GABA-Producing Lactococcus lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Milk Sample Collection
2.3. Isolation of Lactic Acid Bacteria and Phenotypic Characterization
2.4. Molecular Identification of Lactococcus Isolates
2.5. Identification of GABA-Producing Lactococcus in Culture Media
2.6. Whole-Genome Sequencing and Bioinformatic Analyses
2.7. Acidifying Capacity
2.8. Proteolytic Activity
2.9. Dextran Production
2.10. Production of Volatile Compounds
2.11. Production of Antimicrobial Substances
2.12. Production of Biogenic Amines
2.13. In Silico Prediction of Antimicrobial Resistance Genes
2.14. Experimental Cabrales-Like Mini Cheeses
2.15. Statistical Analysis
3. Results
3.1. Phenotypic Characterization and Molecular Identification of LAB Isolates
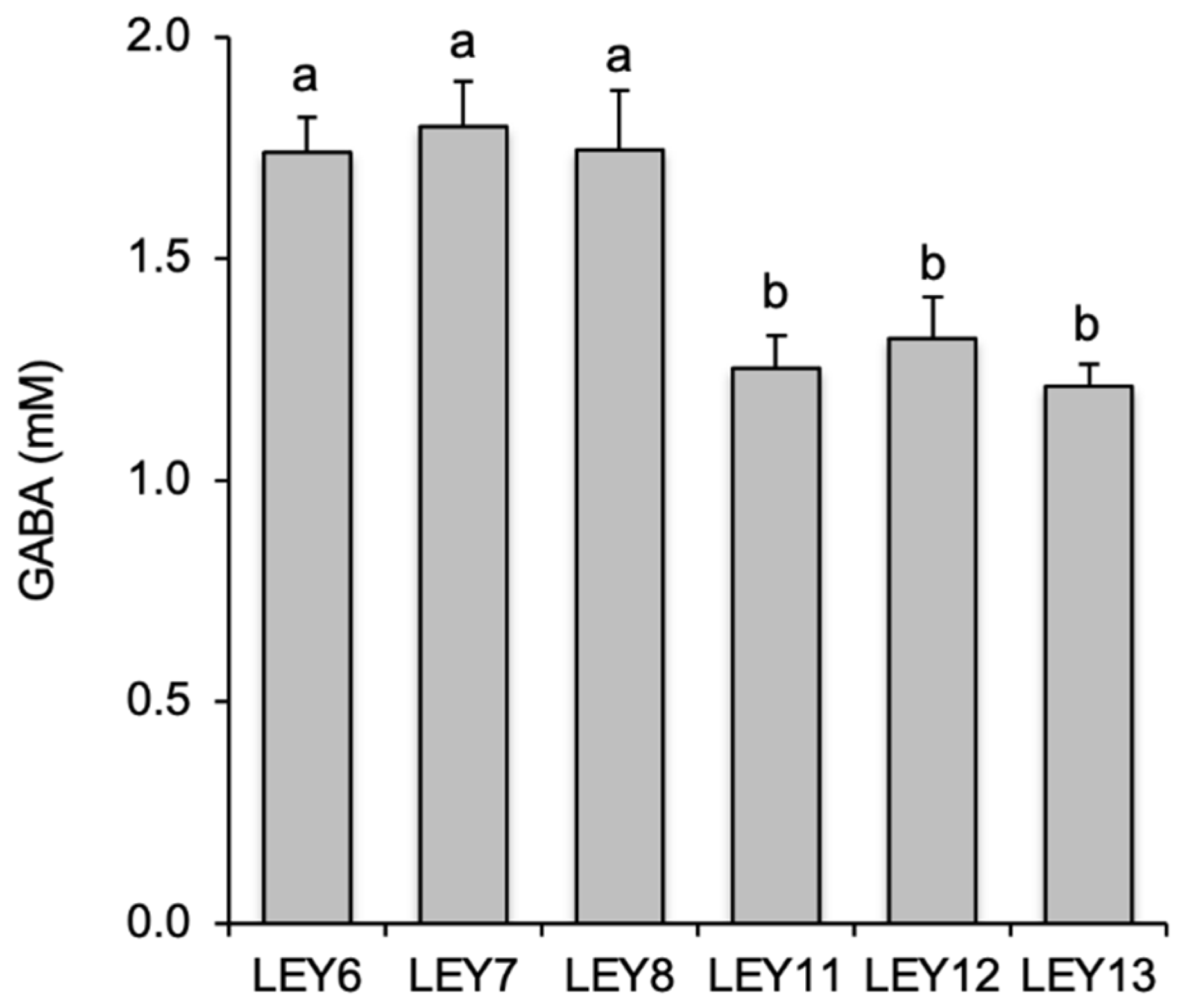
3.2. All six L. lactis Isolates Produced GABA in Culture Media
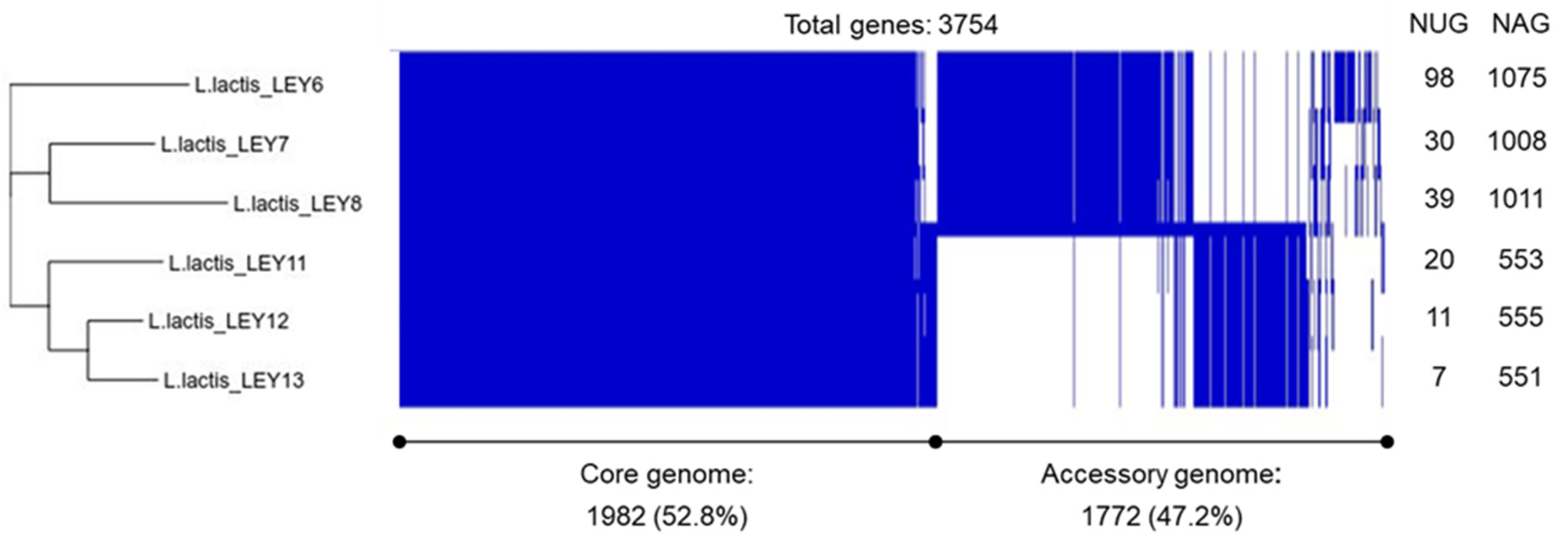
3.3. Whole-Genome Sequencing and Comparison Confirmed the Six GABA-Producing L. lactis to Be Different Strains
3.4. In Silico Identification of the GAD Cluster and Other Technologically Relevant Genes in the Genome of the GABA-Producing L. lactis Strains
3.5. Technological Characterization of the GABA-Producing L. lactis Strains
3.5.1. Acidifying Capacity
3.5.2. Proteolytic Activity
3.5.3. Dextran Production
3.5.4. Production of Volatile Compounds
3.5.5. Production of Antimicrobial Substances
3.6. Safety of GABA-Producing L. lactis Strains
3.6.1. Biogenic Amine Production
3.6.2. Antimicrobial Resistance Genes
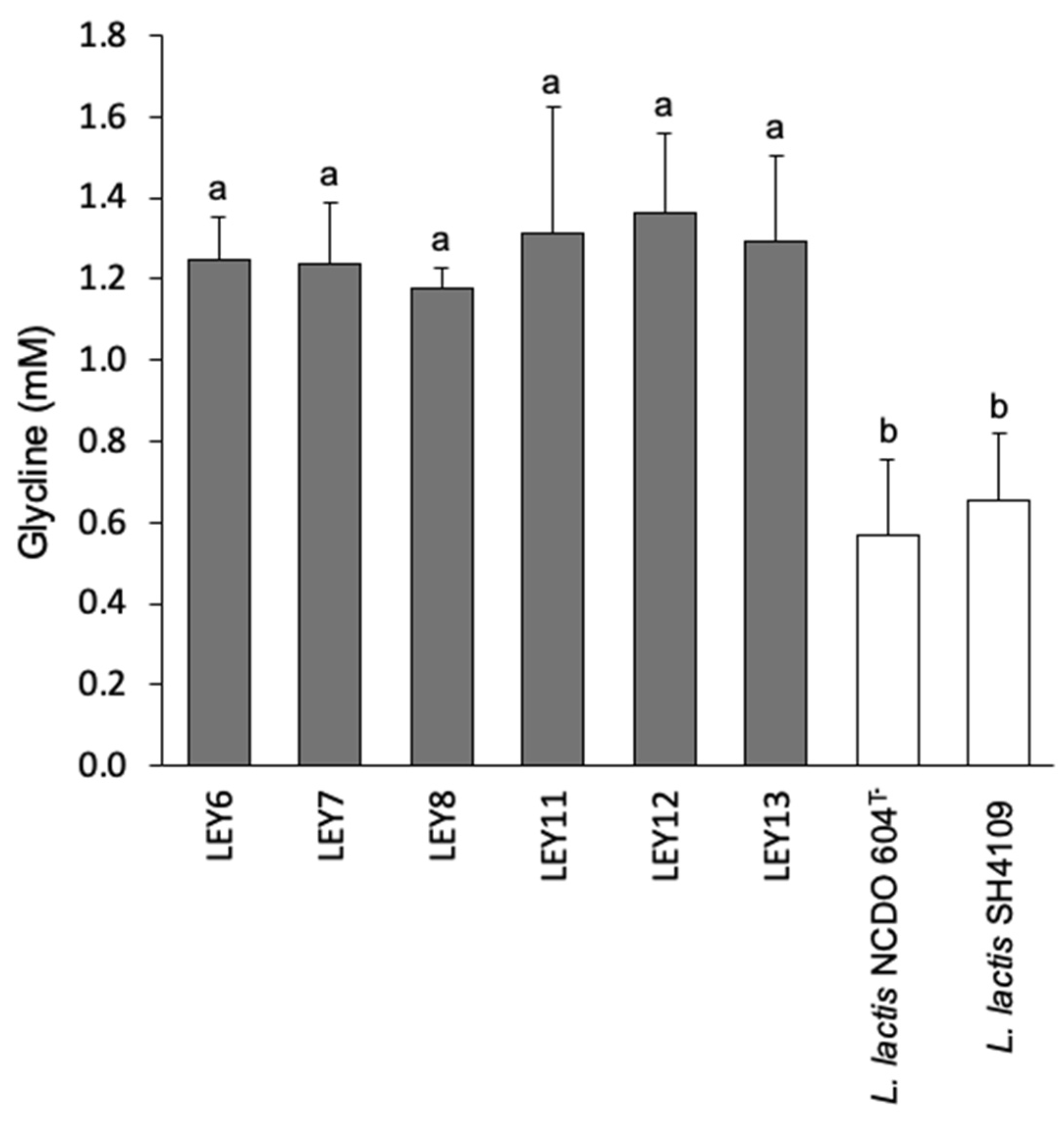
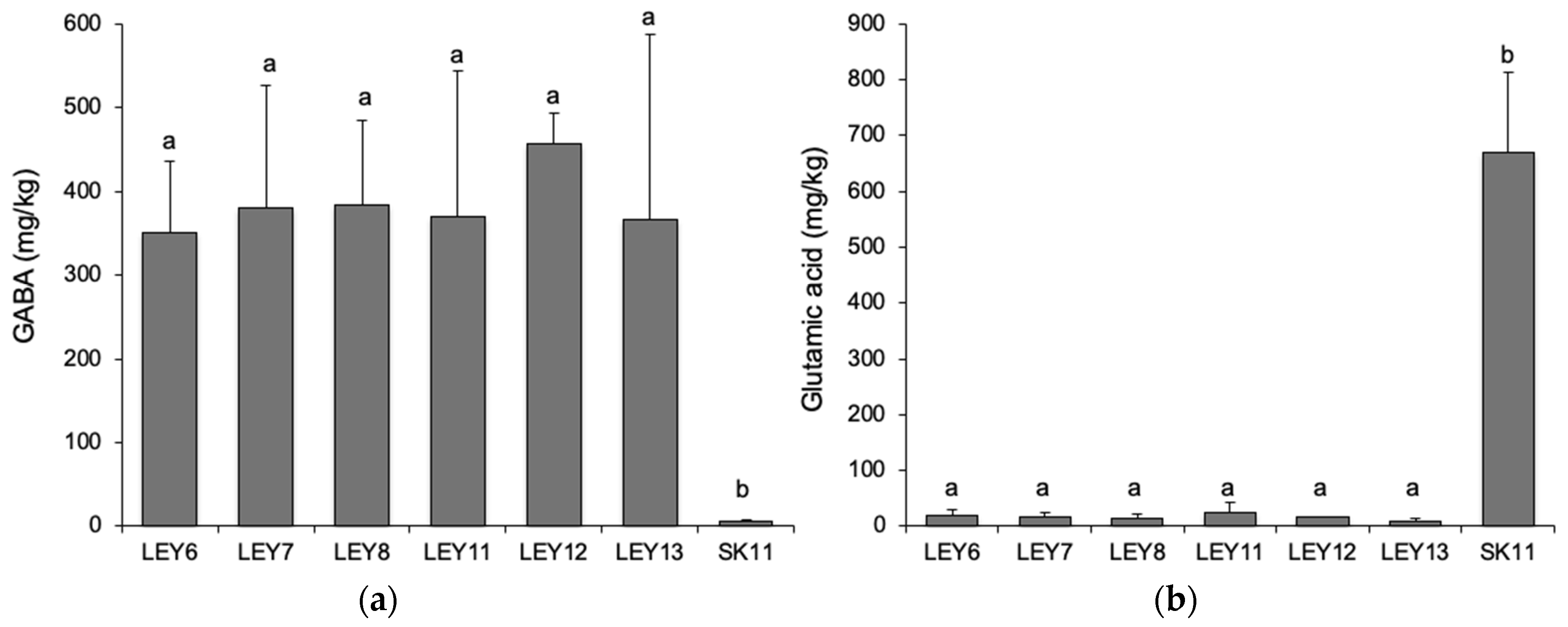
3.7. All Six L. lactis Strains Produced GABA in a Cabrales-Like Mini Cheese Model; None Produced BA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; Hilbert, F.; Lindqvist, R.; Nauta, M.; et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 9: Suitability of taxonomic units notified to EFSA until september 2018. EFSA J. 2019, 17. [Google Scholar] [CrossRef]
- FDA Gamma-Aminobutyric Acid GRAS. Notice Number 595. 2015. Available online: https://www.fda.gov/food/gras-notice-inventory/agency-response-letter-gras-notice-no-grn-000595 (accessed on 27 January 2021).
