Broadening the Horizons of RNA Delivery Strategies in Cancer Therapy
Abstract
1. Introduction
2. The Challenge of RNA Delivery
2.1. Strategies for Using RNA Interference
2.2. Strategies for Using mRNA
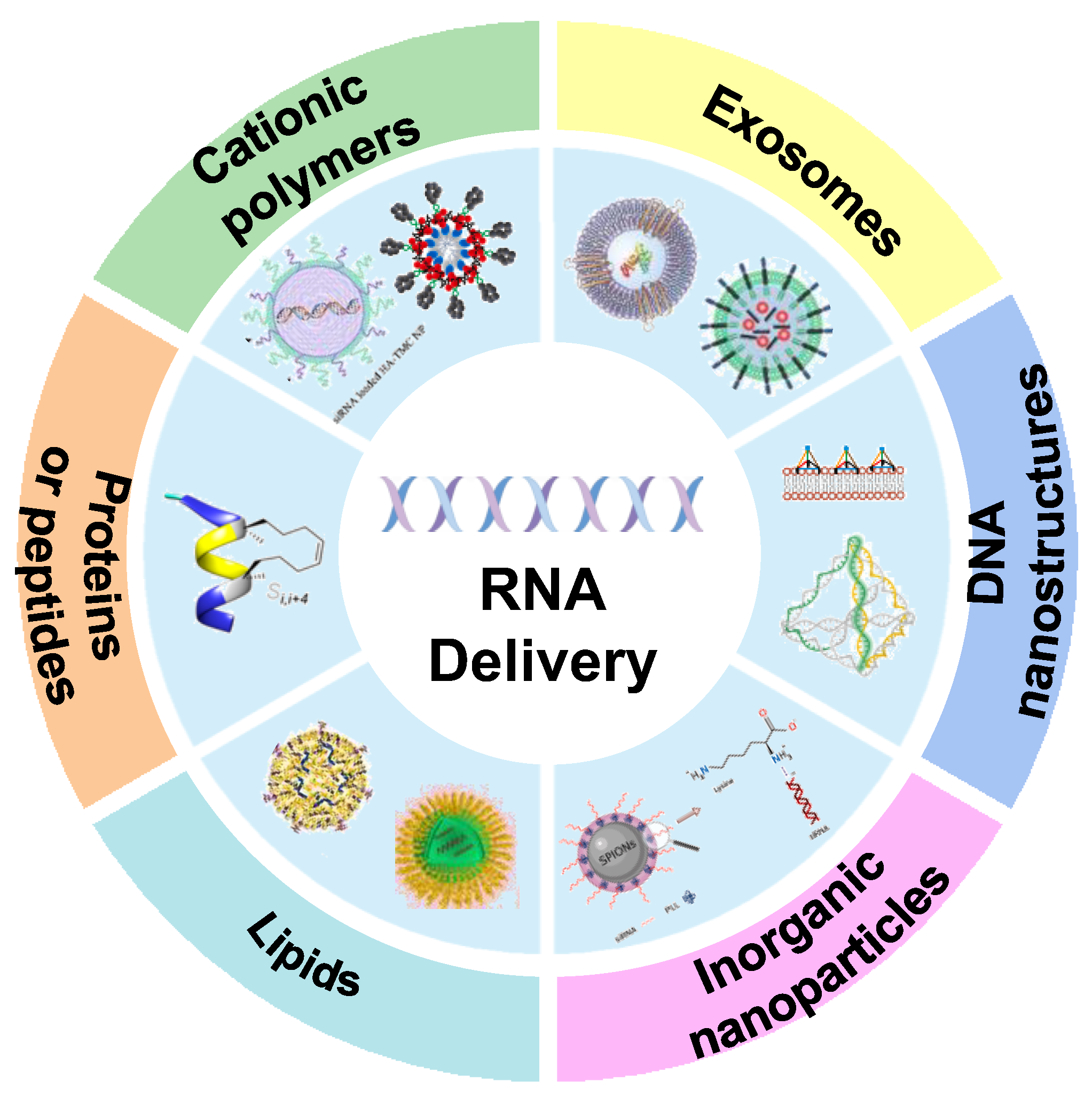
3. Classification of RNA Drug Carrier
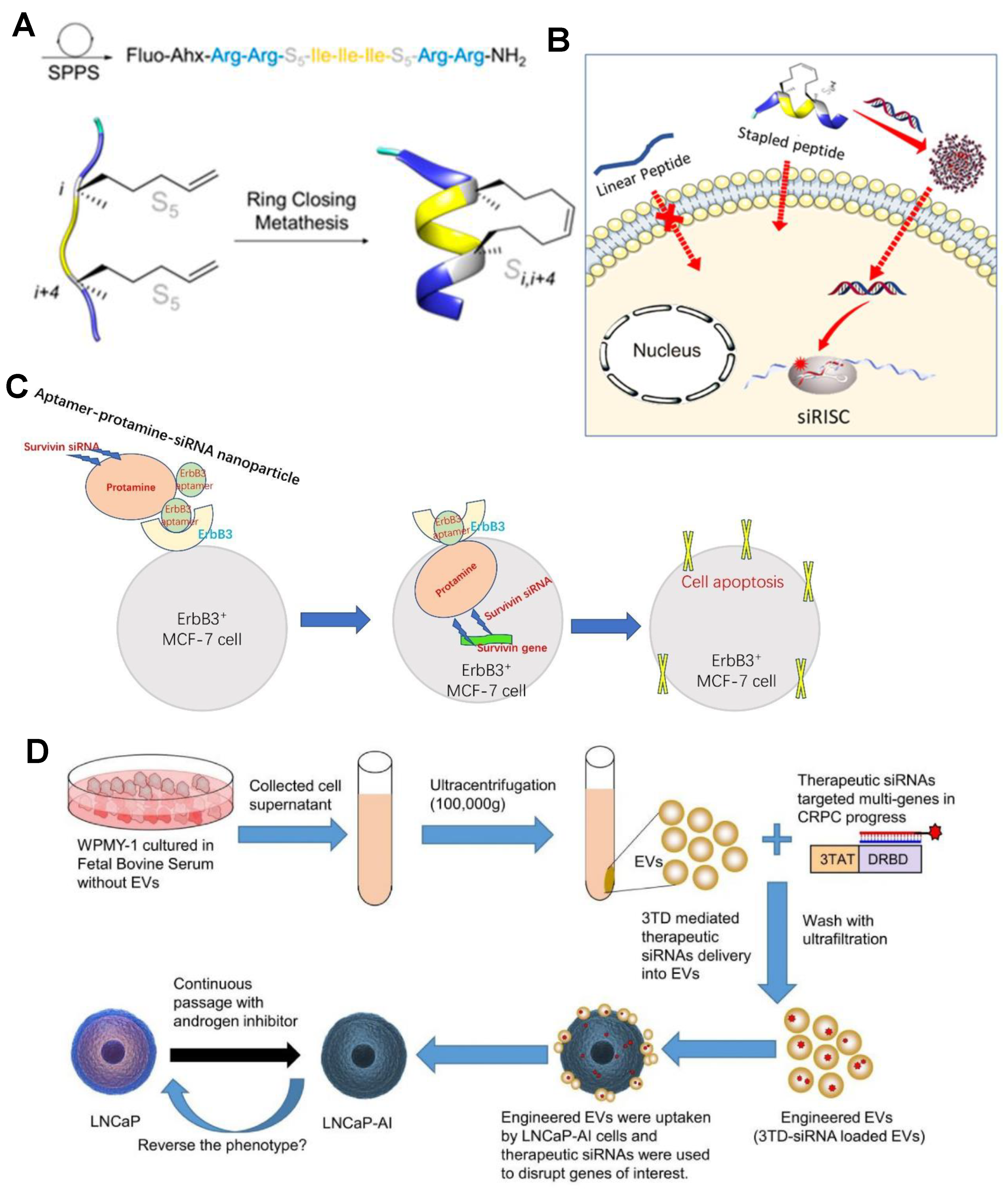
3.1. Cationic Polymers
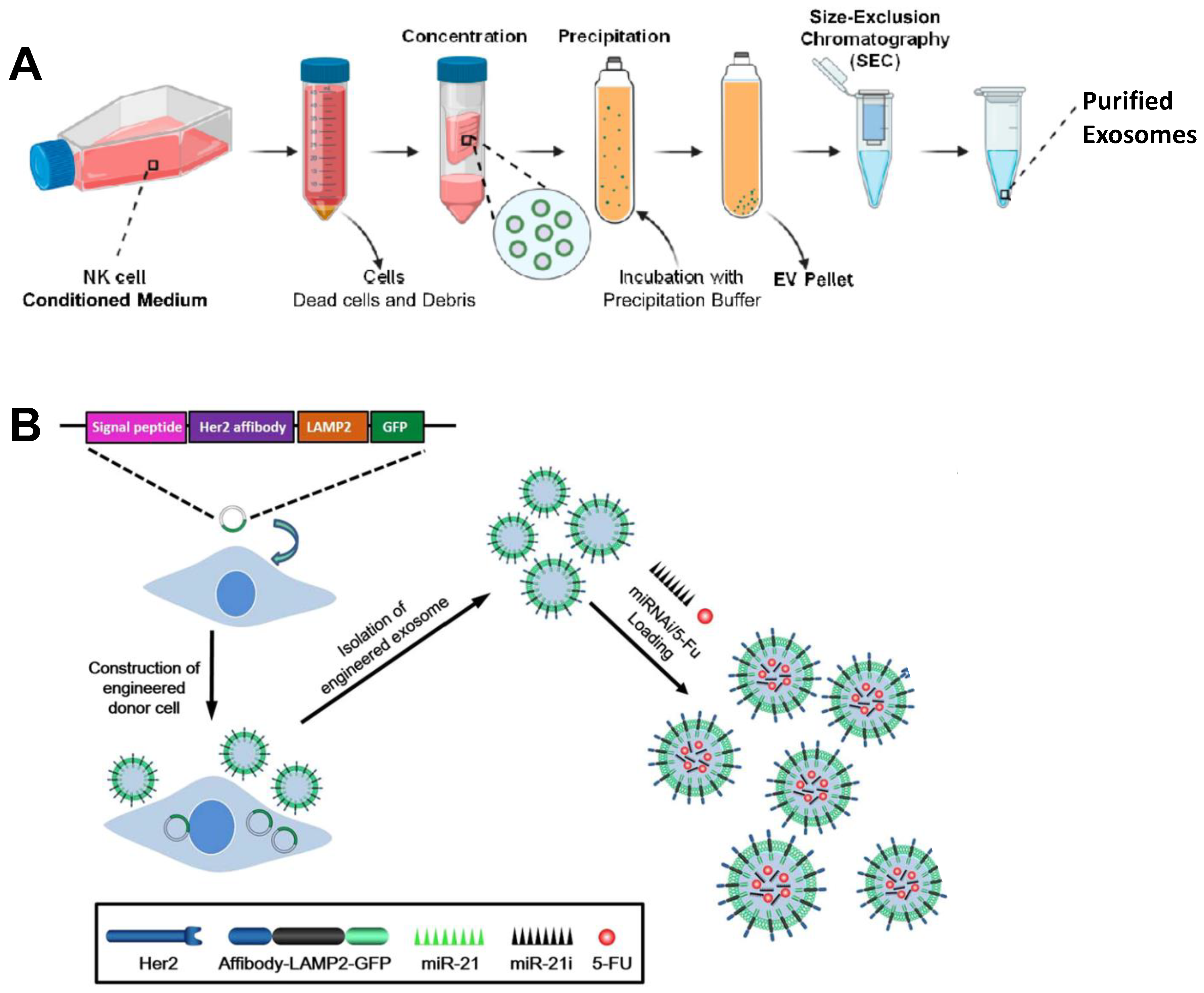
3.2. Exosomes
3.3. DNA Nanostructures
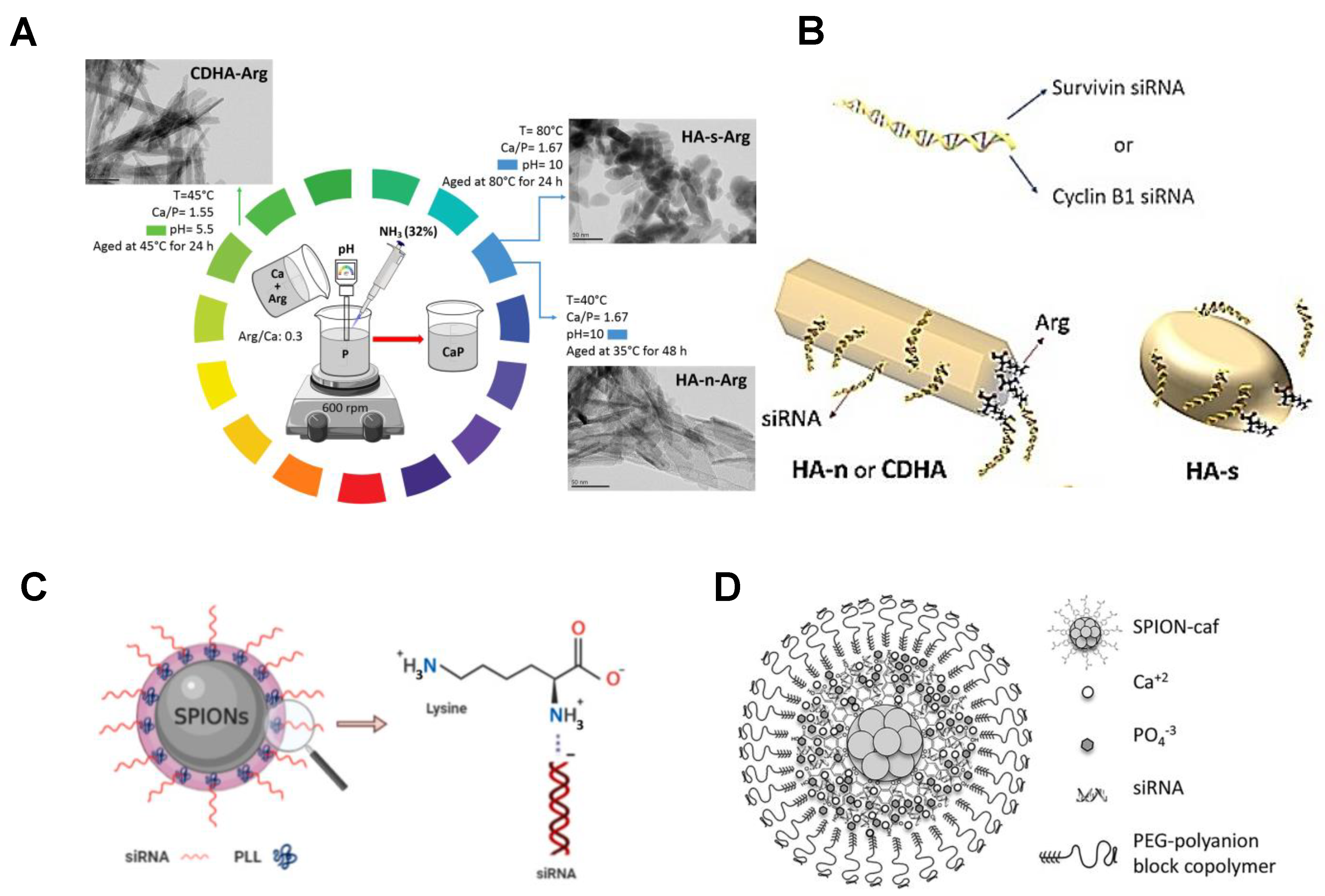
3.4. Inorganic Nanoparticles
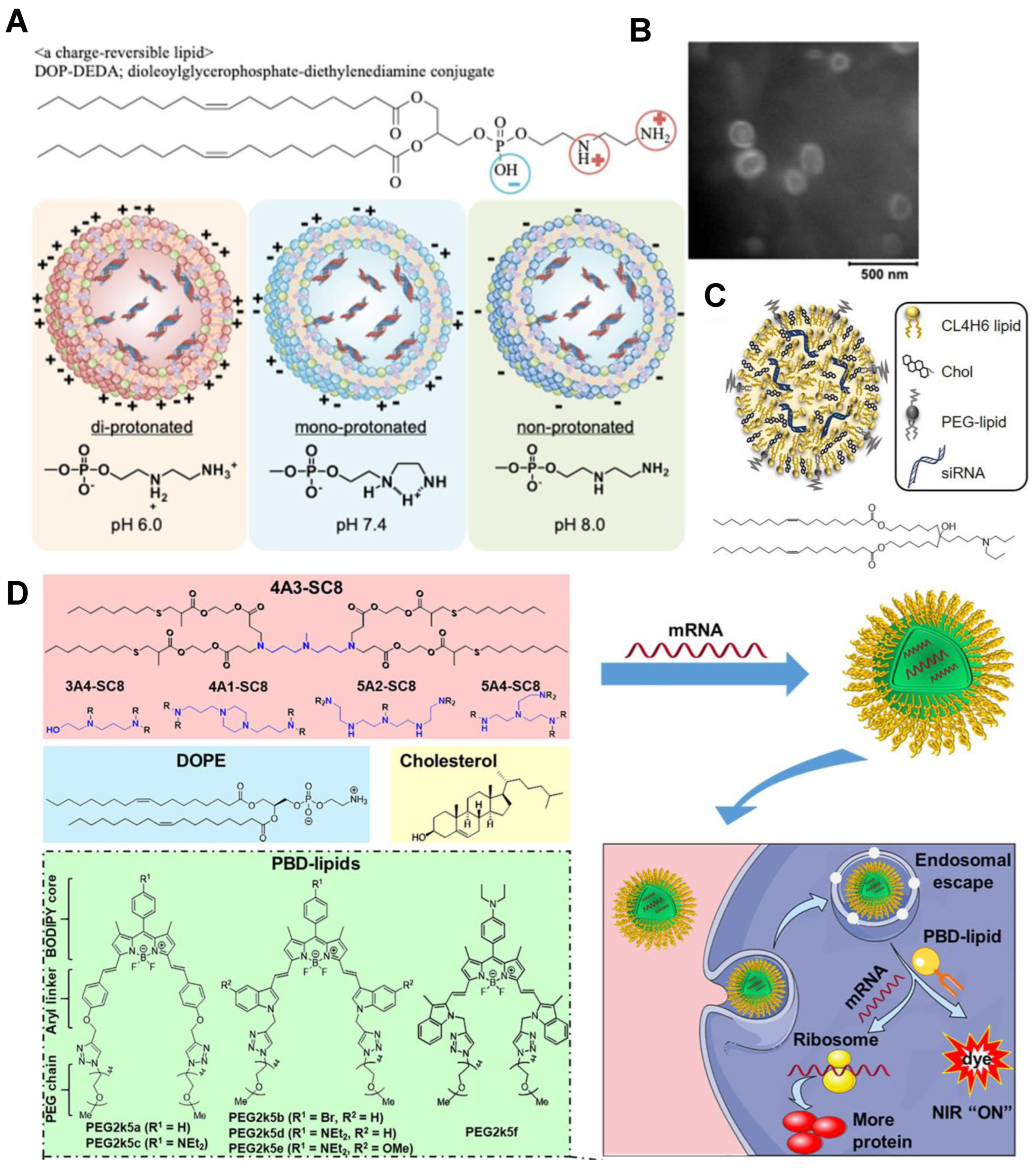
3.5. Lipid-Based Carriers
3.6. Protein or Peptide as Carriers
4. RNA Delivery in Clinical Practice
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, J.; Rao, L.; Yu, G.; Cook, T.R.; Chen, X.; Huang, F. Supramolecular cancer nanotheranostics. Chem. Soc. Rev. 2021, 50, 2839–2891. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Li, M.; Dey, R.; Chen, Y. Nanomaterials for cancer therapy: Current progress and perspectives. J. Hematol. Oncol. 2021, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Anguela, X.M.; High, K.A. Entering the modern era of gene therapy. Annu. Rev. Med. 2019, 70, 273–288. [Google Scholar] [CrossRef]
- Jeon, Y.H.; Jung, Y.T. Production of a replicating retroviral vector expressing reovirus fast protein for cancer gene therapy. J. Virol. Methods 2022, 299, 114332. [Google Scholar] [CrossRef]
- Duan, L.; Xu, L.; Xu, X.; Qin, Z.; Zhou, X.; Xiao, Y.; Liang, Y.; Xia, J. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale 2021, 13, 1387–1397. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Rivas-Garcia, L.; Baptista, P.V.; Fernandes, A.R. Gene therapy in cancer treatment: Why go nano? Pharmaceutics 2020, 12, 233. [Google Scholar] [CrossRef] [PubMed]
- Witzigmann, D.; Kulkarni, J.A.; Leung, J.; Chen, S.; Cullis, P.R.; Van der Meel, R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliver. Rev. 2020, 159, 344–363. [Google Scholar] [CrossRef] [PubMed]
- Van de Vyver, T.; De Smedt, S.C.; Raemdonck, K. Modulating intracellular pathways to improve non-viral delivery of RNA therapeutics. Adv. Drug Deliv Rev. 2022, 181, 114041. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Lin, Y.X.; Wang, Y.; Blake, S.; Yu, M.; Mei, L.; Wang, H.; Shi, J. RNA nanotechnology-mediated cancer immunotherapy. Theranostics 2020, 10, 281–299. [Google Scholar] [CrossRef]
- Phua, K.K.; Staats, H.F.; Leong, K.W.; Nair, S.K. Intranasal mRNA nanoparticle vaccination induces prophylactic and therapeutic anti-tumor immunity. Sci. Rep. 2014, 4, 5128. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar] [PubMed]
- Alexander, R.; David, R.D. A new two stranded helical structure: Polyadenylic acid and polyuridylic acidj. Am. Chem. Soc. 1956, 78, 3548–3549. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Conry, R.M.; LoBuglio, A.F.; Wright, M.; Sumerel, L.; Pike, M.J.; Johanning, F.; Benjamin, R.; Lu, D.; Curiel, D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995, 55, 1397–1400. [Google Scholar] [CrossRef]
- Weide, B.; Carralot, J.P.; Reese, A.; Scheel, B.; Eigentler, T.K.; Hoerr, I.; Rammensee, H.G.; Garbe, C.; Pascolo, S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008, 31, 180–188. [Google Scholar] [CrossRef]
- Kim, Y.K. RNA therapy: Rich history, various applications and unlimited future prospects. Exp. Mol. Med. 2022, 54, 455–465. [Google Scholar] [CrossRef]
- Cox, A.; Lim, S.A.; Chung, E.J. Strategies to deliver RNA by nanoparticles for therapeutic potential. Mol. Asp. Med. 2022, 83, 100991. [Google Scholar] [CrossRef]
- Yoo, Y.J.; Lee, C.H.; Park, S.H.; Lim, Y.T. Nanoparticle-based delivery strategies of multifaceted immunomodulatory RNA for cancer immunotherapy. J. Control. Release 2022, 343, 564–583. [Google Scholar] [CrossRef]
