Development of Adenovirus Containing Liposomes Produced by Extrusion vs. Homogenization: A Comparison for Scale-Up Purposes
Abstract
1. Introduction
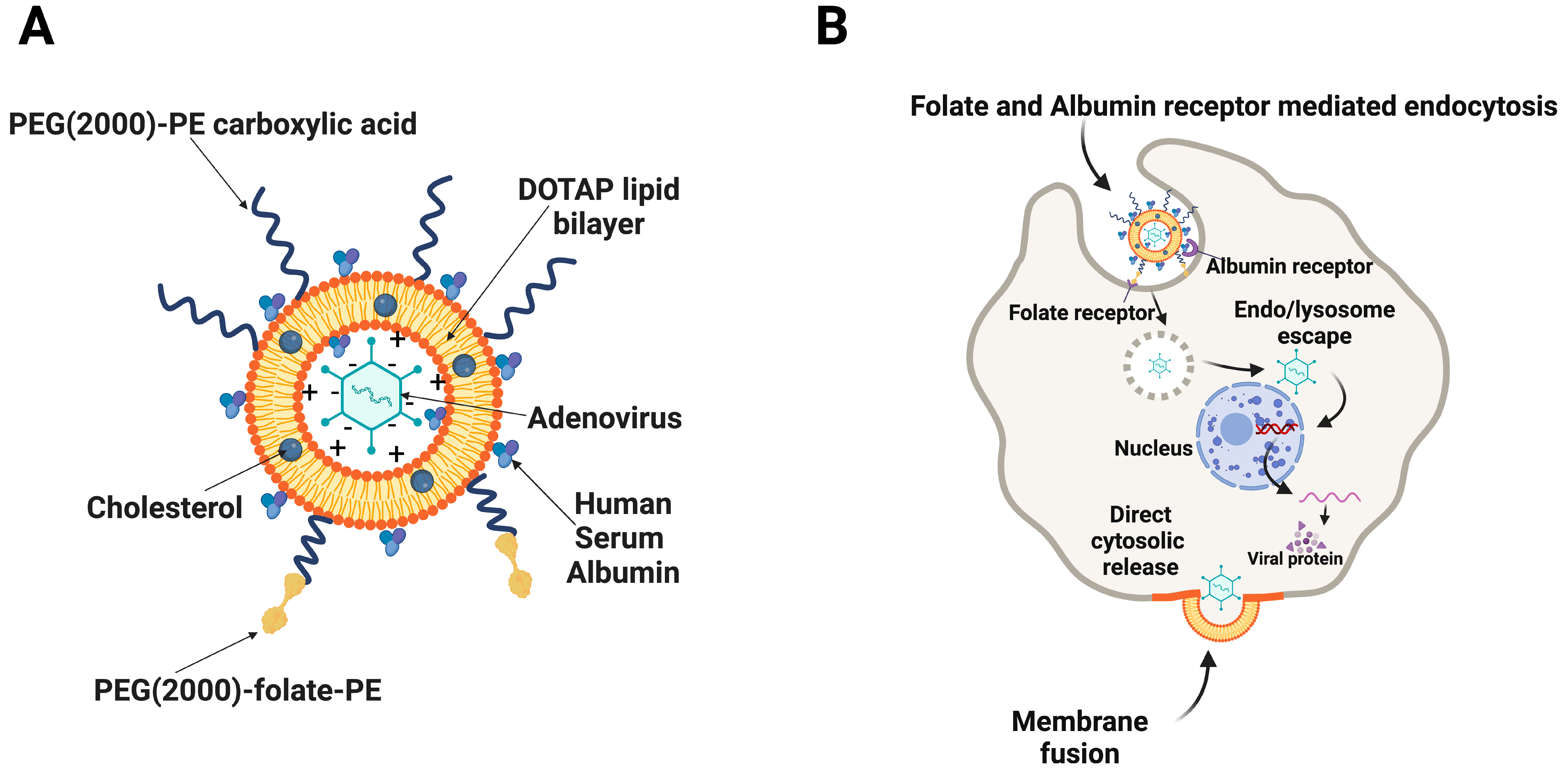
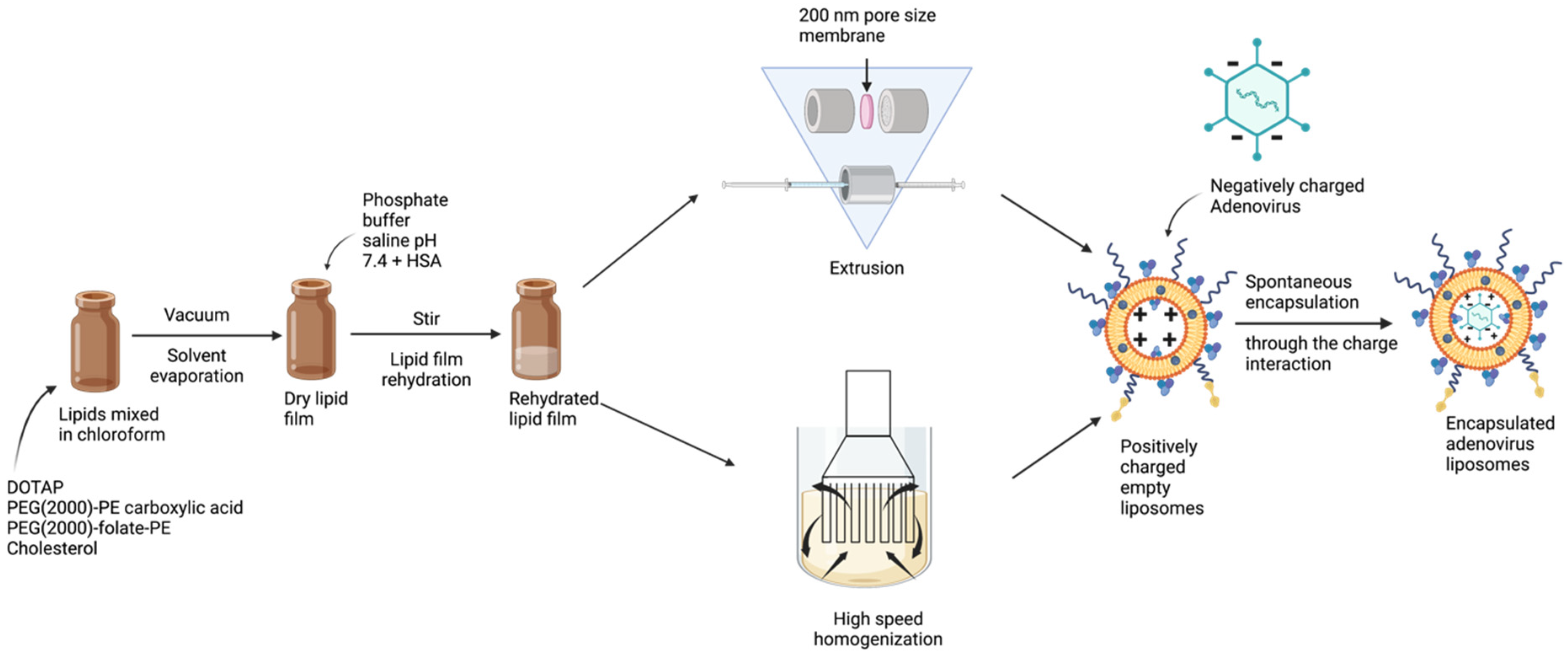
2. Materials and Methods
2.1. Reagents and Cell Lines
2.2. Manufacturing of Liposomes by Extrusion Technique
2.3. Manufacturing of Liposomes by Homogenization Technique
2.4. In Vitro Transduction
2.5. Liposome Formulation Optimization
2.6. Dynamic Light Scattering (DLS) and Zeta Potential Measurements
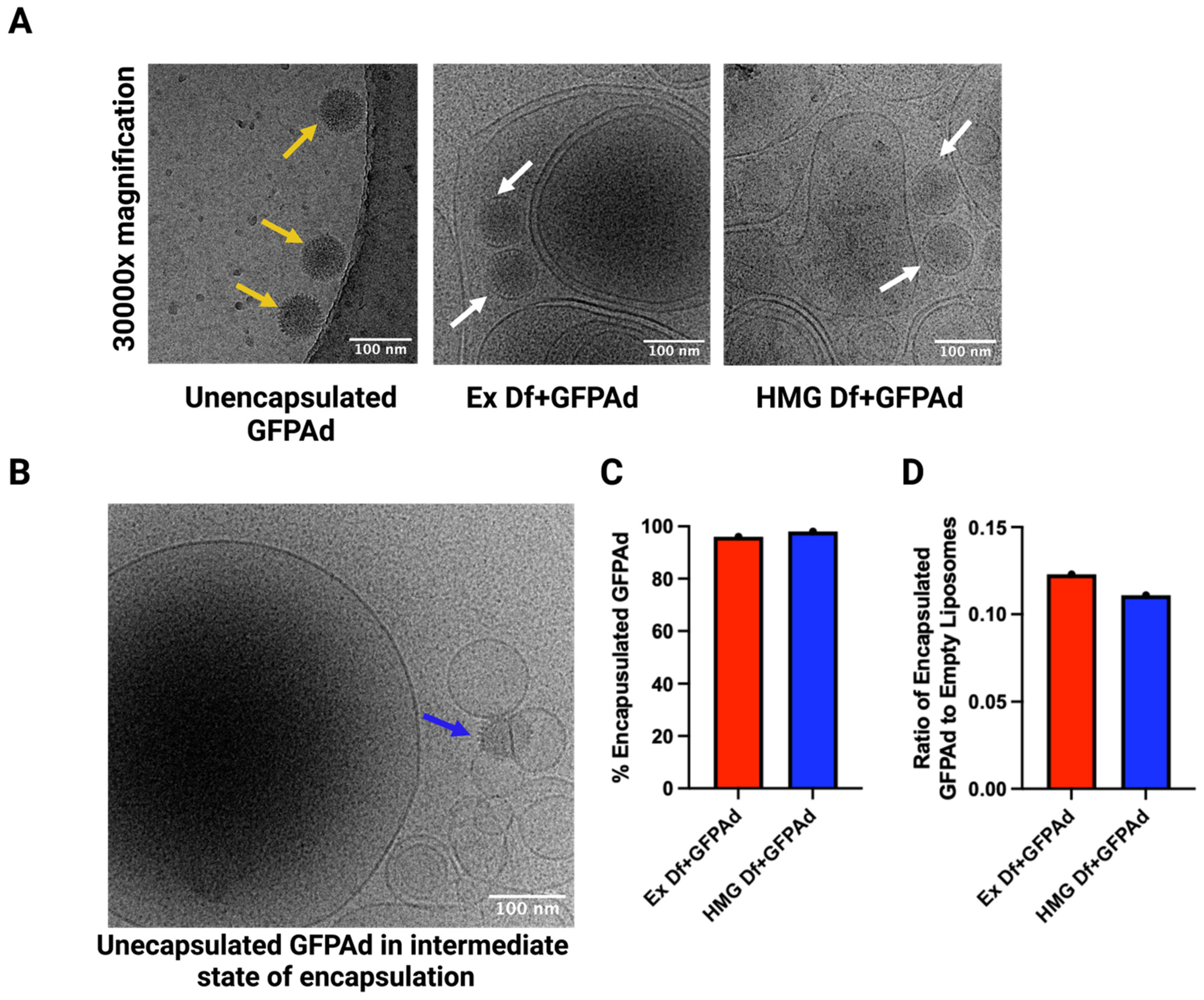
2.7. Cryo-Transmission Electron Microscopy (Cryo-EM)
2.8. Fluorescence Microscopy
2.9. Long-Term Storage Stability of Ad Liposomes
2.10. In Vivo Biodistribution of Ad Liposomes
3. Results
3.1. Liposome Formulation Optimization
3.2. Comparitve Characterization of Ad Liposomes Produced by Extrusion vs. Homogenization
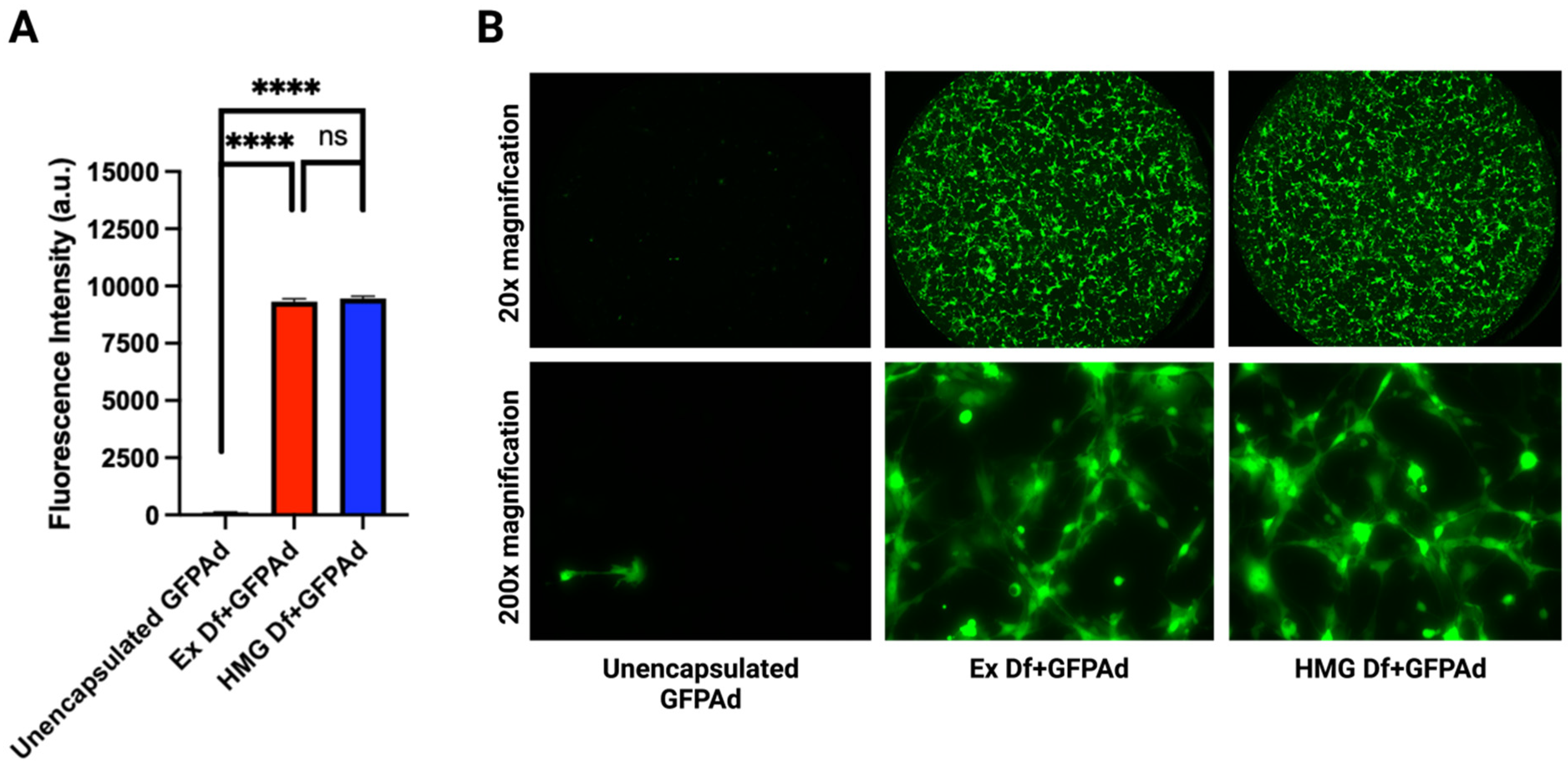
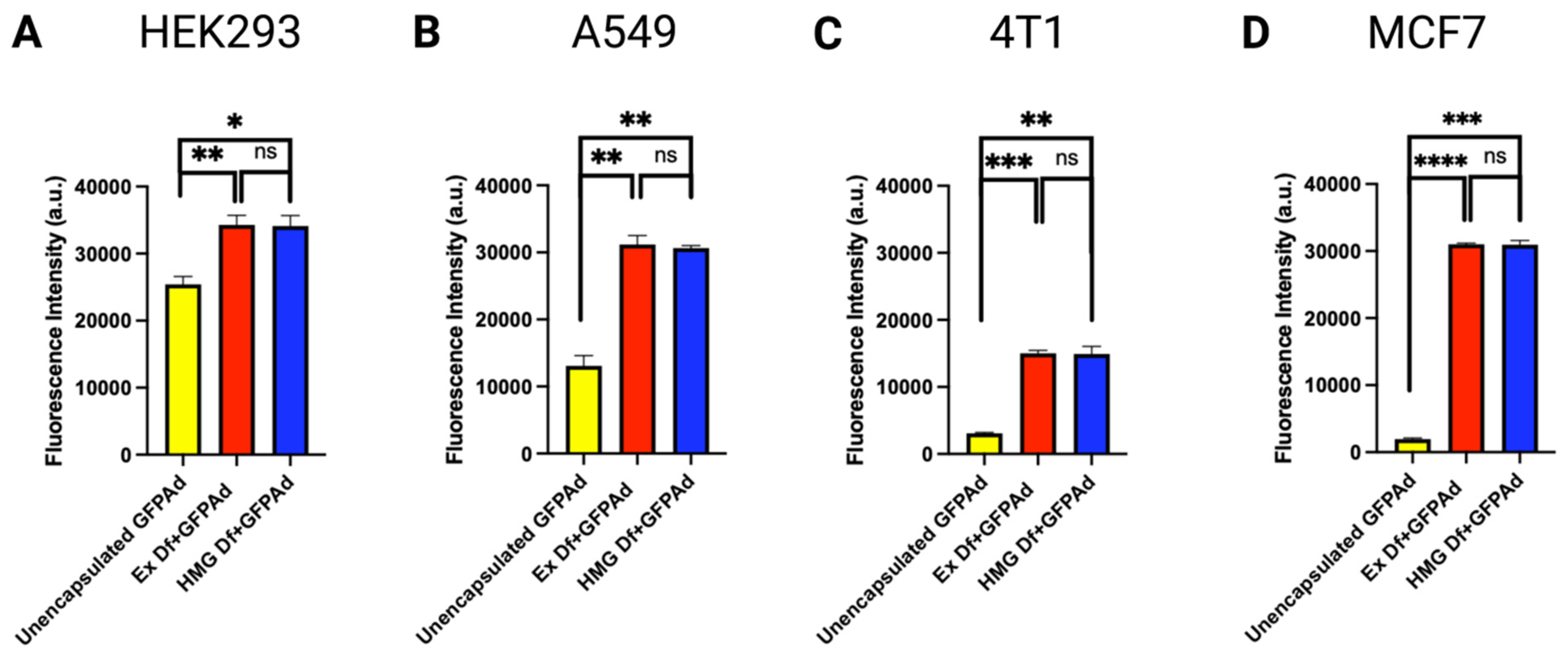
3.3. Comparative In Vitro Transduction of Ad Liposomes Manufactured by Extrusion and Homogenization on CAR Positive and CAR Deficient Cell Lines
3.4. Long-term Storage Stability of Ad Liposomes
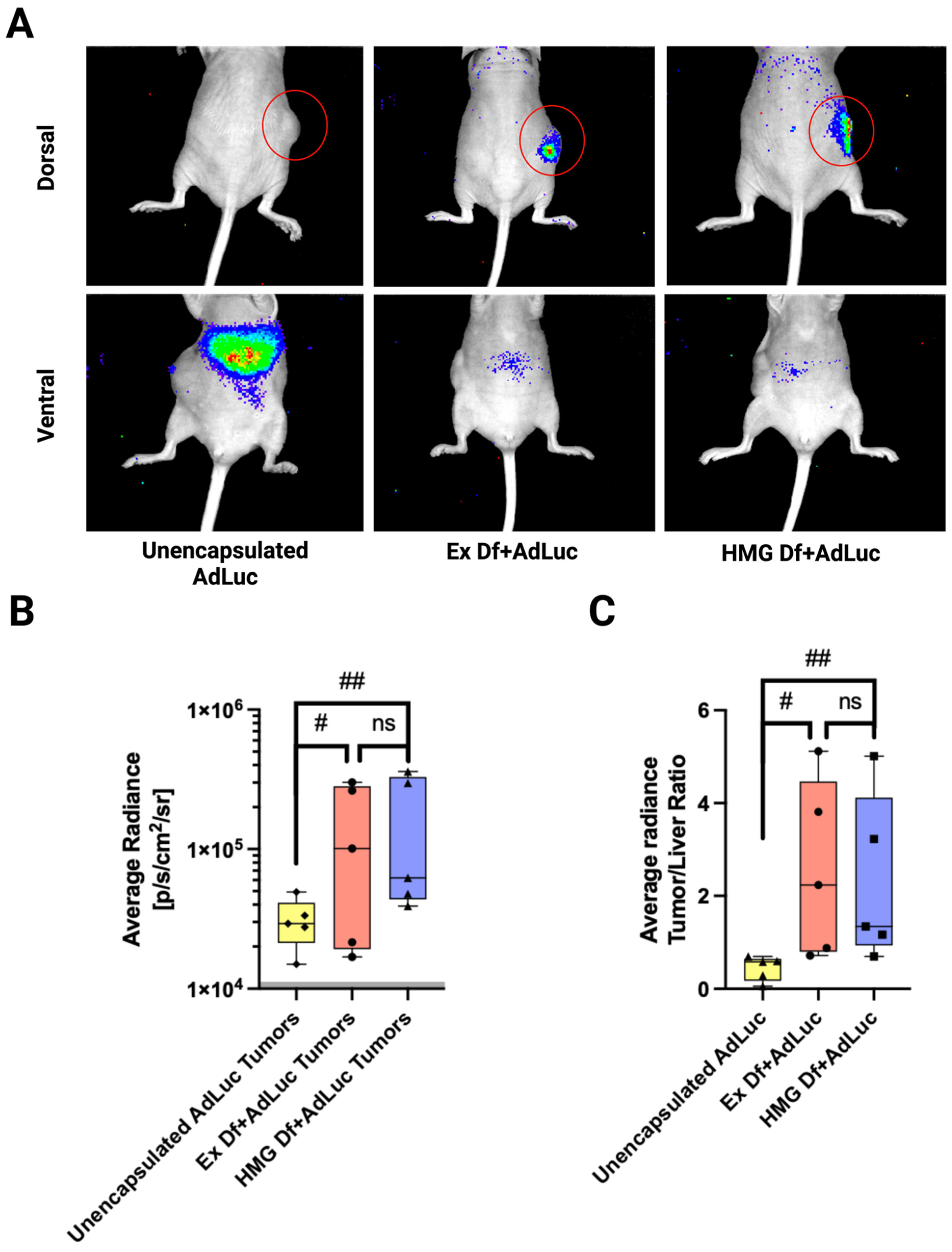
3.5. In Vivo Biodistribution of Ad Liposomes
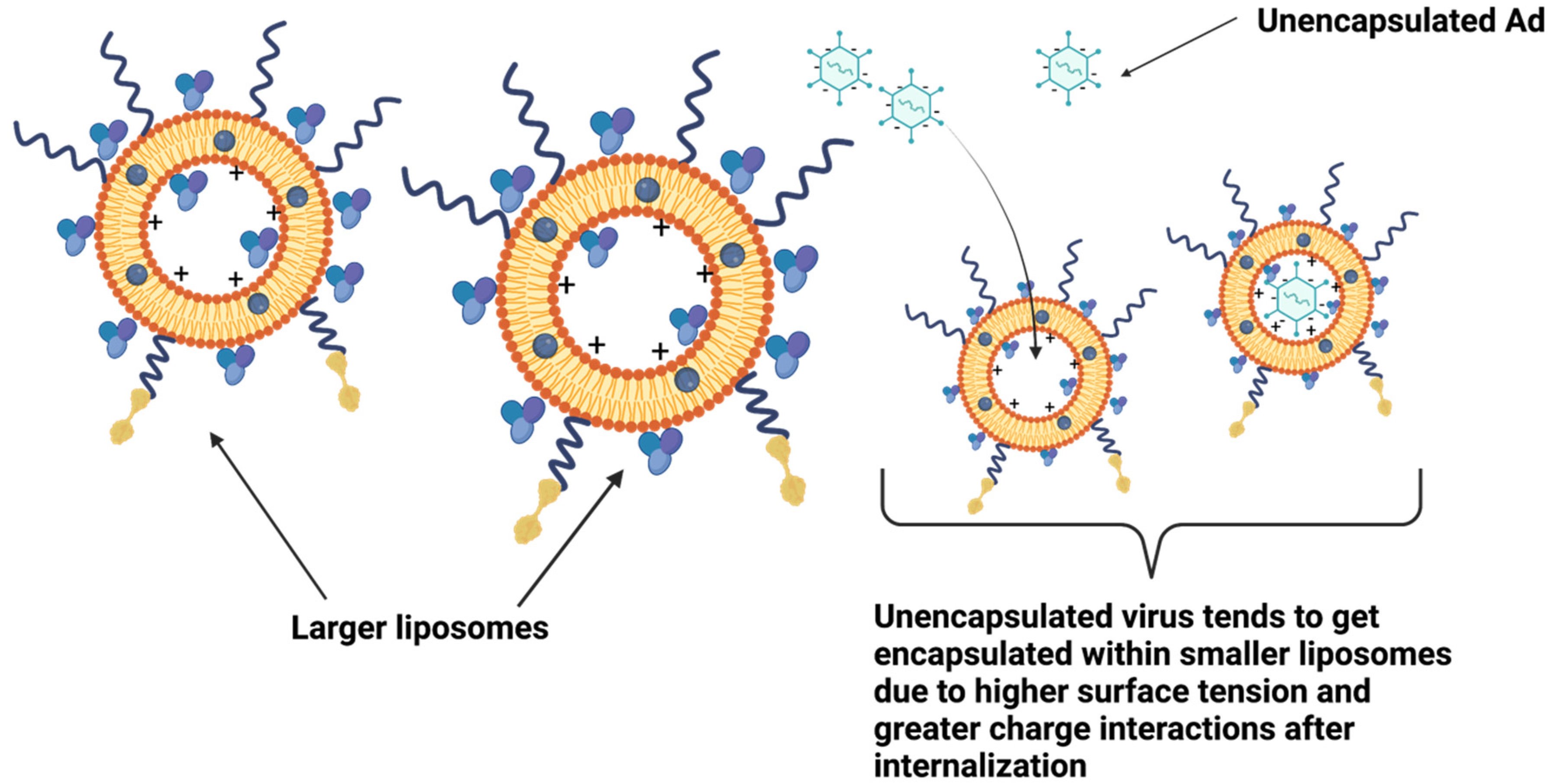
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chiriva-Internati, M.; Bot, A. A new era in cancer immunotherapy: Discovering novel targets and reprogramming the immune system. Int. Rev. Immunol. 2015, 34, 101–103. [Google Scholar] [CrossRef]
- Tazawa, H.; Kagawa, S.; Fujiwara, T. Advances in adenovirus-mediated p53 cancer gene therapy. Expert Opin. Biol. Ther. 2013, 13, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Larson, C.; Oronsky, B.; Abrouk, N.E.; Oronsky, A.; Reid, T.R. Toxicology and biodistribution of AdAPT-001, a replication-competent type 5 adenovirus with a trap for the immunosuppressive cytokine, TGF-beta. Am. J. Cancer Res. 2021, 11, 5184. [Google Scholar] [PubMed]