- Smit, G.; Smit, B.A.; Engels, W.J.M. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef]
- Ayad, E.H.E.; Verheul, A.; Wouters, J.T.M.; Smit, G. Antimicrobial-producing wild lactococci isolated from artisanal and non-dairy origins. Int. Dairy J. 2002, 12, 145–150. [Google Scholar] [CrossRef]
- Nishitani, Y.; Tanoue, T.; Yamada, K.; Ishida, T.; Yoshida, M.; Azuma, T.; Mizuno, M. Lactococcus lactis subsp. cremoris FC alleviates symptoms of colitis induced by dextran sulfate sodium in mice. Int. Immunopharmacol. 2009, 9, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Luerce, T.D.; Gomes-Santos, A.C.; Rocha, C.S.; Moreira, T.G.; Cruz, D.N.; Lemos, L.; Sousa, A.L.; Pereira, V.B.; De Azevedo, M.; Moraes, K.; et al. Anti-inflammatory effects of Lactococcus lactis NCDO 2118 during the remission period of chemically induced colitis. Gut Pathog. 2014, 6, 33. [Google Scholar] [CrossRef]
- Ramalho, J.B.; Soares, M.B.; Spiazzi, C.C.; Bicca, D.F.; Soares, V.M.; Pereira, J.G.; Da Silva, W.P.; Sehn, C.P.; Cibin, F.W.S. In vitro probiotic and antioxidant potential of Lactococcus lactis subsp. cremoris ll95 and its effect in mice behaviour. Nutrients 2019, 11, 901. [Google Scholar] [CrossRef]
- Diez-Gutiérrez, L.; San Vicente, L.; Barrón, L.J.R.; Villarán, M.D.C.; Chávarri, M. Gamma-aminobutyric acid and probiotics: Multiple health benefits and their future in the global functional food and nutraceuticals market. J. Funct. Foods 2020, 64, 103669. [Google Scholar] [CrossRef]
- Inoue, K.; Shirai, T.; Ochiai, H.; Kasao, M.; Hayakawa, K.; Kimura, M.; Sansawa, H. Blood-pressure-lowering effect of a novel fermented milk containing γ-aminobutyric acid (GABA) in mild hypertensives. Eur. J. Clin. Nutr. 2003, 57, 490–495. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, H.; Zhang, C.; Lu, Y.; Zhu, X.; Lu, Z. γ-Aminobutyric acid-rich yogurt fermented by Streptococcus salivarius subsp. thermophiles fmb5 appears to have anti-diabetic effect on streptozotocin-induced diabetic mice. J. Funct. Foods. 2016, 20, 267–275. [Google Scholar] [CrossRef]