- Goswami, R.; Subramanian, G.; Silayeva, L.; Newkirk, I.; Doctor, D.; Chawla, K.; Chattopadhyay, S.; Chandra, D.; Chilukuri, N.; Betapudi, V. Gene therapy leaves a vicious cycle. Front. Oncol. 2019, 9, 297. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wang, J.C.; Zhao, E.Y.; Gao, L.Y.; Feng, Q.; Liu, X.Y.; Zhao, Z.X.; Ma, X.F.; Hou, W.J.; Zhang, L.R.; et al. Self-assembly cationic nanoparticles based on cholesterol-grafted bioreducible poly(amidoamine) for siRNA delivery. Biomaterials 2013, 34, 5303–5316. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Li, Y.; Tian, T.; Zhang, T.; Shi, S.; Lu, B.; Gao, Y.; Qin, X.; Zhang, M.; Wei, W.; et al. Tetrahedral framework nucleic acids loaded with aptamer AS1411 for siRNA delivery and gene silencing in malignant melanoma. ACS Appl. Mater. Interfaces 2021, 13, 6109–6118. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, S.; Koide, H.; Asai, T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020, 154–155, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Kara, G.; Parlar, A.; Cakmak, M.C.; Cokol, M.; Denkbas, E.B.; Bakan, F. Silencing of survivin and cyclin B1 through siRNA-loaded arginine modified calcium phosphate nanoparticles for non-small-cell lung cancer therapy. Colloid Surf. B 2020, 196, 111340. [Google Scholar] [CrossRef]
- Masjedi, A.; Ahmadi, A.; Atyabi, F.; Farhadi, S.; Irandoust, M.; Khazaei-Poul, Y.; Ghasemi Chaleshtari, M.; Edalati Fathabad, M.; Baghaei, M.; Haghnavaz, N.; et al. Silencing of IL-6 and STAT3 by siRNA loaded hyaluronate-N,N,N-trimethyl chitosan nanoparticles potently reduces cancer cell progression. Int. J. Biol. Macromol. 2020, 149, 487–500. [Google Scholar] [CrossRef]
- Liu, J.; Ding, X.; Fu, Y.; Xiang, C.; Yuan, Y.; Zhang, Y.; Yu, P. Cyclodextrins based delivery systems for macro biomolecules. Eur. J. Med. Chem. 2021, 212, 113105. [Google Scholar] [CrossRef]
- Singh, P.; Shah, A.; Shah, J.; Vataliya, J.; Mittal, A.; Chitkara, D. RNA interference nanotherapeutics for treatment of glioblastoma multiforme. Mol. Pharm. 2020, 17, 4040–4066. [Google Scholar] [CrossRef]
- Sayed, N.; Allawadhi, P.; Khurana, A.; Singh, V.; Navik, U.; Pasumarthi, S.K.; Khurana, I.; Banothu, A.K.; Weiskirchen, R.; Bharani, K.K. Gene therapy: Comprehensive overview and therapeutic applications. Life Sci. 2022, 294, 120375. [Google Scholar] [CrossRef]
- Liu, Q.; Garcia, M.; Wang, S.; Chen, C.W. Therapeutic target discovery using high-throughput genetic screens in acute myeloid leukemia. Cells 2020, 9, 1888. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Zureigat, H.; Al Karaki, A.; Al-Kadash, A.; Gharaibeh, L.; Hatmal, M.M.; Aljabali, A.A.A.; Awidi, A. siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur. J. Pharmacol. 2021, 905, 174178. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Bao, C.; Zhao, F.; Yu, H.; Zhong, G.; Xu, L.; Yan, S. Exploring specific miRNA-mRNA axes with relationship to taxanes-resistance in breast cancer. Front. Oncol. 2020, 10, 1397. [Google Scholar] [CrossRef] [PubMed]
- Kotowska-Zimmer, A.; Pewinska, M.; Olejniczak, M. Artificial miRNAs as therapeutic tools: Challenges and opportunities. Wiley Interdiscip. Rev. RNA 2021, 12, e1640. [Google Scholar] [CrossRef] [PubMed]
- Maczuga, P.; Lubelski, J.; van Logtenstein, R.; Borel, F.; Blits, B.; Fakkert, E.; Costessi, A.; Butler, D.; van Deventer, S.; Petry, H.; et al. Embedding siRNA sequences targeting apolipoprotein B100 in shRNA and miRNA scaffolds results in differential processing and in vivo efficacy. Mol. Ther. 2013, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Brummelkamp, T.R.; Bernards, R.; Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002, 296, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Gu, S. Guidelines for the optimal design of miRNA-based shRNAs. Methods 2016, 103, 157–166. [Google Scholar] [CrossRef]
- Fang, J.; Nakamura, H.; Maeda, H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Deliv. Rev. 2011, 63, 136–151. [Google Scholar] [CrossRef]
