Use of Acetic Acid to Partially Replace Lactic Acid for Decontamination against Escherichia coli O157:H7 in Fresh Produce and Mechanism of Action
Abstract
:1. Introduction
2. Materials and Methods
2.1. Time-Kill Curve Analysis
2.2. Sample Preparation
2.3. Inoculation
2.4. Decontamination and Microbiological Analysis
2.5. Sensory Analysis
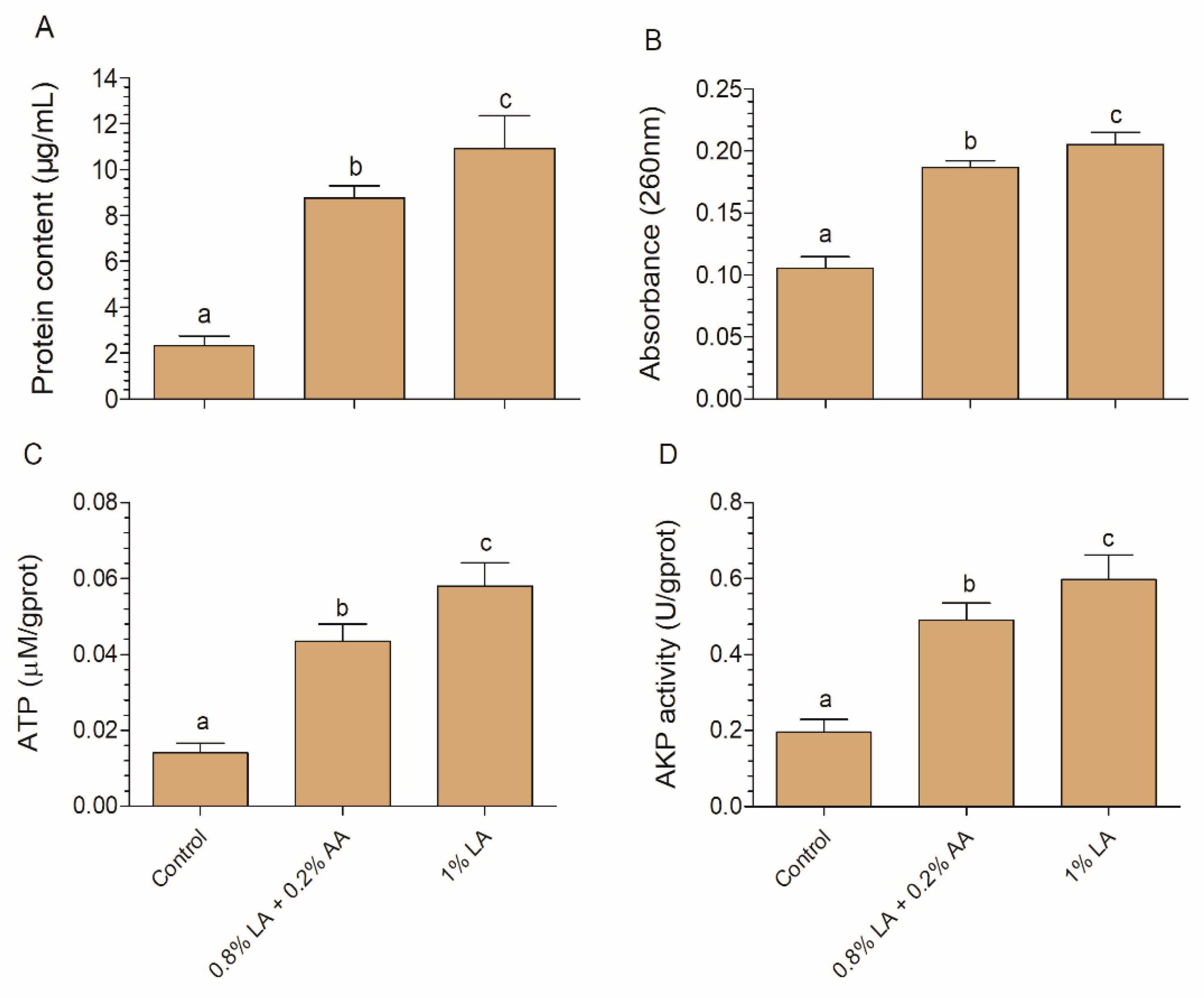
2.6. Protein, Nucleotide, ATP, and AKP Leakage Analysis
2.7. Proteomic Analysis
2.7.1. Protein Preparation
2.7.2. Protein Digestion and Peptide Labeling
2.7.3. Peptide Fractionation
2.7.4. LC–MS/MS