- Renes, E.; Ladero, V.; Tornadijo, M.E.; Fresno, J.M. Production of sheep milk cheese with high γ-aminobutyric acid and ornithine concentration and with reduced biogenic amines level using autochthonous lactic acid bacteria strains. Food Microbiol. 2019, 78, 1–10. [Google Scholar] [CrossRef]
- Pouliot-Mathieu, K.; Gardner-Fortier, C.; Lemieux, S.; St-Gelais, D.; Champagne, C.P.; Vuillemard, J.C. Effect of cheese containing gamma-aminobutyric acid-producing lactic acid bacteria on blood pressure in men. Pharma Nutrition 2013, 1, 141–148. [Google Scholar] [CrossRef]
- Carafa, I.; Stocco, G.; Nardin, T.; Larcher, R.; Bittante, G.; Tuohy, K.; Franciosi, E. Production of naturally γ-aminobutyric acid-enriched cheese using the dairy strains Streptococcus thermophilus 84C and Lactobacillus brevis DSM 32386. Front. Microbiol. 2019, 10, 93. [Google Scholar] [CrossRef]
- Wang, H.K.; Dong, C.; Chen, Y.F.; Cui, L.M.; Zhang, H.P. A new probiotic Cheddar cheese with high ACE-inhibitory activity and γ-aminobutyric acid content produced with koumiss-derived Lactobacillus casei Zhang. Food Technol. Biotechnol. 2010, 48, 62–70. [Google Scholar]
- Irigoyen, A.; Ortigosa, M.; Juansaras, I.; Oneca, M.; Torre, P. Influence of an adjunct culture of Lactobacillus on the free amino acids and volatile compounds in a Roncal-type ewe’s-milk cheese. Food Chem. 2007, 100, 71–80. [Google Scholar] [CrossRef]
- Fröhlich-Wyder, M.T.; Bisig, W.; Guggisberg, D.; Irmler, S.; Jakob, E.; Wechsler, D. Influence of low pH on the metabolic activity of Lactobacillus buchneri and Lactobacillus parabuchneri strains in Tilsit-type model cheese. Dairy Sci. Technol. 2015, 95, 569–585. [Google Scholar] [CrossRef]
- Hagi, T.; Nakagawa, H.; Ohmori, H.; Sasaki, K.; Kobayashi, M.; Narita, T.; Nomura, M. Characterization of unique metabolites in γ-aminobutyric acid-rich cheese by metabolome analysis using liquid chromatography-mass spectrometry. J. Food Biochem. 2019, 43, e13039. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Kimoto, H.; Someya, Y.; Furukawa, S.; Suzuki, I. Production of γ-aminobutyric acid by cheese starters during cheese ripening. J. Dairy Sci. 1998, 81, 1486–1491. [Google Scholar] [CrossRef]