- Ghosh, K.; Thodeti, C.K.; Dudley, A.C.; Mammoto, A.; Klagsbrun, M.; Ingber, D.E. Tumor-derived endothelial cells exhibit aberrant Rho-mediated mechanosensing and abnormal angiogenesis in vitro. Proc. Natl. Acad. Sci. USA 2008, 105, 11305–11310. [Google Scholar] [CrossRef]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Biziaev, N.S.; Egorova, T.V.; Alkalaeva, E.Z. Dynamics of Eukaryotic mRNA Structure during Translation. Mol. Biol. 2022, 52, 382–394. [Google Scholar] [CrossRef]
- Raja, M.A.G.; Katas, H.; Amjad, M.W. Design, mechanism, delivery and therapeutics of canonical and dicer-substrate siRNA. Asian J. Pharm. Sci. 2019, 14, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.A. The centrality of RNA. Cell 2009, 136, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Cao, Y.; Li, L.; Wang, X.; Lu, H.; Yang, J.; Wang, S. Self-assembled polymeric micelle as a novel mRNA delivery carrier. J. Control. Release 2021, 338, 537–547. [Google Scholar] [CrossRef]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Xiong, Q.; Lee, G.Y.; Ding, J.; Li, W.; Shi, J. Biomedical applications of mRNA nanomedicine. Nano Res. 2018, 11, 5281–5309. [Google Scholar] [CrossRef]
- Shuai, Q.; Zhu, F.; Zhao, M.; Yan, Y. mRNA delivery via non-viral carriers for biomedical applications. Int. J. Pharm. 2021, 607, 121020. [Google Scholar] [CrossRef]
- Liu, T.; Wei, Q.; Jin, J.; Luo, Q.; Liu, Y.; Yang, Y.; Cheng, C.; Li, L.; Pi, J.; Si, Y.; et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020, 48, 3816–3831. [Google Scholar] [CrossRef]
- Wang, H.X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.W.; Bao, G.; Leong, K.W. CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 2017, 117, 9874–9906. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Kariko, K.; Tureci, O. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Nikkhoo, A.; Rostami, N.; Farhadi, S.; Esmaily, M.; Moghadaszadeh Ardebili, S.; Atyabi, F.; Baghaei, M.; Haghnavaz, N.; Yousefi, M.; Aliparasti, M.R.; et al. Codelivery of STAT3 siRNA and BV6 by carboxymethyl dextran trimethyl chitosan nanoparticles suppresses cancer cell progression. Int. J. Pharm. 2020, 581, 119236. [Google Scholar] [CrossRef] [PubMed]
- Labala, S.; Jose, A.; Chawla, S.R.; Khan, M.S.; Bhatnagar, S.; Kulkarni, O.P.; Venuganti, V.V.K. Effective melanoma cancer suppression by iontophoretic co-delivery of STAT3 siRNA and imatinib using gold nanoparticles. Int. J. Pharm. 2017, 525, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Ball, R.L.; Hajj, K.A.; Vizelman, J.; Bajaj, P.; Whitehead, K.A. Lipid nanoparticle formulations for enhanced co-delivery of siRNA and mRNA. Nano Lett. 2018, 18, 3814–3822. [Google Scholar] [CrossRef]
- Tseng, Y.C.; Mozumdar, S.; Huang, L. Lipid-based systemic delivery of siRNA. Adv. Drug Deliv. Rev. 2009, 61, 721–731. [Google Scholar] [CrossRef]
- Subhan, M.A.; Attia, S.A.; Torchilin, V.P. Advances in siRNA delivery strategies for the treatment of MDR cancer. Life Sci. 2021, 274, 119337. [Google Scholar] [CrossRef]
- Qin, L.; Yan, P.; Xie, C.; Huang, J.; Ren, Z.; Li, X.; Best, S.; Cai, X.; Han, G. Gold nanorod-assembled ZnGa2O4:Cr nanofibers for LED-amplified gene silencing in cancer cells. Nanascale 2018, 10, 13432–13442. [Google Scholar] [CrossRef]
- Laroui, H.; Theiss, A.L.; Yan, Y.; Dalmasso, G.; Nguyen, H.T.; Sitaraman, S.V.; Merlin, D. Functional TNFalpha gene silencing mediated by polyethyleneimine/TNFalpha siRNA nanocomplexes in inflamed colon. Biomaterials 2011, 32, 1218–1228. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Yin, C.; Tan, S.; Wang, X.; Yang, C.; Zhang, T.D.; Zhang, X.; Ye, F.; Xu, J.; et al. Polyvinylamine with moderate binding affinity as a highly effective vehicle for RNA delivery. J. Control. Release 2022, 345, 20–37. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’H, F.; Zanta, M.A.; Mergny, M.D.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed]
- Aliabadi, H.M.; Bahadur, K.C.R.; Bousoik, E.; Hall, R.; Barbarino, A.; Thapa, B.; Coyle, M.; Mahdipoor, P.; Uludag, H. A systematic comparison of lipopolymers for siRNA delivery to multiple breast cancer cell lines: In vitro studies. Acta Biomater. 2020, 102, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Kircheis, R.; Wightman, L.; Wagner, E. Design and gene delivery activity of modified polyethylenimines. Adv. Drug Deliver. Rev. 2001, 53, 341–358. [Google Scholar] [CrossRef]