2.7.5. Database Searching and Data Analysis
2.8. Bioinformatics and Statistical Analysis
3. Results and Discussion
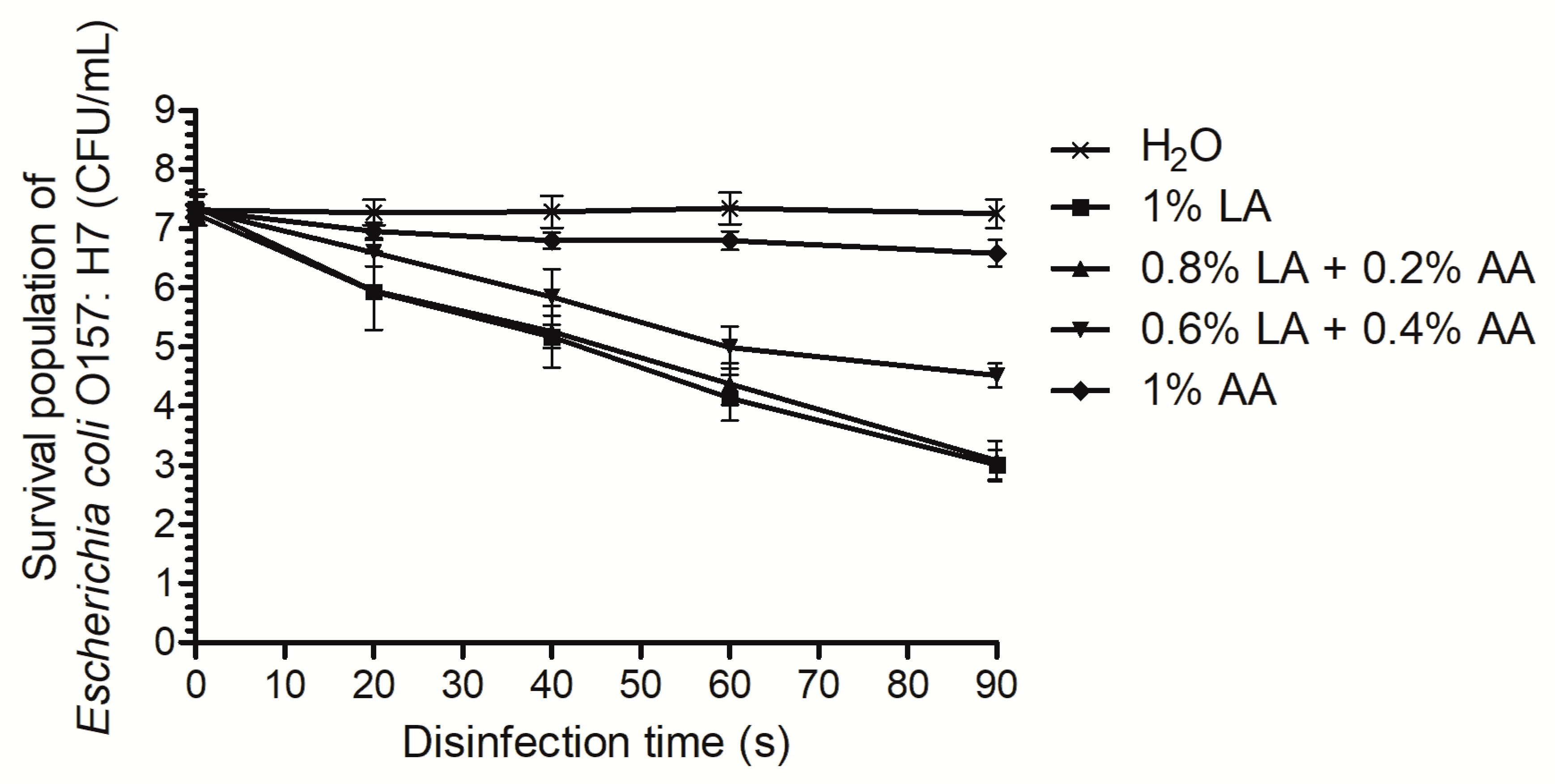
3.1. Time-Kill Curves
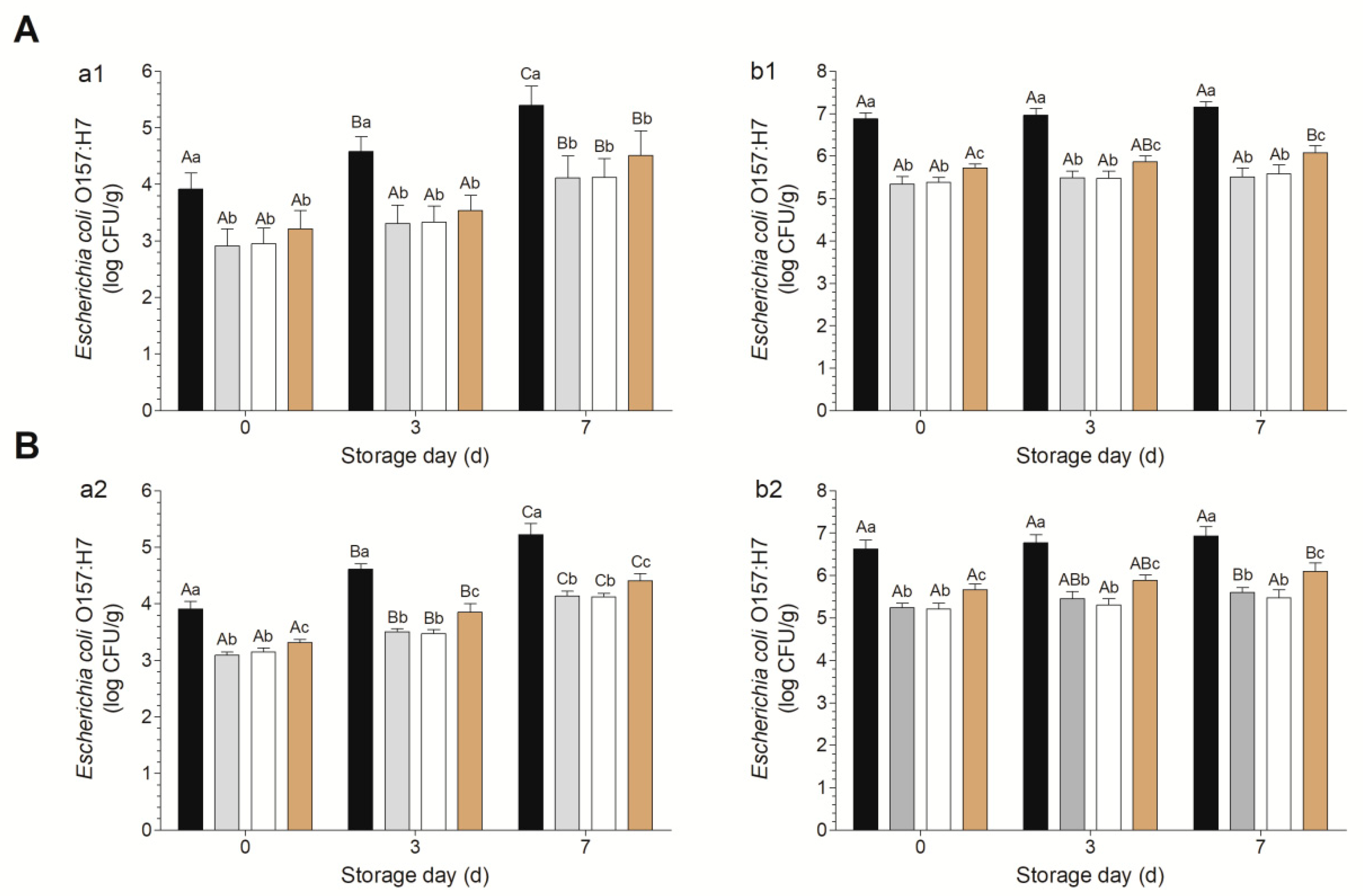
3.2. Decontamination Efficacies of Various Combinations against Escherichia coli O157:H7 on Fresh-Cut Baby Spinach and Arugula
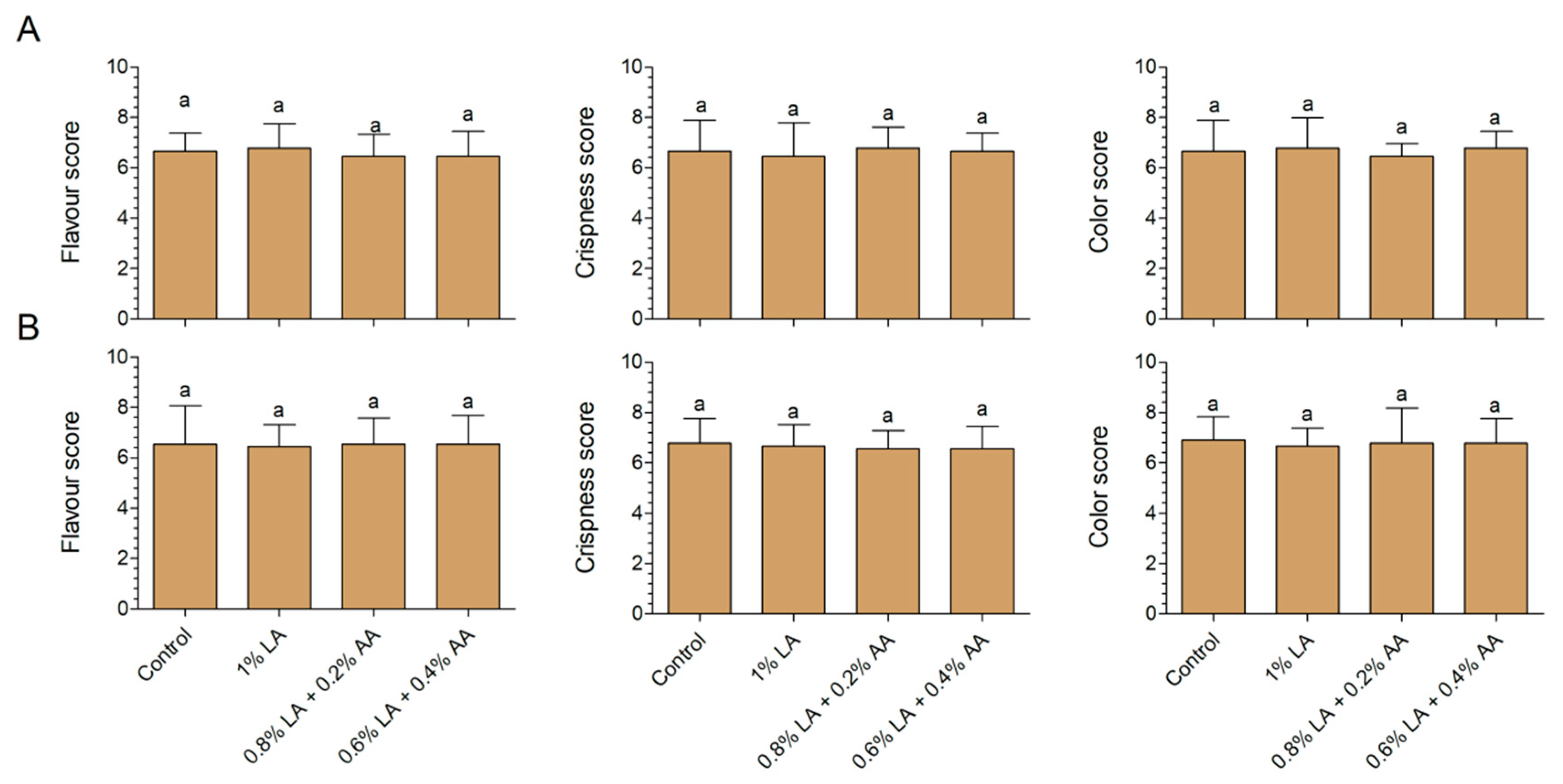
3.3. Effects of Sanitizers on Sensory Characteristics of Fresh-Cut Baby Spinach and Arugula
3.4. Effects of Sanitizers on the Proteome
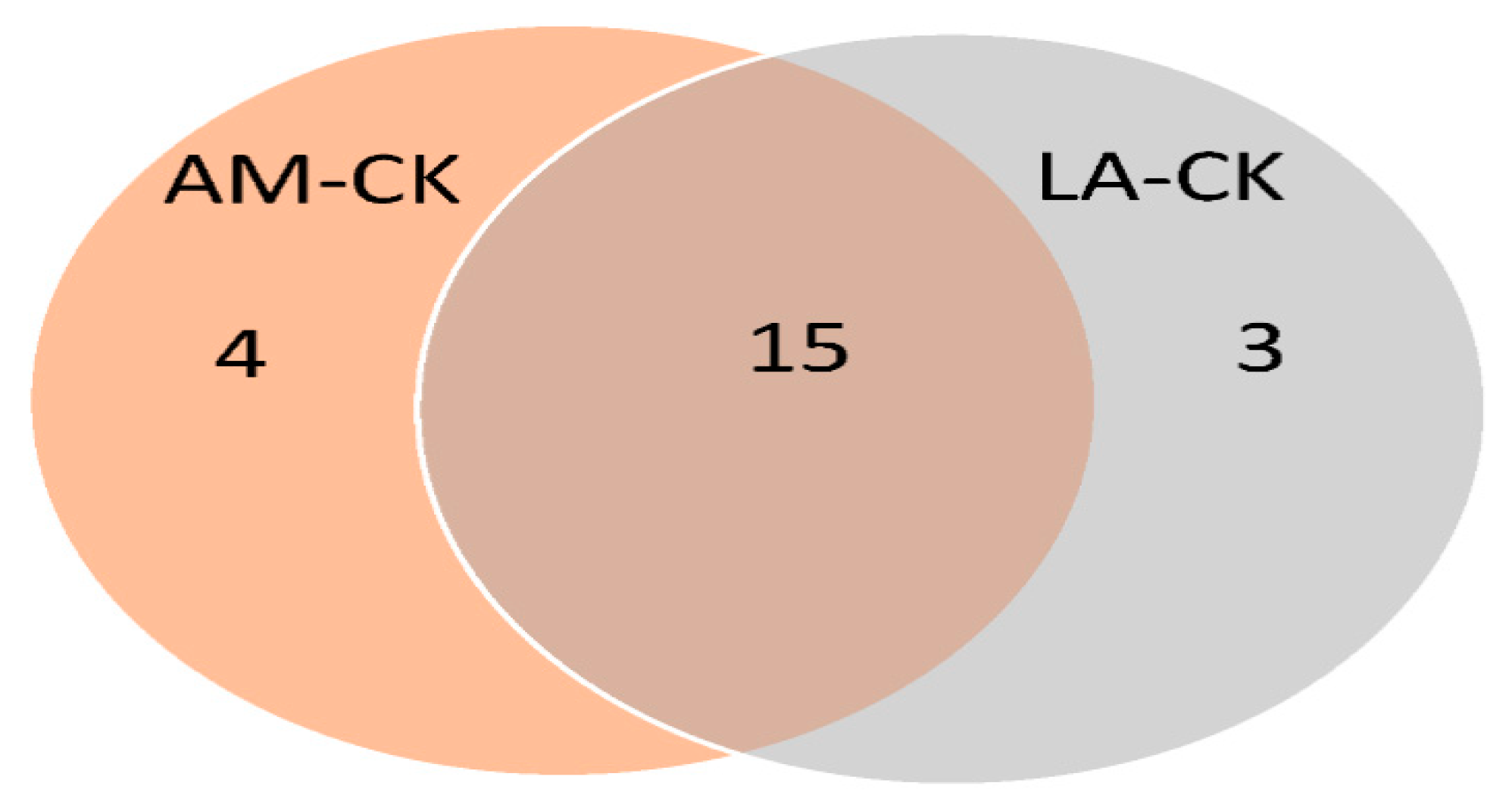
3.5. Venn Diagram Analysis