- Sanders, J.W.; Leenhouts, K.; Burghoorn, J.; Brands, J.R.; Venema, G.; Kok, J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 1998, 27, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Su, M.S.; Schlicht, S.; Gänzle, M.G. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Fact. 2011, 10. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C. Surviving the Acid Test: Responses of Gram-Positive Bacteria to Low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef]
- Limon, A.; Gallegos-Perez, J.L.; Reyes-Ruiz, J.M.; Aljohi, M.A.; Alshanqeeti, A.S.; Miledi, R. The endogenous GABA bioactivity of camel, bovine, goat and human milks. Food Chem. 2014, 145, 481–487. [Google Scholar] [CrossRef]
- Gasson, M.J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 1983, 154, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Visser, S.; Exterkate, F.A.; Slangen, C.J.; de Veer, G.J. Comparative study of action of cell wall proteinases from various strains of Streptococcus cremoris on bovine alpha(s1)-, beta-, and kappa-casein. Appl. Environ. Microbiol. 1986, 52, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- del Rio, B.; Redruello, B.; Ladero, V.; Fernandez, M.; Martin, M.C.; Alvarez, M.A. Putrescine production by Lactococcus lactis subsp. cremoris CECT 8666 is reduced by NaCl via a decrease in bacterial growth and the repression of the genes involved in putrescine production. Int. J. Food Microbiol. 2016, 232, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, L.; Ahrné, S. Lactic Acid Bacteria. In Applied Microbial Systematics; Priest, F.G., Goodfellow, M., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 367–388. ISBN 978-94-011-4020-1. [Google Scholar]
- Marroki, A.; Zúñiga, M.; Kihal, M.; Pérez-Martínez, G. Characterization of Lactobacillus from algerian goat’s milk based on phenotypic, 16S rDNA sequencing and their technological properties. Braz. J. Microbiol. 2011, 42, 158–171. [Google Scholar] [CrossRef]
- Drici, H.; Gilbert, C.; Kihal, M.; Atlan, D. A typical citrate-fermenting Lactococcus lactis strains isolated from dromedary’s milk. J. Appl. Microbiol. 2010, 108, 647–657. [Google Scholar] [CrossRef]
