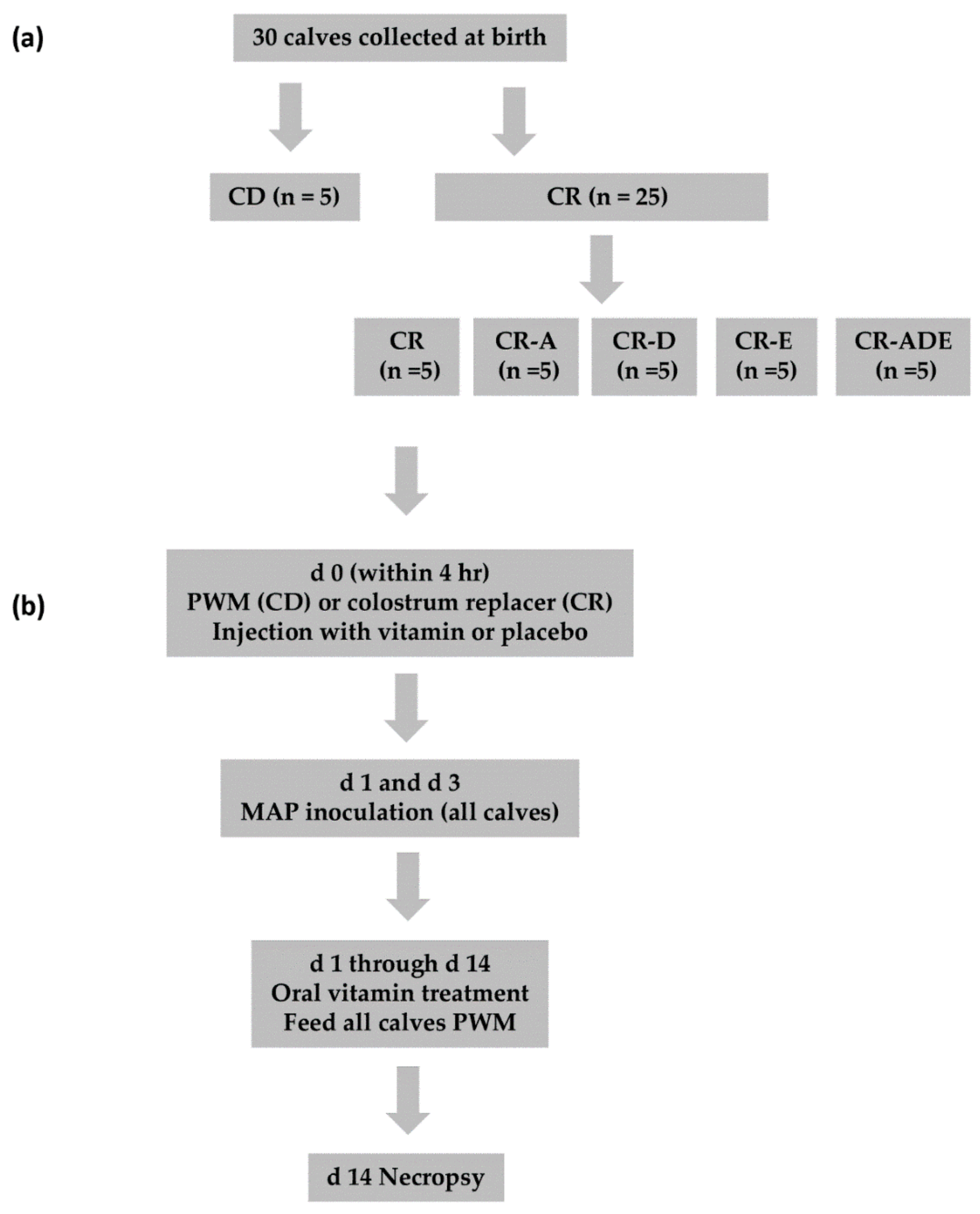
2.1. Study Design
2.2. Animal Care
2.3. Fecal Culture and DNA Extraction
2.4. Tissue Culture, DNA Extraction, and Histopathology
2.5. 16S Sequencing Protocols
2.6. Microbial Community Data Analysis
3. Results
3.1. MAP Infection in Neonatal Calves
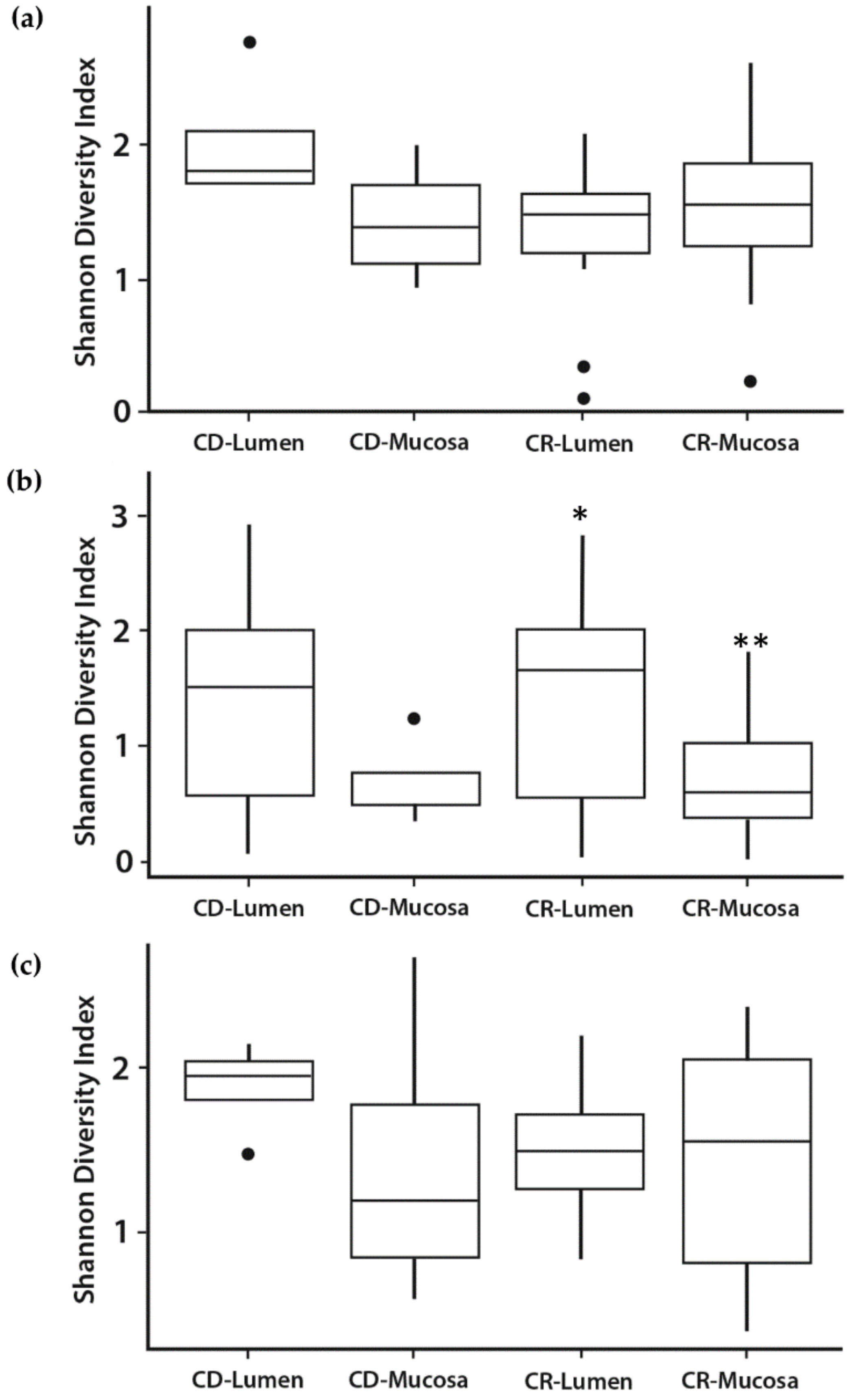
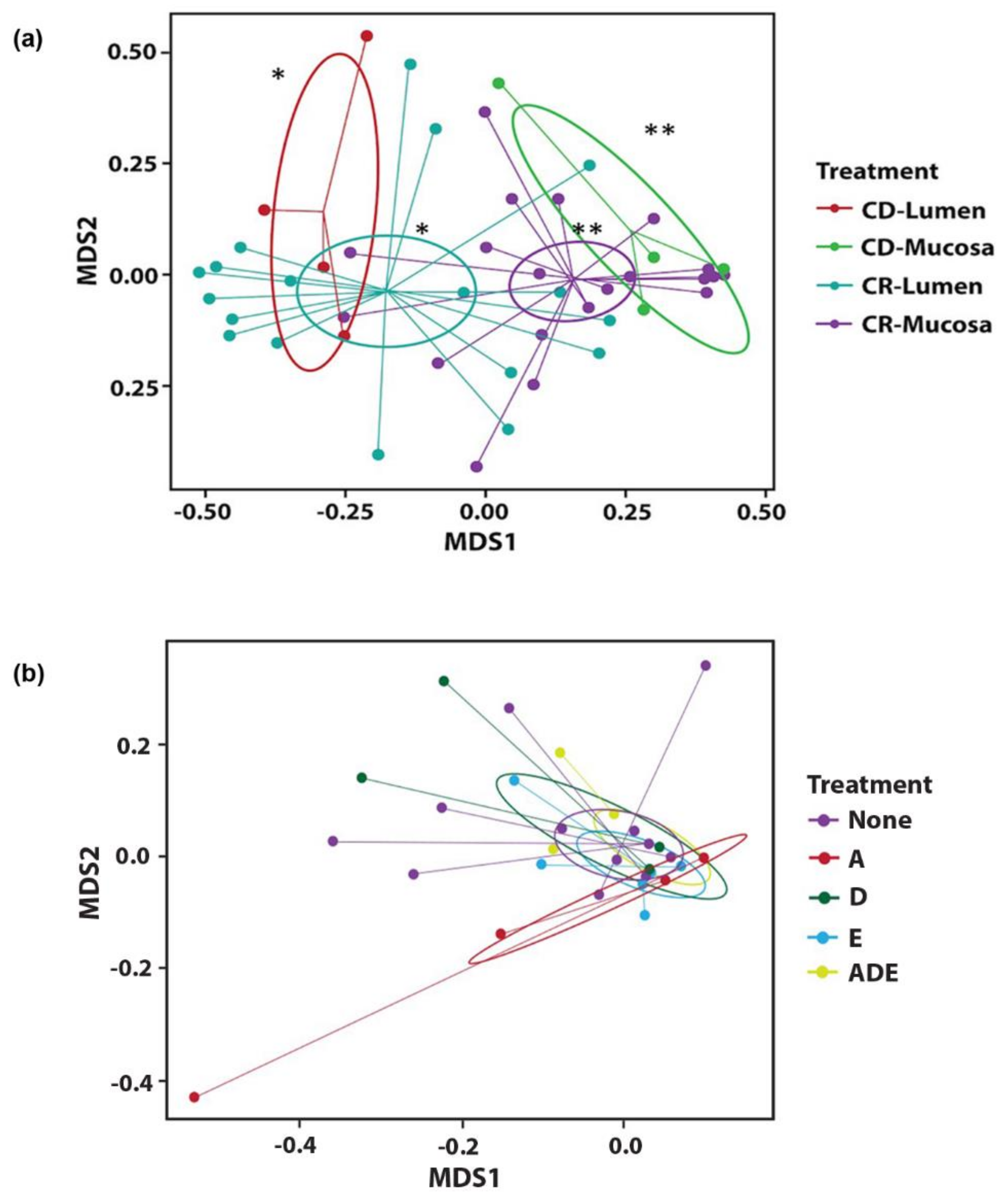
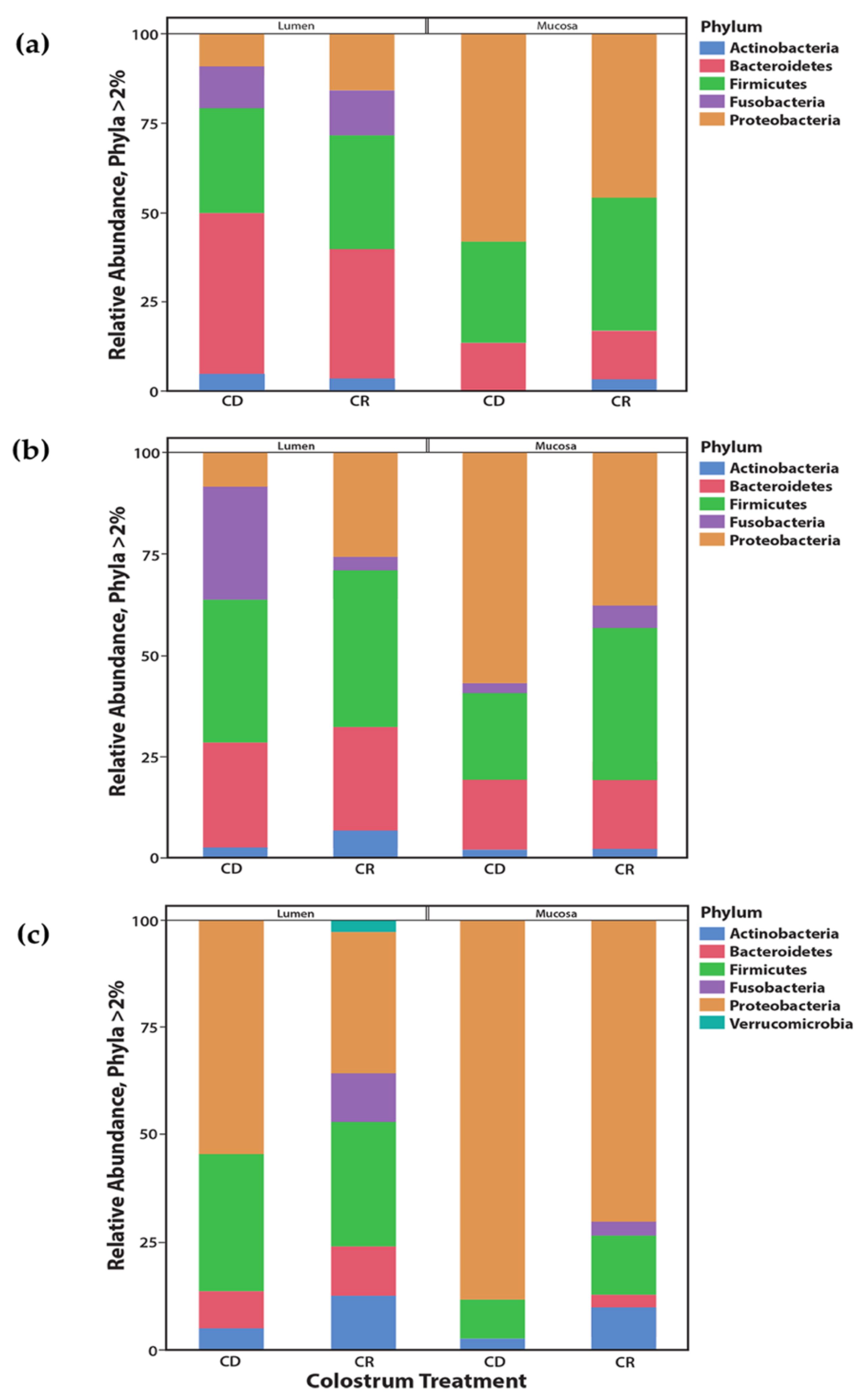
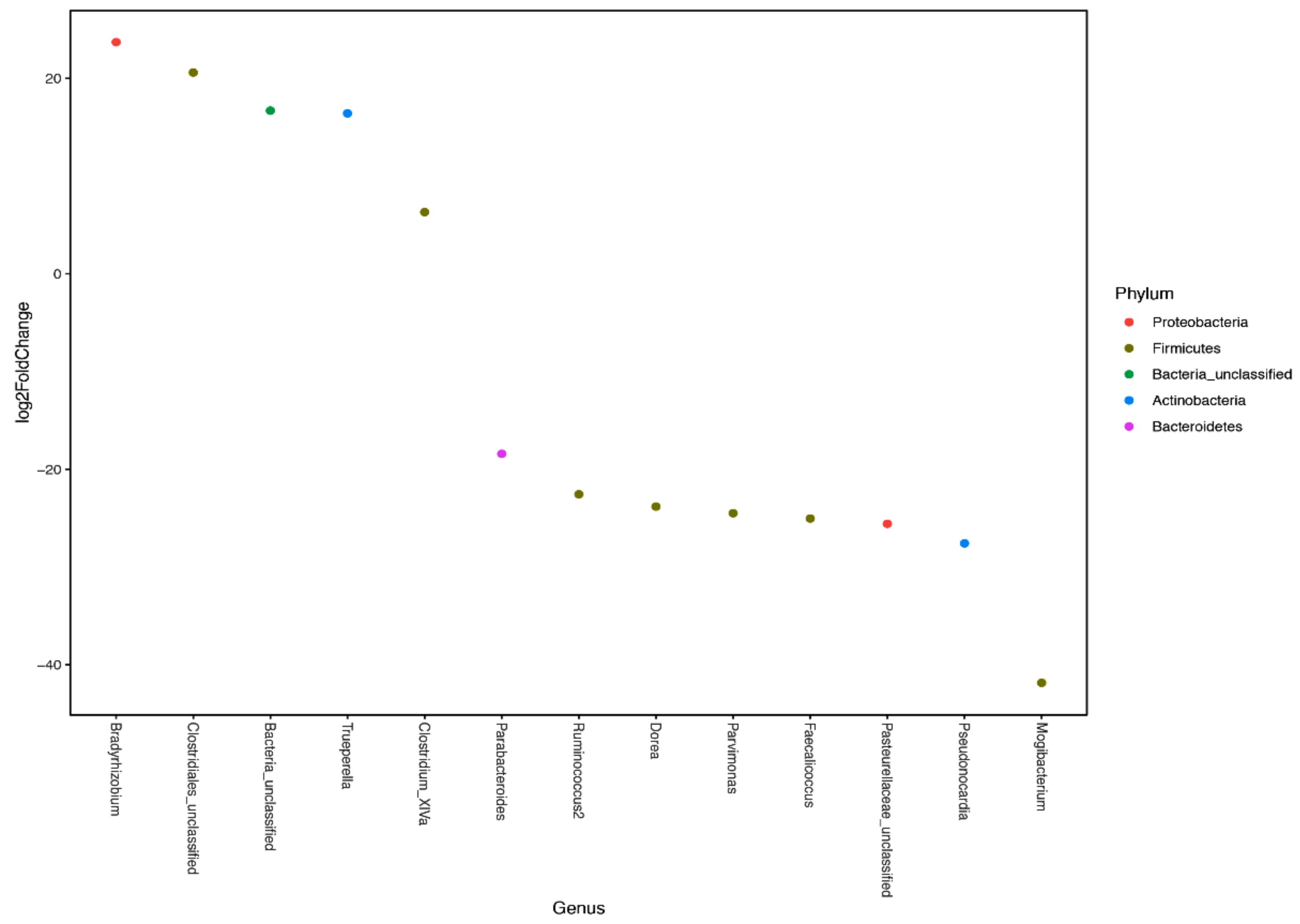
3.2. Microbial Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harris, N.B.; Barletta, R.G. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbio. Rev. 2001, 14, 489–512. [Google Scholar] [CrossRef]
- Stabel, J.R. Pasteurization of colostrum reduces the incidence of paratuberculosis in neonatal dairy calves. J. Dairy Sci. 2008, 91, 3600–3606. [Google Scholar] [CrossRef] [PubMed]
- McGuirk, S.M. Disease management of dairy calves and heifers. Vet. Clin. N. Am. Food Anim. Pract. 