- Senzer, N.N.; Kaufman, H.L.; Amatruda, T.; Nemunaitis, M.; Reid, T.; Daniels, G.; Gonzalez, R.; Glaspy, J.; Whitman, E.; Harrington, K. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009, 27, 5763. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hedjran, F.; Larson, C.; Perez, G.; Reid, T. A novel immunocompetent murine model for replicating oncolytic adenoviral therapy. Cancer Gene Ther. 2015, 22, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Harrington, K.; Freeman, D.J.; Kelly, B.; Harper, J.; Soria, J.C. Optimizing oncolytic virotherapy in cancer treatment. Nat. Rev. Drug Discov. 2019, 18, 689–706. [Google Scholar] [CrossRef]
- Koch, J.; Schober, S.J.; Hindupur, S.V.; Schöning, C.; Klein, F.G.; Mantwill, K.; Ehrenfeld, M.; Schillinger, U.; Hohnecker, T.; Qi, P.; et al. Targeting the Retinoblastoma/E2F repressive complex by CDK4/6 inhibitors amplifies oncolytic potency of an oncolytic adenovirus. Nat. Commun. 2022, 13, 4689. [Google Scholar] [CrossRef]
- Russell, L.; Peng, K.W. The emerging role of oncolytic virus therapy against cancer. Chin. Clin. Oncol. 2018, 7, 16. [Google Scholar] [CrossRef]
- Samson, A.; West, E.J.; Carmichael, J.; Scott, K.J.; Turnbull, S.; Kuszlewicz, B.; Dave, R.V.; Peckham-Cooper, A.; Tidswell, E.; Kingston, J.; et al. Neoadjuvant Intravenous Oncolytic Vaccinia Virus Therapy Promotes Anticancer Immunity in Patients. Cancer Immunol. Res. 2022, 10, 745–756. [Google Scholar] [CrossRef]
- Hedjran, F.; Shantanu, K.; Tony, R. Deletion analysis of Ad5 E1a transcriptional control region: Impact on tumor-selective expression of E1a and E1b. Cancer Gene Ther. 2011, 18, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Reeh, M.; Bockhorn, M.; Görgens, D.; Vieth, M.; Hoffmann, T.; Simon, R.; Izbicki, J.R.; Sauter, G.; Schumacher, U.; Anders, M. Presence of the Coxsackievirus and Adenovirus Receptor (CAR) in human neoplasms: A multitumour array analysis. Br. J. Cancer 2013, 109, 1848–1858. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhan, Z.; Pan, Y.; Li, J. Expression of coxsackie and adenovirurus receptor and its significance in human lung cancer. Chin. J. Clin. Oncol. 2007, 4, 273–276. [Google Scholar] [CrossRef]