- Saidi, Y.; del Rio, B.; Senouci, D.E.; Redruello, B.; Martinez, B.; Ladero, V.; Kihal, M.; Alvarez, M.A. Polyphasic characterisation of non-starter lactic acid bacteria from Algerian raw Camel’s milk and their technological aptitudes. Food Technol. Biotechnol. 2020, 58, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Redruello, B.; Ladero, V.; Cuesta, I.; Álvarez-Buylla, J.R.; Martín, M.C.; Fernández, M.; Alvarez, M.A. A fast, reliable, ultra high performance liquid chromatography method for the simultaneous determination of amino acids, biogenic amines and ammonium ions in cheese, using diethyl ethoxymethylenemalonate as a derivatising agent. Food Chem. 2013, 139, 1029–1035. [Google Scholar] [CrossRef]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Altschul, S. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Rattray, F.P.; Mayo, B.; Martín, M.C.; Fernández, M.; Alvarez, M.A. Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Putrescine-Producing Lactococcus lactis. Appl. Environ. Microbiol. 2011, 77, 6409–6418. [Google Scholar] [CrossRef]
- Redruello, B.; Szwengiel, A.; Ladero, V.; del Rio, B.; Alvarez, M.A. Identification of technological/metabolic/environmental profiles of cheeses with high GABA contents. LWT Food Sci. Technol. 2020, 130, 109603. [Google Scholar] [CrossRef]
- Lu, X.; Chen, Z.; Gu, Z.; Han, Y. Isolation of γ-aminobutyric acid-producing bacteria and optimization of fermentative medium. Biochem. Eng. J. 2008, 41, 48–52. [Google Scholar] [CrossRef]
- Diana, M.; Tres, A.; Quílez, J.; Llombart, M.; Rafecas, M. Spanish cheese screening and selection of lactic acid bacteria with high gamma-aminobutyric acid production. LWT Food Sci. Technol. 2014, 56, 351–355. [Google Scholar] [CrossRef]
- Franciosi, E.; Carafa, I.; Nardin, T.; Schiavon, S.; Poznanski, E.; Cavazza, A.; Larcher, R.; Tuohy, K.M. Biodiversity and γ -aminobutyric acid production by lactic acid bacteria isolated from traditional alpine raw cow’s milk cheeses. Biomed. Res. Int. 2015, 2015, 625740. [Google Scholar] [CrossRef] [PubMed]