3.6. Unique Enriched Biological Processes and Network Clustering of LA–CK
3.7. Unique Enriched Biological Processes and Network Clustering of AM–CK
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Yu, Y.; Dong, Y. Disinfection of Ready-to-Eat Lettuce Using Polyhexamethylene Guanidine Hydrochloride. Microorganisms 2020, 8, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Tao, D.; Wang, S.; Li, C.; Li, Y.; Zheng, F.; Wu, Z. Disinfection of lettuce using organic acids: An ecological analysis using 16S rRNA sequencing. RSC Adv. 2019, 9, 17514–17520. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yu, Y.; Dong, Y. Combination of polyhexamethylene guanidine hydrochloride and potassium peroxymonosulfate to disinfect ready-to-eat lettuce. RSC Adv. 2020, 10, 40316–40320. [Google Scholar] [CrossRef]
- Callejón, R.M.; Rodríguez-Naranjo, M.I.; Ubeda, C.; Ortega, R.H.; Garcia-Parrilla, M.C.; Troncoso, A.M. Reported Foodborne Outbreaks Due to Fresh Produce in the United States and European Union: Trends and Causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef]
- Reports of Selected Escherichia coli Outbreak Investigations. Available online: https://www.cdc.gov/ecoli/outbreaks.html (accessed on 12 June 2021).