2008, 24, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Bartol, F.F.; Wiley, A.A.; Bagnell, C.A. Epigenetic programming of porcine endometrial function and the lactocrine hypothesis. Reprod. Domest. Anim. 2008, 43, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Chase, C.C.; Hurley, D.J.; Reber, D.J. Neonatal immune development in the calf and its impact on vaccine response. Vet. Clin. N. Am. Food Anim. Pract. 2008, 24, 87–104. [Google Scholar] [CrossRef]
- Stelwagen, K.; Carpenter, E.; Haigh, B.; Hodgkinson, A.; Wheeler, T.T. Immune components of bovine colostrum and milk. J. Anim. Sci. 2009, 87, 3–9. [Google Scholar] [CrossRef]
- Robison, J.D.; Stott, G.D.; DeNise, S.K. Effects of passive immunity on growth and survival in the dairy heifer. J. Dairy Sci. 1988, 71, 1283–1287. [Google Scholar] [CrossRef]
- Stillwell, G.; Carvalho, R.C. Clinical outcome of calves with failure of passive transfer as diagnosed by a commercially available IgG quick test kit. Can. Vet. J. 2011, 52, 524–526. [Google Scholar]
- Cho, Y.; Yoon, K. An overview of calf diarrhea—Infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014, 15, 1–17. [Google Scholar] [CrossRef]
- Krueger, L.A.; Beitz, D.C.; Onda, K.; Osman, M.; O’Neil, M.R.; Lei, S.; Wattoo, F.H.; Stuart, R.L.; Tyler, H.D.; Nonnecke, B. Effects of D-α-tocopherol and dietary energy on growth and health of preruminant dairy calves. J. Dairy Sci. 2014, 97, 3715–3727. [Google Scholar] [CrossRef]
- Nonnecke, B.J.; McGill, J.L.; Ridpath, J.F.; Sacco, R.E.; Lippolis, J.D.; Reinhardt, T.A. Acute phase response elicited by experimental bovine diarrhea virus (BVDV) infection is associated with decreased vitamin D and E status of vitamin-replete preruminant calves. J. Dairy Sci. 2014, 97, 5566–5579. [Google Scholar] [CrossRef] [PubMed]
- Traber, M.G.; Atkinson, J. Vitamin E, antioxidant and nothing more. Free Radic. Biol. Med. 2007, 43, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Grainger, J.R.; Spencer, S.P.; Belkaid, Y. The role of retinoic acid in tolerance and immunity. Immunity 