- Siragusa, S.; De Angelis, M.; Di Cagno, R.; Rizzello, C.G.; Coda, R.; Gobbetti, M. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 2007, 73, 7283–7290. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, J.A.; Flórez, A.B.; Vázquez, L.; Vasek, O.M.; Mayo, B. Production of γ-aminobutyric acid (GABA) by lactic acid bacteria strains isolated from traditional, starter-free dairy products made of raw milk. Benef. Microbes. 2019, 10, 579–587. [Google Scholar] [CrossRef]
- Perez, M.; Calles-Enríquez, M.; del Rio, B.; Redruello, B.; de Jong, A.; Kuipers, O.P.; Kok, J.; Martin, M.C.; Ladero, V.; Fernandez, M.; et al. Construction and characterization of a double mutant of Enterococcus faecalis that does not produce biogenic amines. Sci. Rep. 2019, 9, 16881. [Google Scholar] [CrossRef]
- Maldonado-Barragán, A.; Ruiz-Barba, J.L.; Jiménez-Díaz, R. Knockout of three-component regulatory systems reveals that the apparently constitutive plantaricin-production phenotype shown by Lactobacillus plantarum on solid medium is regulated via quorum sensing. Int. J. Food Microbiol. 2009, 130, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gardner-Fortier, C.; St-Gelais, D.; Champagne, C.P.; Vuillemard, J.C. Determination of optimal conditions for γ-aminobutyric acid production by Lactococcus lactis ssp. lactis. Int. Dairy J. 2013, 32, 136–143. [Google Scholar] [CrossRef]
- Laroute, V.; Yasaro, C.; Narin, W.; Mazzoli, R.; Pessione, E.; Cocaign-Bousquet, M.; Loubière, P. GABA production in Lactococcus lactis is enhanced by Arginine and co-addition of malate. Front. Microbiol. 2016, 7, 1050. [Google Scholar] [CrossRef] [PubMed]
| Strains/Isolates | Relevant Features | Reference/Source |
|---|---|---|
| Bacterial strains | ||
| Latilactobacillus sakei CECT 906 T | Microbial indicator for bacteriocin production | CECT a |
| Lactococcus lactis subsp. cremoris MG1363 | Microbial indicator for bacteriocin production | [23] |
| Lactococcus lactis NCDO 604 T | Positive control for proteolytic activity | NCDO b |
| Lactococcus lactis SH4109 | Positive control for proteolytic activity | [23] |
| Listeria innocua CECT 910 T | Microbial indicator for bacteriocin production | CECT a |
| Micrococcus luteus NCIMB 8166 | Microbial indicator for bacteriocin production | NCIMB c |