- Kasala, D.; Hong, J.; Yun, C.-O. Overcoming the barriers to optimization of adenovirus delivery using biomaterials: Current status and future perspective. J. Control. Release 2021, 332, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, B.; Li, Q.; Mei, K.; Xu, J.-R.; Li, H.-X.; Wang, Y.-S.; Wen, Y.-J.; Wang, X.-D.; Yang, H.-S. Gene therapy with recombinant adenovirus encoding endostatin encapsulated in cationic liposome in coxsackievirus and adenovirus receptor-deficient colon carcinoma murine models. Hum. Gene Ther. 2011, 22, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef]
- Nakhaei, P.; Margiana, R.; Bokov, D.O.; Abdelbasset, W.K.; Jadidi Kouhbanani, M.A.; Varma, R.S.; Marofi, F.; Jarahian, M.; Beheshtkhoo, N. Liposomes: Structure, Biomedical Applications, and Stability Parameters With Emphasis on Cholesterol. Front. Bioeng. Biotechnol. 2021, 9, 705886. [Google Scholar] [CrossRef]
- Kim, B.-K.; Hwang, G.-B.; Seu, Y.-B.; Choi, J.-S.; Jin, K.S.; Doh, K.-O. DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2015, 1848, 1996–2001. [Google Scholar] [CrossRef]
- Porteous, D.J.; Dorin, J.R.; McLachlan, G.; Davidson-Smith, H.; Davidson, H.; Stevenson, B.J.; Carothers, A.D.; Wallace, W.A.H.; Moralee, S.; Hoenes, C.; et al. Evidence for safety and efficacy of DOTAP cationic liposome mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Gene Ther. 1997, 4, 210–218. [Google Scholar] [CrossRef]
- Templeton, N.S.; Lasic, D.D.; Frederik, P.M.; Strey, H.H.; Roberts, D.D.; Pavlakis, G.N. Improved DNA: Liposome complexes for increased systemic delivery and gene expression. Nat. Biotechnol. 1997, 15, 647–652. [Google Scholar] [CrossRef]
- Yotnda, P.; Chen, D.-H.; Chiu, W.; Piedra, P.A.; Davis, A.; Templeton, N.S.; Brenner, M.K. Bilamellar cationic liposomes protect adenovectors from preexisting humoral immune responses. Mol. Ther. 2002, 5, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Vupputuri, S.; Tayebi, L.; Hikkaduwa Koralege, R.S.; Nigatu, A.; Mozafari, M.; Mishra, A.; Liu, L.; Ramsey, J.D. Polyethylene glycol–modified DOTAP:cholesterol/adenovirus hybrid vectors have improved transduction efficiency and reduced immunogenicity. J. Nanoparticle Res. 2021, 23, 37. [Google Scholar] [CrossRef]