- Tai, W.; Gao, X. Functional peptides for siRNA delivery. Adv. Drug. Deliv. Rev. 2017, 110–111, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Laroui, N.; Heyraud, M.; Laconde, G.; Ali, L.M.A.; Bourbiaux, K.; Subra, G.; Vezenkov, L.L.; Legrand, B.; Amblard, M.; et al. Hydrocarbon-stapled peptide based-nanoparticles for siRNA delivery. Nanomaterials 2020, 10, 2334. [Google Scholar] [CrossRef]
- Xu, X.; Li, L.; Li, X.; Tao, D.; Zhang, P.; Gong, J. Aptamer-protamine-siRNA nanoparticles in targeted therapy of ErbB3 positive breast cancer cells. Int. J. Pharm. 2020, 590, 119963. [Google Scholar] [CrossRef]
- Ahsan, S.M.; Thomas, M.; Reddy, K.K.; Sooraparaju, S.G.; Asthana, A.; Bhatnagar, I. Chitosan as biomaterial in drug delivery and tissue engineering. Int. J. Biol. Macromol. 2018, 110, 97–109. [Google Scholar] [CrossRef]
- Diao, Y.; Wang, G.; Zhu, B.; Li, Z.; Wang, S.; Yu, L.; Li, R.; Fan, W.; Zhang, Y.; Zhou, L.; et al. Loading of “cocktail siRNAs” into extracellular vesicles via TAT-DRBD peptide for the treatment of castration-resistant prostate cancer. Cancer Biol. Ther. 2022, 23, 163–172. [Google Scholar] [CrossRef]
- Zhang, X.; Pei, Z.; Chen, J.; Ji, C.; Xu, J.; Zhang, X.; Wang, J. Exosomes for immunoregulation and therapeutic intervention in cancer. J. Cancer 2016, 7, 1081–1087. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef]
- Ailincai, D.; Mititelu-Tartau, L.; Marin, L. Citryl-imine-PEG-ylated chitosan hydrogels—Promising materials for drug delivery applications. Int. J. Biol. Macromol. 2020, 162, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Nam, J.-P.; Nah, J.-W. Application of chitosan and chitosan derivatives as biomaterials. J. Ind. Eng. Chem. 2016, 33, 1–10. [Google Scholar] [CrossRef]
- Bakshi, P.S.; Selvakumar, D.; Kadirvelu, K.; Kumar, N.S. Chitosan as an environment friendly biomaterial—A review on recent modifications and applications. Int. J. Biol. Macromol. 2020, 150, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Peers, S.; Montembault, A.; Ladaviere, C. Chitosan hydrogels for sustained drug delivery. J. Control. Release 2020, 326, 150–163. [Google Scholar] [CrossRef]
- Nguyen, M.A.; Wyatt, H.; Susser, L.; Geoffrion, M.; Rasheed, A.; Duchez, A.C.; Cottee, M.L.; Afolayan, E.; Farah, E.; Kahiel, Z.; et al. Delivery of microRNAs by chitosan nanoparticles to functionally alter macrophage cholesterol efflux in vitro and in vivo. ACS Nano 2019, 13, 6491–6505. [Google Scholar] [CrossRef]
- Santos-Carballal, B.; Fernandez Fernandez, E.; Goycoolea, F.M. Chitosan in non-viral gene delivery: Role of structure, characterization methods, and insights in cancer and rare diseases therapies. Polymers 2018, 10, 444. [Google Scholar] [CrossRef]
- Chuan, D.; Jin, T.; Fan, R.; Zhou, L.; Guo, G. Chitosan for gene delivery: Methods for improvement and applications. Adv. Colloid Interface Sci. 2019, 268, 25–38. [Google Scholar] [CrossRef]
- Helmschrodt, C.; Hobel, S.; Schoniger, S.; Bauer, A.; Bonicelli, J.; Gringmuth, M.; Fietz, S.A.; Aigner, A.; Richter, A.; Richter, F. Polyethylenimine nanoparticle-mediated siRNA delivery to reduce alpha-synuclein expression in a model of parkinson’s disease. Mol. Ther. Nucleic Acids 2017, 9, 57–68. [Google Scholar] [CrossRef]
- Wen, J.; Qiu, N.; Zhu, Z.; Bai, P.; Hu, M.; Qi, W.; Liu, Y.; Wei, A.; Chen, L. A size-shrinkable matrix metallopeptidase-2-sensitive delivery nanosystem improves the penetration of human programmed death-ligand 1 siRNA into lung-tumor spheroids. Drug Deliv. 2021, 28, 1055–1066. [Google Scholar] [CrossRef]
- Luo, D.; Xu, X.; Iqbal, M.Z.; Zhao, Q.; Zhao, R.; Farheen, J.; Zhang, Q.; Zhang, P.; Kong, X. siRNA-loaded hydroxyapatite nanoparticles for KRAS gene silencing in anti-pancreatic cancer therapy. Pharmaceutics 2021, 13, 1428. [Google Scholar] [CrossRef]