- De Corato, U. Improving the shelf-life and quality of fresh and minimally-processed fruits and vegetables for a modern food industry: A comprehensive critical review from the traditional technologies into the most promising advancements. Crit. Rev. Food Sci. Nutr. 2019, 60, 940–975. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, M.; Bhandari, B.; Gao, Z. Recent developments in novel shelf life extension technologies of fresh-cut fruits and vegetables. Trends Food Sci. Technol. 2017, 64, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Ölmez, H.; Kretzschmar, U. Potential alternative disinfection methods for organic fresh-cut industry for minimizing water consumption and environmental impact. LWT Food Sci. Technol. 2009, 42, 686–693. [Google Scholar] [CrossRef]
- Gil, M.I.; Selma, M.V.; López-Gálvez, F.; Allende, A. Fresh-cut product sanitation and wash water disinfection: Problems and solutions. Int. J. Food Microbiol. 2009, 134, 37–45. [Google Scholar] [CrossRef]
- Luo, Y.; Zhou, B.; Van Haute, S.; Nou, X.; Zhang, B.; Teng, Z.; Turner, E.R.; Wang, Q.; Millner, P.D. Association between bacterial survival and free chlorine concentration during commercial fresh-cut produce wash operation. Food Microbiol. 2018, 70, 120–128. [Google Scholar] [CrossRef]
- Garrido, Y.; Marín, A.; Tudela, J.A.; Allende, A.; Gil, M.I. Chlorate uptake during washing is influenced by product type and cut piece size, as well as washing time and wash water content. Postharvest Biol. Technol. 2019, 151, 45–52. [Google Scholar] [CrossRef]
- Van Haute, S.; Tryland, I.; Escudero, C.; Vanneste, M.; Sampers, I. Chlorine dioxide as water disinfectant during fresh-cut iceberg lettuce washing: Disinfectant demand, disinfection efficiency, and chlorite formation. LWT 2017, 75, 301–304. [Google Scholar] [CrossRef]
- Van Haute, S.; Tryland, I.; Veys, A.; Sampers, I. Wash water disinfection of a full-scale leafy vegetables washing process with hydrogen peroxide and the use of a commercial metal ion mixture to improve disinfection efficiency. Food Control. 2015, 50, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, Y.; Yu, Y.; Wu, Z.; Wang, H. Combination of ozone and ultrasonic-assisted aerosolization sanitizer as a sanitizing process to disinfect fresh-cut lettuce. Ultrason. Sonochem. 2021, 76, 105622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, H. Effects of potential organic compatible sanitisers on organic and conventional fresh-cut lettuce (Lactuca sativa Var. Crispa L). Food Control 2017, 72, 20–26. [Google Scholar] [CrossRef]
- Velázquez, L.D.C.; Barbini, N.B.; Escudero, M.E.; Estrada, C.S.M.L.; de Guzmán, A.M.S. Evaluation of chlorine, benzalkonium chloride and lactic acid as sanitizers for reducing Escherichia coli O157:H7 and Yersinia enterocolitica on fresh vegetables. Food Control 2009, 20, 262–268. [Google Scholar] [CrossRef]
- Ölmez, H.; Temur, S. Effects of different sanitizing treatments on biofilms and attachment of Escherichia coli and Listeria monocytogenes on green leaf lettuce. LWT 2010, 43, 964–970. [Google Scholar] [CrossRef]
- Neal, J.A.; Marquez-Gonzalez, M.; Cabrera-Diaz, E.; Lucia, L.M.; O’Bryan, C.A.; Crandall, P.G.; Ricke, S.C.; Castillo, A. Comparison of multiple chemical sanitizers for reducing Salmonella and Escherichia coli O157:H7 on spinach (Spinacia oleracea) leaves. Food Res. Int. 2012, 45, 1123–1128. [Google Scholar] [CrossRef]
- Wang, J.; Wang, S.; Sun, Y.; Li, C.; Li, Y.; Zhang, Q.; Wu, Z. Reduction of Escherichia coli O157:H7 and naturally present microbes on fresh-cut lettuce using lactic acid and aqueous ozone. RSC Adv. 2019, 9, 22636–22643. [Google Scholar] [CrossRef] [Green Version]
- Ricke, S. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult. Sci. 2003, 82, 632–639. [Google Scholar] [CrossRef]
- Hui, W.; Colin, O.; Yang, X. Development of a real-time PCR procedure for quantification of viable Escherichia coli in popu-lations of E. coli exposed to lactic acid, and the acid tolerance of verotoxigenic E. coli (VTEC) from cattle hides. Food Control 2014, 43, 104–109. [Google Scholar]