- Berger, N.; Sachse, A.; Bender, J.; Schubert, R.; Brandl, M. Filter extrusion of liposomes using different devices: Comparison of liposome size, encapsulation efficiency, and process characteristics. Int. J. Pharm. 2001, 223, 55–68. [Google Scholar] [CrossRef]
- Zhang, H. Thin-film hydration followed by extrusion method for liposome preparation. In Liposomes; Springer: Berlin/Heidelberg, Germany, 2017; pp. 17–22. [Google Scholar]
- Lincoln, J.E. Overview of the us fda gmps: Good manufacturing practice (gmp)/quality system (qs) regulation (21 CFR part 820). J. Valid. Technol. 2012, 18, 17. [Google Scholar]
- Shah, V.M.; Nguyen, D.X.; Patel, P.; Cote, B.; Al-Fatease, A.; Pham, Y.; Huynh, M.G.; Woo, Y.; Alani, A.W. Liposomes produced by microfluidics and extrusion: A comparison for scale-up purposes. Nanomed. Nanotechnol. Biol. Med. 2019, 18, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Maja, L.; Željko, K.; Mateja, P. Sustainable technologies for liposome preparation. J. Supercrit. Fluids 2020, 165, 104984. [Google Scholar] [CrossRef]
- Martin, F.J.; Morano, J.K. Liposome Extrusion Method. US Patent No. 4,737,323, 1988. U.S.P.T.O.. [Google Scholar]
- Ong, S.G.M.; Chitneni, M.; Lee, K.S.; Ming, L.C.; Yuen, K.H. Evaluation of extrusion technique for nanosizing liposomes. Pharmaceutics 2016, 8, 36. [Google Scholar] [CrossRef]
- Beltrán, J.D.; Ricaurte, L.; Estrada, K.B.; Quintanilla-Carvajal, M.X. Effect of homogenization methods on the physical stability of nutrition grade nanoliposomes used for encapsulating high oleic palm oil. LWT 2020, 118, 108801. [Google Scholar] [CrossRef]
- Jensen, G.M.; Bunch, T.H.; Hu, N.; Eley, C.G. Process development and quality control of injectable liposomal therapeutics. In Liposome Technology; CRC Press: Boca Raton, FL, USA, 2018; pp. 319–332. [Google Scholar]
- Jaradat, E.; Weaver, E.; Meziane, A.; Lamprou, D.A. Microfluidics technology for the design and formulation of nanomedicines. Nanomaterials 2021, 11, 3440. [Google Scholar] [CrossRef]