2011, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.D.; Reinhardt, T.A.; Lippolis, J.D.; Sacco, R.E.; Nonnecke, B.J. Vitamin D signaling in the bovine immune system: A model for understanding human vitamin D requirements. Nutrients 2012, 4, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Rhinn, M.; Dollé, P. Retinoic acid signaling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef]
- Krueger, L.A.; Reinhardt, T.A.; Beitz, D.C.; Stuart, R.L.; Stabel, J.R. Effects of fractionated colostrum replacer and vitamins A, D, and E on haptoglobin and clinical health in neonatal Holstein calves challenged with Mycobacterium avium ssp. paratuberculosis. J. Dairy Sci. 2016, 99, 2884–2895. [Google Scholar] [CrossRef]
- Stabel, J.R. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J. Vet. Diagn. Invest. 1997, 9, 375–380. [Google Scholar] [CrossRef]
- Jenvey, C.J.; Hostetter, J.M.; Shircliff, A.L.; Bannantine, J.P.; Stabel, J.R. Quantification of macrophages and Mycobacterium avium subsp. paratuberculosis in bovine intestinal tissue during different stages of disease. Vet. Pathol. 2019, 56, 671–680. [Google Scholar] [CrossRef]
- Leite, F.L.; Stokes, K.D.; Robbe-Austerman, S.; Stabel, J.R. Comparison of fecal DNA extraction kits for the detection of Mycobacterium avium subsp. paratuberculosis by polymerase chain reaction. J. Vet. Diagn. Invest. 2013, 25, 27–34. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. Available online: https://www.mothur.org/wiki/MiSeq_SOP (accessed on 24 September 2019). [CrossRef]
- Looft, T.; Allen, H.K.; Casey, T.A.; Alt, D.P.; Stanton, T.B. Carbadox has both temporary and lasting effects on the swine gut microbiota. Front. Microbiol. 2014, 5, 276. [Google Scholar] [CrossRef] [PubMed]
- Puente-Sánchez, F.; Aguirre, J.; Parro, V. A novel conceptual approach to read-filtering in high-throughput amplicon sequencing studies. Nucleic Acids Res. 2016, 44, e40. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Bayles, D.O.; Looft, T.; Trachsel, J.; Bass, B.E.; Alt, D.P.; Bearson, S.M.; Nicholson, T.; Casey, T.A. Pipeline for amplifying and analyzing amplicons of the V1–V3 region of the 16S rRNA gene. BMC Res. Notes 2016, 9, 380. [Google Scholar] [CrossRef]
- McMurdie, P.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Henry, M.; Stevens, H.; et al. Vegan: Community Ecology Package. Ordination Methods, Diversity Analysis and Other Functions for Community and Vegetation Ecologists. Version 2.4-5. 2017. Available online: https://CRAN.R-project.org/package=vegan (accessed on 24 September 2019).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2009; Available online: http://ggplot2.org (accessed on 24 September 2019).
- Dinno, A. Dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums. Version 1.3.5. 2017. Available online: https://cran.project.org/web/packages/dunn.test/dunn.test.pdf (accessed on 24 September 2019).
- Trachsel, J. Funfuns. 2018. Available online: https://github.com/Jtrachsel/funfuns (accessed on 24 September 2019).
