Abstract
Introduction: Antimicrobial resistance is a significant public health issue worldwide, associated with limited treatment options and with major consequences for healthcare systems. Our study aims to assess rates and patterns of resistance to five last-resort antimicrobials in a cohort of carbapenemase-producing Klebsiella pneumoniae strains, isolated over a one-year interval. Additionally, we have tested two potentially synergistic combinations for in vitro efficacy. Methods: This prospective observational study evaluated Klebsiella pneumoniae strains with diminished carbapenem susceptibility from patients admitted to the National Institute for Infectious Diseases “Prof. Dr. Matei Balș” in Bucharest between August 2023 and July 2024. Strains presenting a minimum inhibitory concentration to meropenem of >0.125 μg/mL underwent phenotypic enzyme production testing, followed by synergistic testing to identify antimicrobial salvage therapy options. A subset of these strains was analysed for the detection of plasmid-mediated resistance genes, using a custom workflow for DNA extraction and amplification/detection. Results: A total of 139 non-duplicate strains were isolated, with 129 (92.8%) being carbapenemase producers. These 129 strains were phenotypically diverse: 29 (22.5%) were NDM, 12 (9.3%) OXA-48 type, 8 (6.2%) KPC, while most of them (62.0%) were double carbapenemase producers: 79 (61.2%) NDM and OXA-48-type, and one strain was NDM and KPC. Forty-six strains were resistant to cefiderocol (35.7%), 108 (83.7%) to ceftazidime/avibactam, 127 (98.4%) to ceftolozane/tazobactam, 116 (90.0%) to imipenem/relebactam and 127 (98.4%) to aztreonam. The association of aztreonam with ceftazidime/avibactam demonstrated a synergistic effect in 127 (98.5%) strains, while aztreonam with imipenem/relebactam was efficient in vitro against 103 (79.8%) strains. Conclusions: Antimicrobial resistance remains a concerning phenomenon among Enterobacterales, especially when considering the increasing resistance rates even against salvage therapy antimicrobials.
Introduction
Gram-negative pathogens have been notoriously associated with various invasive presentations of infectious diseases, especially in healthcare-associated settings. Klebsiella pneumoniae is a well-studied representative of this group, being involved in urinary tract infections (UTIs), bloodstream infections (BSIs), community-acquired pneumonia (CAP) and various opportunistic infections []. Klebsiella pneumoniae commonly inhabits the human gastrointestinal tract and nasopharynx, and infections occur especially in individuals with immunosuppression. In case of repeated infections with K. pneumoniae, alterations of the digestive and respiratory microbiome have been described [,].
Common mechanisms of Klebsiella pneumoniae antimicrobial resistance include enzyme-mediated antibiotic inactivation, through the production of carbapenemases or beta-lactamases, changes in the structure of antibiotic targets, such as penicillin-binding proteins (PBPs) and alterations of cellular permeability []. Nonetheless, the emergence of multidrug-resistant (MDR), extended-drug-resistant (XDR) or even pandrug-resistant (PDR) strains represents a major threat to public health and healthcare systems.
Carbapenem-resistant Klebsiella pneumoniae (CRKP) strains possess a large number of plasmid-encoded and genome-encoded resistance genes, and use successful interbacterial communication for transmission of these resistance genes []. Rates of susceptibility have geographically heterogeneous patterns; thus, antimicrobial resistance surveillance is an important tool to provide data needed for evidence-based updates of national and regional antibiotic prescribing guidelines [].
This study aims to characterise antimicrobial resistance patterns in non-duplicate, carbapenem-resistant Klebsiella pneumoniae strains isolated from a tertiary care infectious diseases hospital in Romania, a country with one of the highest rates of antibiotic resistance among European states []. The study focused on resistance patterns for last-resort antimicrobials, such as ceftazidime/avibactam (CZA), ceftolozane/tazobactam (C/T), imipenem/relebactam (IMR), cefiderocol (FDC), aztreonam (ATM) and potentially synergistic combinations. Additionally, molecular detection and characterisation for plasmid-mediated antibiotic resistance have been performed on a subset of the strains. Another objective of the study was to assess the rates of resistance to cefiderocol before and after its introduction into clinical practice in this hospital.
Methods
Study design
This prospective study was conducted in the Clinical Medical Microbiology Laboratory of the National Institute for Infectious Diseases “Prof. Dr. Matei Balș” (NIID), a mono-disciplinary tertiary care hospital in Bucharest, Romania, between August 2023 and July 2024. The study was performed in accordance with the Declaration of Helsinki. The institutional review board approved data access without the need for individual consent forms, as the completely anonymised data were extracted from the Institution’s electronic medical database.