- Ferguson, M.S.; Lemoine, N.R.; Wang, Y. Systemic delivery of oncolytic viruses: Hopes and hurdles. Adv. Virol. 2012, 2012, 805629. [Google Scholar] [CrossRef]
- Reid, T.; Galanis, E.; Abbruzzese, J.; Sze, D.; Wein, L.M.; Andrews, J.; Randlev, B.; Heise, C.; Uprichard, M.; Hatfield, M. Hepatic Arterial Infusion of a Replication-selective Oncolytic Adenovirus (dl 1520) Phase II Viral, Immunologic, and Clinical Endpoints. Cancer Res. 2002, 62, 6070–6079. [Google Scholar] [PubMed]
- Yumul, R.; Richter, M.; Lu, Z.Z.; Saydaminova, K.; Wang, H.; Wang, C.H.; Carter, D.; Lieber, A. Epithelial Junction Opener Improves Oncolytic Adenovirus Therapy in Mouse Tumor Models. Hum. Gene Ther. 2016, 27, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.; Bax, H.J.; Josephs, D.H.; Ilieva, K.M.; Pellizzari, G.; Opzoomer, J.; Bloomfield, J.; Fittall, M.; Grigoriadis, A.; Figini, M. Targeting folate receptor alpha for cancer treatment. Oncotarget 2016, 7, 52553. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Low, P.S. Immunotherapy of folate receptor-expressing tumors: Review of recent advances and future prospects. J. Control. Release 2003, 91, 17–29. [Google Scholar] [CrossRef]
- Meier, R.; Henning, T.D.; Boddington, S.; Tavri, S.; Arora, S.; Piontek, G.; Rudelius, M.; Corot, C.; Daldrup-Link, H.E. Breast cancers: MR imaging of folate-receptor expression with the folate-specific nanoparticle P1133. Radiology 2010, 255, 527–535. [Google Scholar] [CrossRef]
- Hassanin, I.; Elzoghby, A. Albumin-based nanoparticles: A promising strategy to overcome cancer drug resistance. Cancer Drug Resist. 2020, 3, 930. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Bell, P.; Vandenberghe, L.H.; Wu, D.; Johnston, J.; Limberis, M.; Wilson, J.M. A comparative analysis of novel fluorescent proteins as reporters for gene transfer studies. J. Histochem. Cytochem. 2007, 55, 931–939. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Tsai, T.-H.; Chang, C.-P.; Chen, S.-F.; Lee, Y.-M.; Shyue, S.-K. Linear correlation between average fluorescence intensity of green fluorescent protein and the multiplicity of infection of recombinant adenovirus. J. Biomed. Sci. 2015, 22, 31. [Google Scholar] [CrossRef]