- Yuan, Z.; Guo, X.; Wei, M.; Xu, Y.; Fang, Z.; Feng, Y.; Yuan, W.-E. Novel fluorinated polycationic delivery of anti-VEGF siRNA for tumor therapy. NPG Asia Mater. 2020, 12, 34. [Google Scholar] [CrossRef]
- Ewe, A.; Hobel, S.; Heine, C.; Merz, L.; Kallendrusch, S.; Bechmann, I.; Merz, F.; Franke, H.; Aigner, A. Optimized polyethylenimine (PEI)-based nanoparticles for siRNA delivery, analyzed in vitro and in an ex vivo tumor tissue slice culture model. Drug Deliv. Transl. Res. 2017, 7, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Zhupanyn, P.; Ewe, A.; Buch, T.; Malek, A.; Rademacher, P.; Muller, C.; Reinert, A.; Jaimes, Y.; Aigner, A. Extracellular vesicle (ECV)-modified polyethylenimine (PEI) complexes for enhanced siRNA delivery in vitro and in vivo. J. Control. Release 2020, 319, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Joshi, U.; Filipczak, N.; Khan, M.M.; Attia, S.A.; Torchilin, V. Hypoxia-sensitive micellar nanoparticles for co-delivery of siRNA and chemotherapeutics to overcome multi-drug resistance in tumor cells. Int. J. Pharm. 2020, 590, 119915. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Zaro, J.; Shen, Y. Advances in exosome-based drug delivery and tumor targeting: From tissue distribution to intracellular fate. Int. J. Nanomed. 2020, 15, 9355–9371. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Lu, Z.X.; Jiang, Y.H.; Shi, X.X.; Xu, D.Z.; Shi, Y.S.; Lin, G.; Liu, C.; Zhang, Y.; Liu, G. Nanotransferrin-based programmable catalysis mediates three-pronged induction of oxidative stress to enhance cancer immunotherapy. ACS Nano 2022, 16, 997–1012. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Zhang, X.; Huang, H.; Tang, S.; Chai, Y.; Xu, Z.; Li, M.; Chen, X.; Liu, J.; et al. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J. Nanobiotechnol. 2022, 20, 279. [Google Scholar] [CrossRef]
- Kim, H.; Rhee, W.J. Exosome-mediated let7c-5p delivery for breast cancer therapeutic development. Biotechnol. Bioproc. Eng. 2020, 25, 513–520. [Google Scholar] [CrossRef]
- Forterre, A.V.; Wang, J.H.; Delcayre, A.; Kim, K.; Green, C.; Pegram, M.D.; Jeffrey, S.S.; Matin, A.C. Extracellular vesicle-mediated in vitro transcribed mRNA delivery for treatment of HER2(+) breast cancer xenografts in mice by prodrug CB1954 without general toxicity. Mol. Cancer Ther. 2020, 19, 858–867. [Google Scholar] [CrossRef]
- Kaban, K.; Hinterleitner, C.; Zhou, Y.; Salva, E.; Kantarci, A.G.; Salih, H.R.; Marklin, M. Therapeutic silencing of BCL-2 using NK cell-derived exosomes as a novel therapeutic approach in breast cancer. Cancers 2021, 13, 2397. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Li, Y.; Zhang, T.; Xiao, D.; Tian, T.; Chen, T.; Zhang, Y.; Li, X.; Lin, Y. Biological effect of differently sized tetrahedral framework nucleic acids: Endocytosis, proliferation, migration, and biodistribution. ACS Appl. Mater. Interfaces 2021, 13, 57067–57074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, N.; Zhou, M.; Li, S.; Cai, X. The application of tetrahedral framework nucleic acids as a drug carrier in biomedicine fields. Curr. Stem. Cell Res. Ther. 2021, 16, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, X.; Tian, T.; Zhang, T.; Gao, S.; Zhang, X.; Yao, Y.; Lin, Y.; Cai, X. A lysosome-activated tetrahedral nanobox for encapsulated siRNA delivery. Adv. Mater. 2022, e2201731. [Google Scholar] [CrossRef]
- Li, K.; Luo, S.; Guan, S.; Situ, B.; Wu, Y.; Ou, Z.; Tao, M.; Zheng, L.; Cai, Z. Tetrahedral framework nucleic acids linked CRISPR/Cas13a signal amplification system for rare tumor cell detection. Talanta 2022, 247, 123531. [Google Scholar] [CrossRef]
- Ogawa, M.; Regino, C.A.S.; Marcelino, B.; Williams, M.; Kosaka, N.; Bryant, L.H., Jr.; Choyke, P.L.; Kobayashi, H. New nanosized biocompatible MR contrast agents based on lysine-dendri-graft macromolecules. Bioconjug. Chem. 2010, 21, 955–960. [Google Scholar] [CrossRef]