- Nakamura, K.; Tanaka, T.; Kuwahara, A.; Takeo, K. Microassay for proteins on nitrocellulose filter using protein dye-staining procedure. Anal. Biochem. 1985, 148, 311–319. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Y.; Zhang, Y.; Wu, R.; Li, P. Antibacterial mechanism of plantaricin LPL-1, a novel class IIa bacteriocin against Listeria monocytogenes. Food Control 2019, 97, 87–93. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, Y.; Wu, R.; Jiang, D.; Bai, B.; Tan, D.; Yan, T.; Sun, X.; Zhang, Q.; Wu, Z. Antibacterial Activity of Juglone against Staphylococcus aureus: From Apparent to Proteomic. Int. J. Mol. Sci. 2016, 17, 965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Gotz, S.; Garciagomez, J.M.; Terol, J.; Talon, M.; Robles, M.J.B. Blast2GO: A universal tool for annotation, visual-ization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Ritter, A.C.; Santi, L.; Vannini, L.; Beys-Da-Silva, W.; Gozzi, G.; Yates, J.; Ragni, L.; Brandelli, A. Comparative proteomic analysis of foodborne Salmonella Enteritidis SE86 subjected to cold plasma treatment. Food Microbiol. 2018, 76, 310–318. [Google Scholar] [CrossRef]
- Ahmed, S.; Zaman, S.; Ahmed, R.; Uddin, N.; Acedo, A.; Bari, L. Effectiveness of non-chlorine sanitizers in improving the safety and quality of fresh betel leaf. LWT 2017, 78, 77–81. [Google Scholar] [CrossRef]
- Finten, G.; Agüero, M.V.; Jagus, R.J. Citric acid as alternative to sodium hypochlorite for washing and disinfection of experimentally-infected spinach leaves. LWT Food Sci. Technol. 2017, 82, 318–325. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, H.; Liu, Q.; Pang, X.; Zhao, X.; Yang, H. Sanitising efficacy of lactic acid combined with low-concentration sodium hypochlorite on Listeria innocua in organic broccoli sprouts. Int. J. Food Microbiol. 2019, 295, 41–48. [Google Scholar] [CrossRef]
- Fan, X.; Sokorai, K.J.; Niemira, B.A.; Mills, R.S.; Zhen, M.Y. Quality of Gamma Ray-irradiated Iceberg Lettuce and Treatments to Minimize Irradiation-induced Disorders. HortScience 2012, 47, 1108–1112. [Google Scholar] [CrossRef] [Green Version]
- Salgado, S.P.; Pearlstein, A.J.; Luo, Y.; Feng, H. Quality of Iceberg (Lactuca sativa L.) and Romaine (L. sativa L. var. longifolial) lettuce treated by combinations of sanitizer, surfactant, and ultrasound. LWT 2014, 56, 261–268. [Google Scholar] [CrossRef]
- Wang, J.; Sun, Y.; Tao, D.; Li, C.; Zheng, F.; Wu, Z. Reduction of Escherichia coli O157:H7, Listeria monocytogenes, and Naturally Present Microbe Counts on Lettuce using an Acid Mixture of Acetic and Lactic Acid. Microorganisms 2019, 7, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayakumar, C.; Wolf-Hall, C.E. Evaluation of Household Sanitizers for Reducing Levels of Escherichia coli on Iceberg Lettuce. J. Food Prot. 2002, 65, 1646–1650. [Google Scholar] [CrossRef] [PubMed]
- Poimenidou, S.V.; Bikouli, V.C.; Gardeli, C.; Mitsi, C.; Tarantilis, P.A.; Nychas, G.J.; Skandamis, P.N. Effect of single or com-bined chemical and natural antimicrobial interventions on Escherichia coli O157:H7, total microbiota and color of packaged spinach and lettuce. Int. J. Food Microbiol. 2016, 220, 6–18. [Google Scholar] [CrossRef]
- Wang, J.; Liu, D.; Sun, X.; Bai, B.; Jiang, D.; Wu, Z. Label-free quantitative proteomic analysis of the inhibitory activities of juglone against translation and energy metabolism in Escherichia coli. Phytochem. Lett. 2016, 18, 55–58. [Google Scholar] [CrossRef]
- Yang, X.Y.; Zhang, L.; Liu, J.; Li, N.; Yu, G.; Cao, K.; Han, J.; Zeng, G.; Pan, Y.; Sun, X.; et al. Proteomic analysis on the anti-bacterial activity of a Ru(II) complex against Streptococcus pneumoniae. J. Proteomics 2015, 115, 107–116. [Google Scholar] [CrossRef]
- Yang, X.-Y.; Xu, J.-Y.; Meng, M.; Li, N.; Liu, C.-Y.; He, Q.-Y. Dirhodium (II) complex interferes with iron-transport system to exert antibacterial action against Streptococcus pneumoniae. J. Proteom. 2018, 194, 160–167. [Google Scholar] [CrossRef]
- Solis, N.; Parker, B.L.; Kwong, S.M.; Robinson, G.; Firth, N.; Cordwell, S.J. Staphylococcus aureus Surface Proteins Involved in Adaptation to Oxacillin Identified Using a Novel Cell Shaving Approach. J. Proteome Res. 2014, 13, 2954–2972. [Google Scholar] [CrossRef]