| Streptococcus thermophilus CNRZ 1066 | Microbial indicator for bacteriocin production | CNRZ d |
| Streptococcus thermophilus LMD9 | Microbial indicator for bacteriocin production | NBCC e |
| Lactococcus lactis subsp. cremoris SK11 | Non-GABA producer starter culture | [24] |
| Mold strain | ||
| Penicillium roqueforti 1AM8 | Proteolytic mold from Cabrales cheese | [25] |
| Lactococcus lactis isolates | ||
| L. lactis subsp. lactis LEY6 | GABA producer | This work |
| L. lactis subsp. lactis LEY7 | GABA producer | This work |
| L. lactis subsp. lactis LEY8 | GABA producer | This work |
| L. lactis subsp. lactis LEY11 | GABA producer | This work |
| L. lactis subsp. lactis LEY12 | GABA producer | This work |
| L. lactis subsp. lactis LEY13 | GABA producer | This work |
| Lactococcus Milk Isolates | Geographic Location of Milk Samples | Growth at | |||
|---|---|---|---|---|---|
| 45 °C | pH 9.6 | 4% NaCl | 6.5% NaCl | ||
| LEY6 | Saida | + | + | + | − |
| LEY7 | Saida | − | + | + | − |
| LEY8 | Saida | − | + | + | − |
| LEY11 | Bechar | + | + | + | − |
| LEY12 | Ghardaia | + | − | + | − |
| LEY13 | Tindouf | + | − | + | − |
| L. lactis Isolates | Incubation in UHT Skimmed Milk * | ||
|---|---|---|---|
| pH after 6 h | pH after 18 h | Milk Clotting after 18 h | |
| LEY6 | 5.12 ± 0.26 a,a | 4.21 ± 0.13 a,b | + |
| LEY7 | 5.33 ± 0.54 a,a | 4.32 ± 0.31 a,b | + |
| LEY8 | 5.14 ± 0.31 a,a | 4.14 ± 0.07 a,b | + |
| LEY11 | 5.63 ± 0.33 a,a | 4.22 ± 0.12 a,b | + |
| LEY12 | 5.71 ± 0.50 a,a | 4.18 ± 0.09 a,b | + |
| LEY13 | 5.42 ± 0.13 a,a | 4.12 ± 0.03 a,b | + |
| Strains | ||||||
|---|---|---|---|---|---|---|
| Volatile Compound † | LEY6 | LEY7 | LEY8 | LEY11 | LEY12 | LEY13 |
| Acetaldehyde | 76.9 ± 7.5 a | 25.7 ± 44.5 a | 57.7 ± 52.4 a | 23.9 ± 4.3 a | 21.9 ± 3.6 a | 26.2 ± 8.6 a |
| 2-Methyl propanal | 179.1 ± 15.5 a | 123.5 ± 43.2 a | 180.0 ± 35.3 a | 121.3 ± 22.3 a | 99.8 ± 30.9 a | 131.8 ± 21.9 a |
| Methyl acetate | − b | − b | − b | 26.1 ± 7.5 a | 25.2 ± 9.4 a | 26.9 ± 2.3 a |
| 2-Methyl butanal | 72.6 ± 12.9 a | 80.1 ± 10.6 a | 61.8 ± 16.9 a | − b | − b | − b |
| 3-Methyl butanal | 877.7 ± 70.3 a | 796.1 ± 197.7 a | 797.0 ± 50.3 a | 852.8 ± 244.5 a | 588.8 ± 248.1 a | 897.9 ± 154.6 a |