- Pankhurst, Q.A.; Thanh, N.T.K.; Jones, S.K.; Dobson, J. Progress in applications of magnetic nanoparticles in biomedicine. J. Phys. D 2009, 42, 224001. [Google Scholar] [CrossRef]
- Yang, P.; Gai, S.; Lin, J. Functionalized mesoporous silica materials for controlled drug delivery. Chem. Soc. Rev. 2012, 41, 3679–3698. [Google Scholar] [CrossRef]
- Lin, G.; Revia, R.A.; Zhang, M. Inorganic nanomaterial-mediated gene therapy in combination with other antitumor treatment modalities. Adv. Funct. Mater. 2021, 31, 2007096. [Google Scholar] [CrossRef]
- Kara, G.; Malekghasemi, S.; Ozpolat, B.; Denkbas, E.B. Development of novel poly-l-lysine-modified sericin-coated superparamagnetic iron oxide nanoparticles as siRNA carrier. Colloid Surf. A 2021, 630, 127622. [Google Scholar] [CrossRef]
- Cristofolini, T.; Dalmina, M.; Sierra, J.A.; Silva, A.H.; Pasa, A.A.; Pittella, F.; Creczynski-Pasa, T.B. Multifunctional hybrid nanoparticles as magnetic delivery systems for siRNA targeting the HER2 gene in breast cancer cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 109, 110555. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Okabe, N.; Note, Y.; Hashiba, K.; Maeki, M.; Tokeshi, M.; Harashima, H. Hydrophobic scaffolds of pH-sensitive cationic lipids contribute to miscibility with phospholipids and improve the efficiency of delivering short interfering RNA by small-sized lipid nanoparticles. Acta Biomater. 2020, 102, 341–350. [Google Scholar] [CrossRef]
- Sayers, E.J.; Peel, S.E.; Schantz, A.; England, R.M.; Beano, M.; Bates, S.M.; Desai, A.S.; Puri, S.; Ashford, M.B.; Jones, A.T. Endocytic profiling of cancer cell models reveals critical factors influencing LNP-mediated mRNA delivery and protein expression. Mol. Ther. 2019, 27, 1950–1962. [Google Scholar] [CrossRef] [PubMed]
- Hirai, Y.; Saeki, R.; Song, F.; Koide, H.; Fukata, N.; Tomita, K.; Maeda, N.; Oku, N.; Asai, T. Charge-reversible lipid derivative: A novel type of pH-responsive lipid for nanoparticle-mediated siRNA delivery. Int. J. Pharm. 2020, 585, 119479. [Google Scholar] [CrossRef] [PubMed]
- Shobaki, N.; Sato, Y.; Suzuki, Y.; Okabe, N.; Harashima, H. Manipulating the function of tumor-associated macrophages by siRNA-loaded lipid nanoparticles for cancer immunotherapy. J. Control. Release 2020, 325, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Liu, S.; Wei, T.; Cheng, Q.; Siegwart, D.J. Theranostic dendrimer-based lipid nanoparticles containing PEGylated BODIPY dyes for tumor imaging and systemic mRNA delivery in vivo. J. Control. Release 2020, 325, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Katas, H.; Alpar, H.O. Development and characterisation of chitosan nanoparticles for siRNA delivery. J. Control. Release 2006, 115, 216–225. [Google Scholar] [CrossRef]
- Soltani, F.; Parhiz, H.; Mokhtarzadeh, A.; Ramezani, M. Synthetic and biological vesicular nano-carriers designed for gene delivery. Curr. Pharm. Des. 2015, 21, 6214–6235. [Google Scholar] [CrossRef]
- Zheng, Q.C.; Jiang, S.; Wu, Y.Z.; Shang, D.; Zhang, Y.; Hu, S.B.; Cheng, X.; Zhang, C.; Sun, P.; Gao, Y.; et al. Corrigendum: Dual-targeting nanoparticle-mediated gene therapy strategy for hepatocellular carcinoma by delivering small interfering RNA. Front. Bioeng. Biotechnol. 2021, 9, 656268. [Google Scholar] [CrossRef]
- Kulkarni, A.D.; Patel, H.M.; Surana, S.J.; Vanjari, Y.H.; Belgamwar, V.S.; Pardeshi, C.V. N,N,N-Trimethyl chitosan: An advanced polymer with myriad of opportunities in nanomedicine. Carbohydr. Polym. 2017, 157, 875–902. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, Y.; Wang, L.; Liang, Z.; Li, D.; Xu, X.; Chen, Y.; Yang, X.; Zhang, H.; Niu, H. Self-crosslinkable chitosan-hyaluronic acid dialdehyde nanoparticles for CD44-targeted siRNA delivery to treat bladder cancer. Bioact. Mater. 2021, 6, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Liu, P. pH-responsive surface charge reversal carboxymethyl chitosan-based drug delivery system for pH and reduction dual-responsive triggered DOX release. Carbohydr. Polym. 2020, 236, 116093. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Qiao, L.; Shan, G.; Gao, M.; Zhang, R.; Yi, X.; He, X. Stepwise responsive carboxymethyl chitosan-based nanoplatform for effective drug-resistant breast cancer suppression. Carbohydr. Polym. 2022, 291, 119554. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.C.; Wang, Z.L.; Xu, T.; He, Z.Y.; Wei, Y.Q. The approved gene therapy drugs worldwide: From 1998 to 2019. Biotechnol. Adv. 2020, 40, 107502. [Google Scholar] [CrossRef]