- Natale, P.; Brüser, T.; Driessen, A.J.M. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim. Biophys. Acta 2008, 1778, 1735–1756. [Google Scholar] [CrossRef] [Green Version]
- Luirink, J.; Hagen-Jongman, C.T.; van der Weijden, C.; Oudega, B.; High, S.; Dobberstein, B.; Kusters, R. An alternative protein targeting pathway in Escherichia coli: Studies on the role of FtsY. EMBO J. 1994, 13, 2289–2296. [Google Scholar] [CrossRef] [Green Version]
- Diao, M.; Qi, D.; Xu, M.; Lu, Z.; Lv, F.; Bie, X.; Zhang, C.; Zhao, H. Antibacterial activity and mechanism of monolauroyl-galactosylglycerol against Bacillus cereus. Food Control 2018, 85, 339–344. [Google Scholar] [CrossRef]
- Kang, S.; Kong, F.; Shi, X.; Han, H.; Li, M.; Guan, B.; Yang, M.; Cao, X.; Tao, D.; Zheng, Y.; et al. Antibacterial activity and mechanism of lactobionic acid against Pseudomonas fluorescens and methicillin-resistant Staphylococcus aureus and its application on whole milk. Food Control 2020, 108, 106876. [Google Scholar] [CrossRef]
- Jeckelmann, J.-M.; Harder, D.; Mari, S.A.; Meury, M.; Ucurum, Z.; Muller, D.J.; Erni, B.; Fotiadis, D. Structure and function of the glucose PTS transporter from Escherichia coli. J. Struct. Biol. 2011, 176, 395–403. [Google Scholar] [CrossRef]
- Yamashita, T.; Kawada-Matsuo, M.; Katsumata, T.; Watanabe, A.; Oogai, Y.; Nishitani, Y.; Miyawaki, S.; Komatsuzawa, H. Antibacterial activity of disodium succinoyl glycyrrhetinate, a derivative of glycyrrhetinic acid againstStreptococcus mutans. Microbiol. Immunol. 2019, 63, 251–260. [Google Scholar] [CrossRef]
- Tchieu, J.H.; Norris, V.; Edwards, J.S.; Saier, M.H. The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001, 3, 329–346. [Google Scholar]
- Liu, Y.; Ceruso, M.; Jiang, Y.; Datta, A.R.; Carter, L.; Strain, E.; Pepe, T.; Anastasi, A.; Fratamico, P. Construction ofListeria monocytogenesMutants with In-Frame Deletions in the Phosphotransferase Transport System (PTS) and Analysis of Their Growth under Stress Conditions. J. Food Sci. 2013, 78, M1392–M1398. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Membrez, M.; Morin-Rivron, D.; Siddharth, J.; Chou, C.J.; Henry, H.; Bruce, S.; Metairon, S.; Raymond, F.; Betrisey, B.; et al. Transcriptomics-driven lipidomics (TDL) identifies the microbiome-regulated targets of ileal lipid metabolism. Npj Syst. Biol. Appl. 2017, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Benga, G. On the definition, nomenclature and classification of water channel proteins (aquaporins and relatives). Mol. Asp. Med. 2012, 33, 514–517. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhang, A.-H.; Sun, H.; Yan, G.-L.; Wang, X.-J. Recent advances and effective strategies in the discovery and applications of natural products. RSC Adv. 2018, 8, 812–824. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Chen, Y.; Lu, Q.; Qin, H.; Ou, H.; He, B.; Ye, J. Cellular metabolism network of Bacillus thuringiensis related to erythromycin stress and degradation. Ecotoxicol. Environ. Saf. 2018, 160, 328–341. [Google Scholar] [CrossRef]
- Sassanfar, M.; Roberts, J.W. Nature of the SOS-inducing signal in Escherichia coli: The involvement of DNA replication. J. Mol. Biol. 1990, 212, 79–96. [Google Scholar] [CrossRef]
- Kim, E.A.; Panushka, J.; Meyer, T.; Carlisle, R.; Baker, S.; Ide, N.; Lynch, M.; Crane, B.R.; Blair, D.F. Architecture of the Flagellar Switch Complex of Escherichia coli: Conformational Plasticity of FliG and Implications for Adaptive Remodeling. J. Mol. Biol. 2017, 429, 1305–1320. [Google Scholar] [CrossRef] [PubMed]
- Ford, K.M.; Antani, J.; Nagarajan, A.; Johnson, M.M.; Lele, P.P. Switching and Torque Generation in Swarming E. coli. Front. Microbiol. 2018, 9, 2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Hu, X.; Gao, Z.; Li, G.; Fu, F.; Shang, X.; Liang, Z.; Shan, Y. Global transcriptomic response of Listeria monocytogenes exposed to Fingered Citron (Citrus medica L. var. sarcodactylis Swingle) essential oil. Food Res. Int. 2021, 143, 110274. [Google Scholar] [CrossRef] [PubMed]
| UniProt Accession Number | Gene ID | Protein Name | Fold Change |
|---|---|---|---|