| Ethanol | 396.5 ± 32.5 a | 353.5 ± 107.6 a | 340.5 ± 18.8 a | 351.5 ± 59.8 a | 315.3 ± 32.7 a | 362.6 ± 58.5 a |
| 2,2,4,6,6 PMH ‡ | − b | 95.0 ± 164.6 a | 66.4 ± 115.1 a | − b | 51.1 ± 88.5 a | 83.6 ± 144.9 a |
| Methyl butanoate | − b | − b | − b | 2.3 ± 4.0 a | 9.1 ± 8.0 a | − b |
| 2-Methyl-1-propanol | 55.6 ± 2.2 a | 51.4 ± 48.6 a | 41.4 ± 7.6 b | 30.7 ± 1.8 b | 35.7 ± 6.4 b | 31.7 ± 7.6 b |
| Methyl hexanoate | 23.9 ± 1.3 a | − b | − b | 3.6 ± 6.3 a | 2.8 ± 4.8 a | 9.6 ± 8.3 a |
| 2 or 3-Methyl-1-butanol | 322.9 ± 8.5 a | 367.6 ± 28.1 a | 298.7 ± 22.8 a | 198.4 ± 36.2 b | 213.2 ± 42.2 b | 205.0 ± 21.5 b |
| Acetoin | − b | − b | − b | − b | 2.6 ± 4.6 a | − b |
| Acetic acid | − b | − b | − b | 21.9 ± 5.7 a | 22.1 ± 3.0 a | 17.7 ± 2.0 a |
| L. lactis Isolates | Tyramine | Histamine | Putrescine (AGDI 1) | Putrescine (ODC 2) | Cadaverine |
|---|---|---|---|---|---|
| LEY6 | − | − | 0.785 ± 0.02 a | − | − |
| LEY7 | − | − | 0.758 ± 0.02 a | − | − |
| LEY8 | − | − | 0.763 ± 0.02 a | − | − |
| LEY11 | − | − | 0.791 ± 0.02 a | − | − |
| LEY12 | − | − | 0.776 ± 0.01 a | − | − |
| LEY13 | − | − | 0.803 ± 0.01 a | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Redruello, B.; Saidi, Y.; Sampedro, L.; Ladero, V.; del Rio, B.; Alvarez, M.A. GABA-Producing Lactococcus lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese. Foods 2021, 10, 633. https://doi.org/10.3390/foods10030633
Redruello B, Saidi Y, Sampedro L, Ladero V, del Rio B, Alvarez MA. GABA-Producing Lactococcus lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese. Foods. 2021; 10(3):633. https://doi.org/10.3390/foods10030633
Chicago/Turabian StyleRedruello, Begoña, Yasmine Saidi, Lorena Sampedro, Victor Ladero, Beatriz del Rio, and Miguel A. Alvarez. 2021. "GABA-Producing Lactococcus lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese" Foods 10, no. 3: 633. https://doi.org/10.3390/foods10030633
APA StyleRedruello, B., Saidi, Y., Sampedro, L., Ladero, V., del Rio, B., & Alvarez, M. A. (2021). GABA-Producing Lactococcus lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese. Foods, 10(3), 633. https://doi.org/10.3390/foods10030633