- Golan, T.; Khvalevsky, E.Z.; Hubert, A.; Gabai, R.M.; Hen, N.; Segal, A.; Domb, A.; Harari, G.; David, E.B.; Raskin, S.; et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 2015, 6, 24560–24570. [Google Scholar] [CrossRef]
- Bartoszewski, R.; Sikorski, A.F. Editorial focus: Understanding off-target effects as the key to successful RNAi therapy. Cell Mol. Biol. Lett. 2019, 24, 69. [Google Scholar] [CrossRef]
- Schultheis, B.; Strumberg, D.; Santel, A.; Vank, C.; Gebhardt, F.; Keil, O.; Lange, C.; Giese, K.; Kaufmann, J.; Khan, M.; et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J. Clin. Oncol. 2014, 32, 4141–4148. [Google Scholar] [CrossRef]
- Beate, S.; Dirk, S.; Jan, K.; Martin, W.; Karin, L.; Thomas, S.; Joerg, K.; Frank, G.; Nico, B.; Uwe, P. A phase Ib/IIa study of combination therapy with gemcitabine and Atu027 in patients with locally advanced or metastatic pancreatic adenocarcinoma. J. Clin. Oncol. 2016, 34, 385. [Google Scholar] [CrossRef]
- Varghese, A.M.; Ang, C.; Dimaio, C.J.; Javle, M.M.; Gutierrez, M.; Yarom, N.; Stemmer, S.M.; Golan, T.; Geva, R.; Semenisty, V.; et al. A phase II study of siG12D-LODER in combination with chemotherapy in patients with locally advanced pancreatic cancer (PROTACT). J. Clin. Oncol. 2020, 38, TPS4672. [Google Scholar] [CrossRef]
- Nakamura, Y.; Shitara, K.; Lee, J. The right treatment of the right patient: Integrating genetic profiling into clinical decision making in advanced gastric cancer in asia. Am. Soc. Clin. Oncol. Educ. Book 2021, 41, e166–e173. [Google Scholar] [CrossRef]
- Sun, W.; Su, Q.; Cao, X.; Shang, B.; Chen, A.; Yin, H.; Liu, B. High expression of polo-like kinase 1 is associated with early development of hepatocellular carcinoma. Int. J. Genom. 2014, 2014, 312130. [Google Scholar] [CrossRef] [PubMed]
- El Dika, I.; Lim, H.Y.; Yong, W.P.; Lin, C.C.; Yoon, J.H.; Modiano, M.; Freilich, B.; Choi, H.J.; Chao, T.Y.; Kelley, R.K.; et al. An open-label, multicenter, phase I, dose escalation study with phase II expansion cohort to determine the safety, pharmacokinetics, and preliminary antitumoractivity of intravenous TKM-080301 in subjects with advanced hepatocellular carcinoma. Oncologist 2019, 24, 747–e218. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.; Lopez-Berestein, G.; Fu, S.; Tsimberidou, A.M.; Pant, S.; Piha-Paul, S.A.; Janku, F.; Hong, D.S.; Sulovic, S.; Meng, X.; et al. EphA2 gene targeting using neutral liposomal small interfering RNA (EPHARNA) delivery: A phase I clinical trial. J. Clin. Oncol. 2017, 35, TPS2604. [Google Scholar] [CrossRef]
- Kumthekar, P.; Ko, C.H.; Paunesku, T.; Dixit, K.; Sonabend, A.M.; Bloch, O.; Tate, M.; Schwartz, M.; Zuckerman, L.; Lezon, R.; et al. A first-in-human phase 0 clinical study of RNA interference-based spherical nucleic acids in patients with recurrent glioblastoma. Sci. Transl. Med. 2021, 13, 584. [Google Scholar] [CrossRef]
- Tabernero, J.; Shapiro, G.I.; LoRusso, P.M.; Cervantes, A.; Schwartz, G.K.; Weiss, G.J.; Paz-Ares, L.; Cho, D.C.; Infante, J.R.; Alsina, M.; et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013, 3, 406–417. [Google Scholar] [CrossRef]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Bauman, J.; Burris, H.; Clarke, J.; Patel, M.; Cho, D.; Gutierrez, M.; Julian, R.; Scott, A.; Cohen, P.; Frederick, J.; et al. Safety, tolerabilifty, and immunogenicity of mRNA-4157 in combination with pembrolizumab in subjects with unresectable solid tumoble solid tumors (keynote-603): An update. J. Immunother. Cancer 2020, 8, A477. [Google Scholar] [CrossRef]