- Kaddah, S.; Khreich, N.; Kaddah, F.; Charcosset, C.; Greige-Gerges, H. Cholesterol modulates the liposome membrane fluidity and permeability for a hydrophilic molecule. Food Chem. Toxicol. 2018, 113, 40–48. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, Q.; Sun, J.; Gu, A.; Jin, M.; Shen, Z.; Qiu, Z.; Wang, J.; Wang, X.; Zhan, Z. Expression of the coxsackie and adenovirus receptor in human lung cancers. Tumor Biol. 2013, 34, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Houri, N.; Huang, K.-C.; Nalbantoglu, J. The Coxsackievirus and Adenovirus Receptor (CAR) undergoes ectodomain shedding and regulated intramembrane proteolysis (RIP). PLoS ONE 2013, 8, e73296. [Google Scholar] [CrossRef] [PubMed]
- Auer, D.; Reimer, D.; Porto, V.; Fleischer, M.; Roessler, J.; Wiedemair, A.; Marth, C.; Müller-Holzner, E.; Daxenbichler, G.; Zeimet, A.G. Expression of coxsackie-adenovirus receptor is related to estrogen sensitivity in breast cancer. Breast Cancer Res. Treat. 2009, 116, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, X.; Zhao, J.; Shi, W.; Zhang, H.; Wang, Y.; Kan, B.; Du, L.; Wang, B.; Wei, Y. A tumor-selective biotherapy with prolonged impact on established metastases based on cytokine gene-engineered MSCs. Mol. Ther. 2008, 16, 749–756. [Google Scholar] [CrossRef]
- Das, S.K.; Menezes, M.E.; Bhatia, S.; Wang, X.Y.; Emdad, L.; Sarkar, D.; Fisher, P.B. Gene therapies for cancer: Strategies, challenges and successes. J. Cell. Physiol. 2015, 230, 259–271. [Google Scholar] [CrossRef]
- Goverdhana, S.; Puntel, M.; Xiong, W.; Zirger, J.; Barcia, C.; Curtin, J.; Soffer, E.; Mondkar, S.; King, G.; Hu, J. Regulatable gene expression systems for gene therapy applications: Progress and future challenges. Mol. Ther. 2005, 12, 189–211. [Google Scholar] [CrossRef]
- Phillips, A.J. The challenge of gene therapy and DNA delivery. J. Pharm. Pharmacol. 2001, 53, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Aurelian, L. Oncolytic viruses as immunotherapy: Progress and remaining challenges. OncoTargets Ther. 2016, 9, 2627. [Google Scholar] [CrossRef] [PubMed]