Bacterial culture
Clinical samples (urine, sputum/bronchial aspirates/bronchoalveolar lavage, blood, and wound) collected from patients admitted to NIID with a suspicion of infection were sent to the laboratory and processed according to the specific Standard Operating Procedures. The specimens were inoculated on Columbia sheep blood agar (ThermoFisher Scientific™—Oxoid, Wesel, Germany), chocolate agar with Polivitex (bioMérieux S.A., Marcy-l’Etoile, France), and lactose agar (CLED, ThermoFisher Scientific™—Oxoid, Wesel, Germany), respectively. The inoculated plates were incubated for 18-24 hours at 37 °C in an aerobic atmosphere, and then were visually read and interpreted.
Strain identification
All bacterial strains were identified using Matrix-Assisted Laser Desorption Ionisation Time-of-Flight mass spectrometry (MALDI-TOF MS) Biotyper (Bruker Daltonik GmbH, Bremen, Germany), which characterise bacterial proteins in whole-cell extracts. Bacterial spectra were analysed using the Biotyper software version 3.1.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) was performed using MICRONAUT-S Gram-negative (customised plate) Romania GN 2 EUCAST (Bruker Daltonik GmbH, Bremen, Germany), followed by interpretation according to the EUCAST (European Committee on Antimicrobial Susceptibility Testing) guidelines versions 13.1 and 14.0, respectively. Klebsiella pneumoniae strains with a minimum inhibitory concentration (MIC) to meropenem of >0.125 µg/mL were assigned for extended-spectrum beta-lactamase and carbapenemase detection. All carbapenemase-producing strains were tested against cefiderocol using UMIC (Bruker Daltonik GmbH, Bremen, Germany).
Enzyme production testing
Phenotypic testing for extended-spectrum beta-lactamase and carbapenemase production has been performed following the EUCAST guideline on detection of resistance mechanisms version 2.0, using lateral flow assays []. NG Test/CTX-M Multi (NG Biotech, France) was performed to identify CTX-M production, and Resist-3 O.K.N. K-SeT (Coris BioConcept, Belgium) to detect OXA-48 type, KPC, and NDM carbapenemases.
Synergistic activity assessment
Synergistic activity between aztreonam 30 µg, ceftazidime/avibactam 14 µg (Oxoid Ltd, UK), ceftolozane/tazobactam (Oxoid Ltd, UK) and imipenem/relebactam 35 µg (MastGroup Ltd, UK) was assessed using the double disk diffusion assay, placing the disks 20 mm centre-to-centre apart from each other on Mueller-Hinton agar plates (bioMérieux S.A., Marcy-l’Etoile, France), previously inoculated with the Klebsiella pneumoniae strain to be tested []. Synergic activity was considered positive if the inhibition zones around any of the cephalosporin or carbapenem disks were augmented or in the presence of an inhibition zone between aztreonam and any of the two disks (Figure 1).
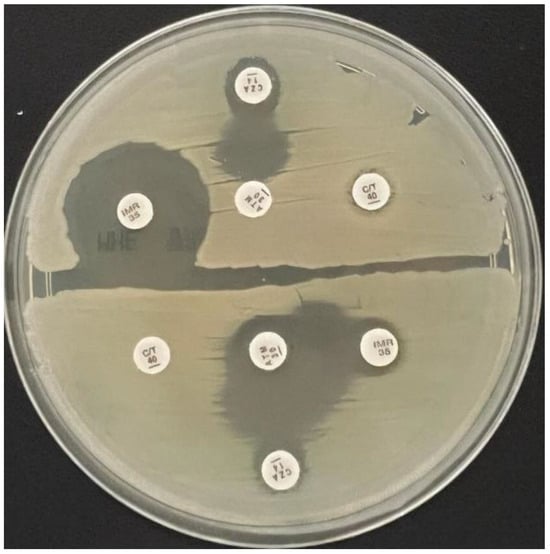
Figure 1.
Synergistic activity assessment between ceftazidime/avibactam and aztreonam (superior half of the plate), respectively ceftazidime/avibactam and aztreonam and imipenem/relebactam and aztreonam (lower half of the plate) in two tested strains on Mueller-Hinton agar medium.
Molecular detection and characterisation of plasmid-mediated antibiotic resistance
A total of 64 isolates were selected for molecular detection and characterisation of plasmid-mediated antibiotic resistance. Colonies were harvested using the eNAT System for Molecular Specimens (Copan, Italy) and stored in the freezer until further processing. A previously described protocol, based on Qiagen chemistry and magnetic beads, was used for DNA extraction []. Briefly, a volume of 0.2 mL of the sample was incubated at 56 °C with constant shaking along with 20 µL Proteinase K and lysis buffer, then washed and eluted in 150 µL nuclease-free buffer. Eluates were stored at -80 °C.
For nucleic acid amplification/detection, the QuantiNova Pathogen+IC kit (Qiagen, Hilden, Germany) was used, according to the manufacturer’s recommendations. Experiments were conducted on Q instruments (Quantabio, Beverly, USA).