- Whitlock, R.H.; Sweeney, R.W.; Fyock, T.L.; Smith, J. MAP super-shedders: Another factor in the control of Johne’s disease. In Proceedings of the Eighth International Colloquium on Paratuberculosis, Copenhagen, Denmark, WI, USA, 14–17 August 2005; Manning, E.J., Nielsen, S.S., Eds.; International Association for Paratuberculosis: Madison, WI, USA; p. 42. [Google Scholar]
- Stabel, J.R.; Bradner, L.; Robbe-Austerman, S.; Beitz, D.C. Clinical disease and stage of lactation influence shedding of Mycobacterium avium subspecies paratuberculosis into milk and colostrum of naturally infected dairy cows. J. Dairy Sci. 2014, 97, 6296–6304. [Google Scholar] [CrossRef]
- Mortier, R.A.; Barkema, H.W.; Orsel, K.; Wolf, R.; De Buck, J. Shedding patterns of dairy calves experimentally infected with Mycobacterium avium subspecies paratuberculosis. Vet. Res. 2014, 45, 71. [Google Scholar] [CrossRef]
- Corbett, C.S.; Barkema, H.W.; De Buck, J. Quantifying fecal shedding of Mycobacterium avium spp. paratuberculosis from calves after experimental infection and exposure. J. Dairy Sci. 2018, 101, 1478–1487. [Google Scholar] [CrossRef]
- Stabel, J.R.; Palmer, M.V.; Harris, B.; Plattner, B.; Hostetter, J.; Robbe-Austerman, S. Pathogenesis of Mycobacterium avium subsp. paratuberculosis in neonatal calves after oral or intraperitoneal experimental infection. Vet. Microbiol. 2009, 136, 306–313. [Google Scholar] [CrossRef]
- Wolf, R.; Barkema, H.W.; De Buck, J.; Orsel, K. Factors affecting management changes on farms participating in a Johne’s disease control program. J. Dairy Sci. 2015, 98, 7831–7845. [Google Scholar] [CrossRef]
- Whittington, R.J.; Whittington, A.; Waldron, A.; Begg, D.J.; de Silva, K.; Purdie, A.C.; Plain, K.M. Development and validation of a liquid medium (M7H9C) for routine culture of Mycobacterium avium subsp. paratuberculosis to replace modified Bactec 12B medium. J. Clin. Microbiol. 2013, 51, 3993–4000. [Google Scholar] [CrossRef] [PubMed]
- Arsenault, R.J.; Maattanen, P.; Daigle, J.; Potter, A.; Griebel, P.; Napper, S. From mouth to macrophage: Mechanisms of innate immune subversion by Mycobacterium avium subsp. paratuberculosis. Vet. Res. 2014, 45, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J. Comp. Pathol. 1997, 116, 217–261. [Google Scholar] [CrossRef]
- Gonzalez, J.; Geijo, M.V.; Garcia-Pariente, C.; Verna, A.; Corpa, J.M.; Reyes, L.E.; Ferreras, M.C.; Juste, R.A.; Marin, J.G.; Perez, V. Histopathological classification of lesions associated with natural paratuberculosis infection in cattle. J. Comp. Pathol. 2005, 133, 184–196. [Google Scholar] [CrossRef]
- Politis, I.; Hidiroglou, M.; Batra, T.R.; Gilmore, J.A.; Gorewit, R.C.; Scherf, H. Effects of vitamin E on immune function of dairy cows. Am. J. Vet. Res. 1995, 56, 179–184. [Google Scholar]
- Lee, G.Y.; Han, S.N. The role of vitamin E in immunity. Nutrients 2018, 10, 1614. [Google Scholar] [CrossRef]
- Politis, I. Reevaluation of vitamin E supplementation of dairy cows: Bioavailability, animal health and milk quality. Animal 2012, 6, 1427–1434. [Google Scholar] [CrossRef]
- Pakkanen, R.; Aalto, J. Growth factors and antimicrobial factors of bovine colostrum. Int. Dairy J. 1997, 7, 285–297. [Google Scholar] [CrossRef]
- Boudry, C.; Dehoux, J.P.; Wavreille, J.; Portetelle, D.; Thewis, A.; Buldgen, A. Effect of a bovine colostrum whey supplementation on growth performance, faecal Escherichia coli population and systemic immune response of piglets at weaning. Animal 2008, 2, 730–737. [Google Scholar] [CrossRef]
- Krueger, L.A.; Beitz, D.C.; Humphrey, S.B.; Stabel, J.R. Gamma delta T cells are early responders to Mycobacterium avium ssp. paratuberculosis in colostrum-replete Holstein calves. J. Dairy Sci. 2016, 99, 9040–9050. [Google Scholar] [CrossRef]