- Jimeno, A.; Gupta, S.; Sullivan, R.; Do, K.T.; Akerley, W.L.; Wang, D.; Teoh, D.; Schalper, K.; Zacharek, S.J.; Sun, J.; et al. A phase 1/2, open-label, multicenter, dose escalation and efficacy study of mRNA-2416, a lipid nanoparticle encapsulated mRNA encoding human OX40L, for intratumoral injection alone or in combination with durvalumab for patients with advanced malignancies. Cancer Res. 2020, 80, CT032. [Google Scholar] [CrossRef]
- Jabulowsky, R.A.; Loquai, C.; Mitzel-Rink, H.; Utikal, J.; Gebhardt, C.; Hassel, J.C.; Kaufmann, R.; Pinter, A.; Derhovanessian, E.; Anft, C.; et al. A first-in-human phase I/II clinical trial assessing novel mRNA-lipoplex nanoparticles encoding shared tumor antigens for immunotherapy of malignant melanoma. Cancer Res. 2018, 78, CT156. [Google Scholar] [CrossRef]
- Sarker, D.; Plummer, R.; Meyer, T.; Sodergren, M.H.; Basu, B.; Chee, C.E.; Huang, K.W.; Palmer, D.H.; Ma, Y.T.; Evans, T.R.J.; et al. MTL-CEBPA, a small activating RNA therapeutic upregulating C/EBP-α, in patients with advanced liver cancer: A first-in-human, multicenter, open-label, phase I trial. Clin. Cancer Res. 2020, 26, 3936–3946. [Google Scholar] [CrossRef]


| RNA Drug Name | Target | Type of RNA | Delivery System | Cancer Types | Phase | ClinicalTrials.gov Identifier | Reference |
|---|---|---|---|---|---|---|---|
| Atu027 | PKN3 | siRNA | Cationic LNPs | Advanced or metastatic pancreatic cancer | I/II | NCT00938574 /NCT01808638 | [117,118] |
| siG12D LODER | KRAS | siRNA | Polymeric NPs (LODER) | Pancreatic ductal adenocarcinoma, pancreatic cancer | I/II | NCT01188785/ NCT01676259 | [119] |
| Mesenchymal stromal cells-derived exosomes with KRAS G12D siRNA | KRAS G12D | siRNA | Exosomes | Pancreatic cancer | I | NCT03608631 | [120] |
| TKM-080301 | PLK1 | siRNA | LNPs | Adrenal cortical carcinoma, neuroendocrine tumor, hepatocellular carcinoma | I/II | NCT01262235 /NCT01437007 /NCT02191878 | [121,122] |
| EPHARNA | EphA2 | siRNA | LNPs | Advanced malignant solid neoplasm | I | NCT01591356 | [123] |
| NU-0129 | BCL2L12 | siRNA | Au NPs | Gliosarcoma, recurrent glioblastoma | I | NCT03020017 | [124] |
| ALN-VSP02 | VEGF and KSP | siRNA | SNALPs | Solid tumors | I | NCT01158079 | [125] |
| TargomiRs | Mir 16 | miRNA | nonliving bacterial minicells (nanoparticles)(EDVs) | Malignant pleural mesothelioma, Non-small cell lung cancer | I | NCT02369198 | [126] |
| mRNA-4157 and Pembrolizumab | Individually designed mRNA coding for tumor neoantigens | mRNA | LNP | High-risk melanoma | II | NCT03897881 /NCT03313778 | [127] |
| mRNA-2416 and Durvalumab | OX40 | mRNA | Liposomes (SM-102, DSPC, PEG2000-DMG, cholesterol) | cancer | I/II | NCT03323398 | [128] |
| Lipo-MERIT | Antigen-specific differentiated clusters of four selected malignant melanoma-associated antigens | mRNA | LNP | Melanoma | I | NCT02410733 | [129] |
| MTL-CEBPA | C/EBPα | saRNA | LNP | Liver cancer | I | NCT02716012 | [130] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Liu, C.; Bai, S.; Lu, Z.; Liu, G. Broadening the Horizons of RNA Delivery Strategies in Cancer Therapy. Bioengineering 2022, 9, 576. https://doi.org/10.3390/bioengineering9100576
Wu S, Liu C, Bai S, Lu Z, Liu G. Broadening the Horizons of RNA Delivery Strategies in Cancer Therapy. Bioengineering. 2022; 9(10):576. https://doi.org/10.3390/bioengineering9100576
Chicago/Turabian StyleWu, Shuaiying, Chao Liu, Shuang Bai, Zhixiang Lu, and Gang Liu. 2022. "Broadening the Horizons of RNA Delivery Strategies in Cancer Therapy" Bioengineering 9, no. 10: 576. https://doi.org/10.3390/bioengineering9100576
APA StyleWu, S., Liu, C., Bai, S., Lu, Z., & Liu, G. (2022). Broadening the Horizons of RNA Delivery Strategies in Cancer Therapy. Bioengineering, 9(10), 576. https://doi.org/10.3390/bioengineering9100576