- Chaurasiya, S.; Fong, Y.; Warner, S.G. Optimizing oncolytic viral design to enhance antitumor efficacy: Progress and challenges. Cancers 2020, 12, 1699. [Google Scholar] [CrossRef]
- Gazolu-Rusanova, D.; Lesov, I.; Tcholakova, S.; Denkov, N.; Ahtchi, B. Food grade nanoemulsions preparation by rotor-stator homogenization. Food Hydrocoll. 2020, 102, 105579. [Google Scholar] [CrossRef]
- Wagner, A.; Platzgummer, M.; Kreismayr, G.; Quendler, H.; Stiegler, G.; Ferko, B.; Vecera, G.; Vorauer-Uhl, K.; Katinger, H. GMP production of liposomes—A new industrial approach. J. Liposome Res. 2006, 16, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-n.; Wei, Q.; Zhao, L.-J.; Fan, C.-Q.; Guo, L.-R.; Ye, J.-F.; Li, Y. Hydrogel-based co-delivery of CIK cells and oncolytic adenovirus armed with IL12 and IL15 for cancer immunotherapy. Biomed. Pharmacother. 2022, 151, 113110. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhao, H.; Shan, M.; Chen, H.; Xu, B.; He, Y.; Zhao, Y.; Liu, Z.; Chen, J.; Xu, Q. Oncolytic adenovirus H101 ameliorate the efficacy of anti-PD-1 monotherapy in colorectal cancer. Cancer Med. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sangro, B.; Mazzolini, G.; Ruiz, J.; Herraiz, M.; Quiroga, J.; Herrero, I.; Benito, A.; Larrache, J.; Pueyo, J.; Subtil, J.C. Phase I trial of intratumoral injection of an adenovirus encoding interleukin-12 for advanced digestive tumors. J. Clin. Oncol. 2004, 22, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
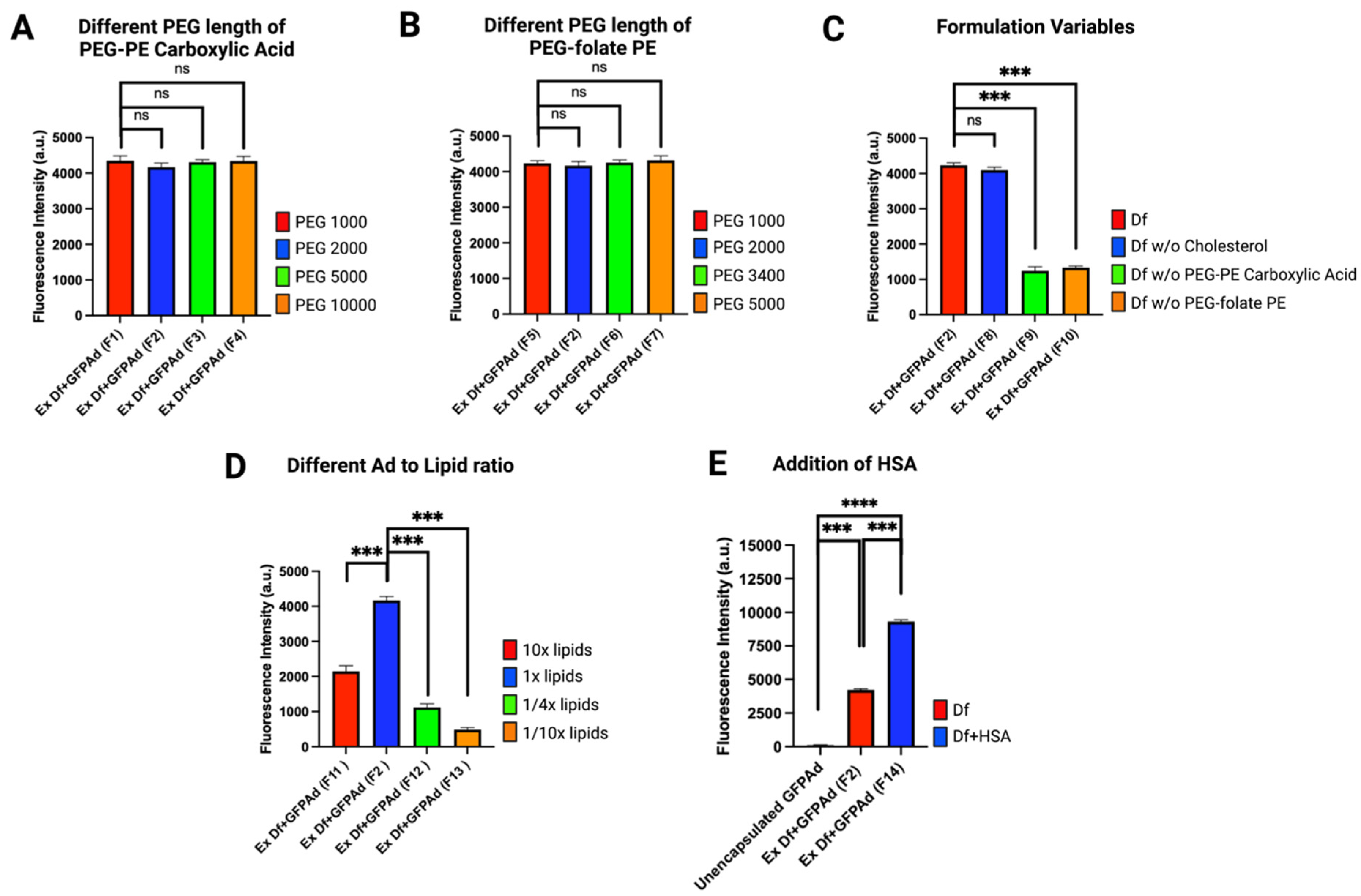

| Formulation 1 | Lipid Film Composition (Molar Ratio) (DOTAP:Cholesterol: PEG-PE Carboxylic Acid: PEG-Folate-PE) | Ingredient Used/Removed |
|---|---|---|
| F1 | 1:0.26:0.02:0.01 | PEG(1000)-PE carboxylic acid |
| F2 | 1:0.26:0.02:0.01 | PEG(2000)-PE carboxylic acid |
| F3 | 1:0.26:0.02:0.01 | PEG(5000)-PE carboxylic acid |
| F4 | 1:0.26:0.02:0.01 | PEG(10000)-PE carboxylic acid |
| F5 | 1:0.26:0.02:0.01 | PEG(1000)-folate-PE |
| F6 | 1:0.26:0.02:0.01 | PEG(3400)-folate-PE |
| F7 | 1:0.26:0.02:0.01 | PEG(5000)-folate-PE |
| F8 | 1:0:0.02:0.01 | F2-w/o cholesterol |
| F9 | 1:0.26:0:0.01 | F2-w/o PEG-PE carboxylic acid |