| Protein export | |||
| P0AG88 | secB | Protein-export protein SecB | 2.35 |
| P0AGA1 | secG | Protein-export membrane protein SecG | 2.06 |
| P0AGA4 | secY | Protein translocase subunit SecY | 0.53 |
| P0AG91 | secD | Protein translocase subunit SecD | 0.65 |
| P0AG95 | secF | Protein translocase subunit SecF | 1.24 |
| P65625 | yidC | Membrane protein insertase YidC | 0.43 |
| P0AGD9 | ffh | Signal recognition particle protein | 0.38 |
| Outer membrane and gram-negative porins | |||
| P61318 | lolA | Outer-membrane lipoprotein carrier protein | 0.82 |
| P0ADC5 | lolC | Lipoprotein-releasing system transmembrane protein LolC | 0.37 |
| P0ABV8 | tolR | Tol-Pal system protein TolR | 2.07 |
| P0ABV0 | tolQ | Tol-Pal system protein TolQ | 0.46 |
| Q8X8E2 | lolE | Lipoprotein-releasing system transmembrane protein | 0.45 |
| P0AC04 | bamD | Outer membrane protein assembly factor BamD | 2.45 |
| P0AEU9 | skp | Chaperone protein Skp | 2.25 |
| P0A9V3 | lptB | Lipopolysaccharide export system ATP-binding protein LptB | 1.42 |
| UniProt Accession Number | Gene ID | Protein Name | Fold Change |
|---|---|---|---|
| Starch and sucrose metabolism, and phosphotransferase system | |||
| Q8XDG9 | mtlD | Mannitol-1-phosphate 5-dehydrogenase | 1.27 |
| Q8X677 | manA | Mannose-6-phosphate isomerase | 0.27 |
| Q8XE22 | pfkB | Phosphofructokinase | 0.59 |
| A0A0H3JCR2 | pgm | Phosphoglucomutase | 0.35 |
| P0AB72 | fbaA | Fructose-bisphosphate aldolase class 2 | 0.57 |
| P62709 | gpmA | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | 2.34 |
| P0A6V9 | glk | Glucokinase | 0.60 |
| Q8XCE1 | treC | Trehalose-6-P hydrolase | 0.38 |
| Q8X710 | malQ | 4-alpha-glucanotransferase | 0.37 |
| Q8X6Y1 | glgP | Alpha-1,4 glucan phosphorylase | 0.41 |
| Q8X6X8 | glgX | Glycogen debranching enzyme | 0.34 |
| Glycerophospholipid metabolism and major intrinsic proteins | |||
| P0A997 | glpC | Anaerobic glycerol-3-phosphate dehydrogenase subunit C | 0.59 |
| P0A6F4 | glpK | Glycerol kinase | 0.55 |
| P0A9C1 | glpA | Anaerobic glycerol-3-phosphate dehydrogenase subunit A | 0.69 |
| A0A0H3JI74 | glpD | Glycerol-3-phosphate dehydrogenase | 0.60 |
| P0A6S9 | gpsA | Glycerol-3-phosphate dehydrogenase [NAD(P)+] | 0.38 |
| Flagellar assembly and bacterial chemotaxis | |||
| P0A966 | cheW | Chemotaxis protein CheW | 2.83 |
| P0AE68 | cheY | Chemotaxis protein CheY | 2.80 |
| Q8XCF9 | cheB | Protein-glutamate methylesterase/protein-glutamine glutaminase | 0.60 |
| P0ABZ3 | fliG | Flagellar motor switch protein FliG | 1.67 |
| P0ABY0 | fliL | Flagellar protein FliL | 2.00 |
| Homologous recombination and DNA repair | |||
| Q8X8H1 | polA | DNA polymerase I | 0.78 |
| P0A7G8 | recA | Protein RecA | 1.41 |
| P0A7H2 | recF | DNA replication and repair protein RecF | 0.67 |
| Q8XDN4 | mutL | DNA mismatch repair protein MutL | 0.40 |
| Q8X8P5 | uvrD | DNA helicase | 0.58 |
| P0A811 | ruvA | Holliday junction ATP-dependent DNA helicase RuvA | 0.69 |
| Q8X5H9 | ftsK | DNA translocase FtsK | 0.21 |
| P0A7C4 | lexA | LexA repressor | 2.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Lei, Y.; Yu, Y.; Yin, L.; Zhang, Y. Use of Acetic Acid to Partially Replace Lactic Acid for Decontamination against Escherichia coli O157:H7 in Fresh Produce and Mechanism of Action. Foods 2021, 10, 2406. https://doi.org/10.3390/foods10102406
Wang J, Lei Y, Yu Y, Yin L, Zhang Y. Use of Acetic Acid to Partially Replace Lactic Acid for Decontamination against Escherichia coli O157:H7 in Fresh Produce and Mechanism of Action. Foods. 2021; 10(10):2406. https://doi.org/10.3390/foods10102406
Chicago/Turabian StyleWang, Jiayi, Yue Lei, Yougui Yu, Lebin Yin, and Yangyang Zhang. 2021. "Use of Acetic Acid to Partially Replace Lactic Acid for Decontamination against Escherichia coli O157:H7 in Fresh Produce and Mechanism of Action" Foods 10, no. 10: 2406. https://doi.org/10.3390/foods10102406
APA StyleWang, J., Lei, Y., Yu, Y., Yin, L., & Zhang, Y. (2021). Use of Acetic Acid to Partially Replace Lactic Acid for Decontamination against Escherichia coli O157:H7 in Fresh Produce and Mechanism of Action. Foods, 10(10), 2406. https://doi.org/10.3390/foods10102406