- Williams, B.L.; Hornig, M.; Buie, T.; Bauman, M.L.; Paik, M.C.; Wick, I.; Bennett, A.; Jabado, O.; Hirschberg, D.L.; Lipkin, W.I. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS ONE 2011, 6, e24585. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Griebel, P.J.; le Guan, L. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front. Vet. Sci. 2015, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Malmuthuge, N.; Li, F.; le Guan, L. Colostrum feeding shapes the hindgut microbiota of dairy calves during the first 12 h of life. FEMS Microbiol. Ecol. 2018, 95, fiy203. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, H.; De Buck, J.; Mortier, R.; Barkema, H.W.; Krause, D.O.; Khafipour, E. The features of fecal and ileal mucosa-associated microbiota in dairy calves during early infection with Mycobacterium avium subspecies paratuberculosis. Front. Microbiol. 2016, 7, 426. [Google Scholar] [CrossRef]
- Alipour, M.J.; Jalanka, J.; Pessa-Morikawa, T.; Kokkonen, T.; Satokari, R.; Hynönen, U.; Iivanainen, A.; Niku, M. The composition of the perinatal intestinal microbiota in cattle. Sci. Rep. 2018, 8, 10437. [Google Scholar] [CrossRef]
- Looft, T.; Allen, H.K.; Cantarel, B.L.; Levine, U.Y.; Bayles, D.O.; Alt, D.P.; Henrissat, B.; Stanton, T.B. Bacteria, phages and pigs: The effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014, 8, 1566–1576. [Google Scholar] [CrossRef]
- Kapsenberg, M.L. Dendritic-cell control of pathogen-driven T-cell polarization. Nat. Rev. Immunol. 2003, 3, 984–993. [Google Scholar] [CrossRef]
- Dahan, S.; Roth-Walter, F.; Arnaboldi, P.; Agarwal, S.; Mayer, L. Epithelia: Lymphocyte interactions in the gut. Immunol. Rev. 2007, 215, 243–253. [Google Scholar] [CrossRef]
- Oikonomou, G.; Teixeira, A.G.; Foditsch, C.; Bicalho, M.L.; Machado, V.S.; Bicalho, R.C. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS ONE 2013, 8, e63157. [Google Scholar] [CrossRef]
- Zivkovic, A.M.; Barile, D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv. Nutr. 2011, 2, 284–289. [Google Scholar] [CrossRef]
- Chichlowski, M.; De Lartigue, G.; German, J.B.; Raybould, H.E.; Mills, D.A. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Britton, R.A.; Roos, S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc. Natl. Acad. Sci. USA 2011, 108, 4645–4652. [Google Scholar] [CrossRef] [PubMed]

| Treatment | Culture (cfu/g) a |
|---|---|
| CD | 1072 ± 728 |
| CR | 521 ± 240 |
| CR-A | 1053 ± 691 |
| CF-D | 1972 ± 1094 |
| CR-E | 1635 ± 890 |
| CR-ADE | 153 ± 106 |
| Treatment | DJ | IL | ICV |
|---|---|---|---|
| CD | 0 | 0 | 0 |
| CR | 1 | 1 | 2 |
| CR-A | 2 | 1 | 2 |
| CF-D | 1 | 1 | 0 |
| CR-E | 1 | 2 | 2 |
| CR-ADE | 2 | 3 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stabel, J.; Krueger, L.; Jenvey, C.; Wherry, T.; Hostetter, J.; Beitz, D. Influence of Colostrum and Vitamins A, D3, and E on Early Intestinal Colonization of Neonatal Holstein Calves Infected with Mycobacterium avium subsp. paratuberculosis. Vet. Sci. 2019, 6, 93. https://doi.org/10.3390/vetsci6040093
Stabel J, Krueger L, Jenvey C, Wherry T, Hostetter J, Beitz D. Influence of Colostrum and Vitamins A, D3, and E on Early Intestinal Colonization of Neonatal Holstein Calves Infected with Mycobacterium avium subsp. paratuberculosis. Veterinary Sciences. 2019; 6(4):93. https://doi.org/10.3390/vetsci6040093
Chicago/Turabian StyleStabel, Judith, Lucas Krueger, Caitlin Jenvey, Taylor Wherry, Jesse Hostetter, and Donald Beitz. 2019. "Influence of Colostrum and Vitamins A, D3, and E on Early Intestinal Colonization of Neonatal Holstein Calves Infected with Mycobacterium avium subsp. paratuberculosis" Veterinary Sciences 6, no. 4: 93. https://doi.org/10.3390/vetsci6040093
APA StyleStabel, J., Krueger, L., Jenvey, C., Wherry, T., Hostetter, J., & Beitz, D. (2019). Influence of Colostrum and Vitamins A, D3, and E on Early Intestinal Colonization of Neonatal Holstein Calves Infected with Mycobacterium avium subsp. paratuberculosis. Veterinary Sciences, 6(4), 93. https://doi.org/10.3390/vetsci6040093