| F10 | 1:0.26:0.02:0 | F2-w/o PEG-folate PE |
| F11 | 1:0.26:0.02:0.01 | 10× lipid concentration |
| F12 | 1:0.26:0.02:0.01 | 1/4× lipid concentration |
| F13 | 1:0.26:0.02:0.01 | 1/10× lipid concentration |
| F14 | 1:0.26:0.02:0.01 | F2-with HSA |
| Formulation | z-Average (nm) | Polydispersity Index (PDI) | Zeta Potential (mV) |
|---|---|---|---|
| Ex Df (Empty Liposomes) | 119 ± 5 | 0.63 ± 0.06 | 3.26 ± 2.84 |
| Ex Df + GFPAd | 140 ± 1 | 0.53 ± 0.01 | −6.05 ± 0.83 |
| HMG Df (Empty Liposomes) | 113 ± 1 | 0.71 ± 0.00 | 3.80 ± 1.55 |
| HMG Df + GFPAd | 135.9 ± 3.6 | 0.40 ± 0.08 | −5.19 ± 1.11 |
| Unencapsulated GFPAd | 118 ± 0 | 0.09 ± 0.00 | −2.58 ± 0.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, J.R.; Dong, T.; Phung, A.T.; Reid, T.; Larson, C.; Sanchez, A.B.; Oronsky, B.; Blair, S.L.; Aisagbonhi, O.; Trogler, W.C.; et al. Development of Adenovirus Containing Liposomes Produced by Extrusion vs. Homogenization: A Comparison for Scale-Up Purposes. Bioengineering 2022, 9, 620. https://doi.org/10.3390/bioengineering9110620
Shah JR, Dong T, Phung AT, Reid T, Larson C, Sanchez AB, Oronsky B, Blair SL, Aisagbonhi O, Trogler WC, et al. Development of Adenovirus Containing Liposomes Produced by Extrusion vs. Homogenization: A Comparison for Scale-Up Purposes. Bioengineering. 2022; 9(11):620. https://doi.org/10.3390/bioengineering9110620
Chicago/Turabian StyleShah, Jaimin R., Tao Dong, Abraham T. Phung, Tony Reid, Christopher Larson, Ana B. Sanchez, Bryan Oronsky, Sarah L. Blair, Omonigho Aisagbonhi, William C. Trogler, and et al. 2022. "Development of Adenovirus Containing Liposomes Produced by Extrusion vs. Homogenization: A Comparison for Scale-Up Purposes" Bioengineering 9, no. 11: 620. https://doi.org/10.3390/bioengineering9110620
APA StyleShah, J. R., Dong, T., Phung, A. T., Reid, T., Larson, C., Sanchez, A. B., Oronsky, B., Blair, S. L., Aisagbonhi, O., Trogler, W. C., & Kummel, A. C. (2022). Development of Adenovirus Containing Liposomes Produced by Extrusion vs. Homogenization: A Comparison for Scale-Up Purposes. Bioengineering, 9(11), 620. https://doi.org/10.3390/bioengineering9110620