Assays for Class 1 integrons (amplifying a fragment of intI1 gene), ESBL (extended spectrum beta lactamase—blaTEM, blaCTX-M, blaSHV), broad spectrum beta lactamase (AmpC) and carbapenemases (KPC, VIM, NDM, OXA-48, OXA-58) were employed, using primers and probes commercially available (Omnigen, Bucharest, Romania). Due to the high number of samples and assays performed, DNA extracts were amplified using bidirectional matrices, with pools consisting of eight samples (16 pools in total, eight row pools and eight column pools)—Table 1 [].

Table 1.
Bi-directional matrix for the 64 samples, yielding 8 row (R) pools and 8 column (C) pools. As shown, each sample was analysed at least twice in different pools.
Quality control
The quality control strains for the AST (Micronaut-S and UMIC Cefiderocol) were Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853. The quality control strains were Klebsiella pneumoniae ATCC 700603 for the lateral flow assay NG Test/CTX-M Multi and Klebsiella pneumoniae ATCC BAA-1705, Klebsiella pneumoniae NCTC 13442 and Klebsiella pneumoniae NCTC 13442 for Resist-3 O.K.N. K-SeT. All molecular assays were performed using Escherichia coli ATCC 25922 as a negative control.
Data storage and analysis
Preliminary data were extracted from the laboratory’s digital database and further stored in an individual, project-assigned Microsoft Excel database, together with all additional information, in addition to the study results. Data analysis was performed using Microsoft Excel version 16.66.1 (2022 Microsoft). The Chi square test (two tailed) was used for categorical variables and the results were considered to be statistically significant at p value <0.05.
Results
Throughout the aforementioned twelve-month interval, 139 non-duplicate Klebsiella pneumoniae strains with an MIC to meropenem of >0.125 µg/mL were isolated from patients with infections admitted to NIID. Most of these strains were isolated from urinary tract infections (UTIs) (98 out of 139 cases), the others were from lower respiratory tract, bloodstream, and wounds (Table 2).

Table 2.
The site of infection for the 139 bacterial strains with an MIC to meropenem of >0.125 µg/mL.
Carbapenemase and ESBL production
The phenotypic analysis demonstrated that 129 strains were carbapenemase producers, and 109 of them were also extended-spectrum beta-lactamase producers. Forty-nine strains were single-carbapenemase producers, with NDM being the most common (n=29, 22.5%), followed by OXA-48-like (n=12, 9.3%) and KPC (n=8, 6.2%). The remaining 80 strains were double carbapenemase producers, 79 producing NDM plus OXA-48-like and one strain producing NDM plus KPC.
Molecular detection of resistance genes
The initial screening for Class 1 integrons (intI1 gene) was positive for all 64 samples, suggesting the bacterial capability to acquire resistance genes. Subsequently, molecular detection and characterisation of plasmid-mediated antimicrobial resistance genes partially confirmed the results of phenotypic analysis. The blaKPC-3 was identified in 4/4 KPC positive isolates, and the blaOXA-48 assay was positive in 42/42 OXA-48-like or strains producing both OXA-48-like and NDM. Additionally, blaOXA-58 was found in two OXA-48-like producers and three strains which produced both OXA-48-like and NDM. Moreover, two KPC-positive strains have also tested positive for blaOXA-48, with the same results in 13/14 NDM-only isolates.
Resistance rates to last-resort antimicrobials
Among the 129 carbapenemase-producing strains, the resistance rates to the five tested last-resort antimicrobials ranged from 35.7% (n=46) for cefiderocol to 98.4% (n=127) for ceftolozane/tazobactam. The patterns of resistance according to the carbapenemase type are presented in Table 3. Strains producing OXA-48-like carbapenemase had the smallest resistance to these five agents, and double carbapenemase producers (NDM and OXA-48-like) had the highest.

Table 3.
Patterns of resistance to last-resort antimicrobials in correlation with the type of carbapenemase produced. n indicates the number of resistant strains.
Temporal changes in antimicrobial resistance rates
One objective of the study was to assess the rates of resistance to cefiderocol before and after its introduction into clinical practice in NIID on March 1st 2024. Thus, the isolates were divided into two subgroups: Subgroup 1, including isolates from August 2023 to the end of February 2024 (n=67), and Subgroup 2, including strains isolated from March 2024 to the end of July 2024 (n=62). Results have shown a slight but not statistically significant increase in resistance rates between the two subgroups (n=23, 34.32% vs. n=23, 37.09%, Chi-square test=0.107 and p=0.742).
In vitro efficacy of synergistic combinations
All 129 carbapenemase-producing strains were tested for resistance against two potentially efficient combinations: ceftazidime/avibactam plus aztreonam (CZA+ATM) and imipenem/relebactam plus aztreonam (IMR+ATM), respectively. Both antimicrobial combinations showed encouraging results, with CZA+ATM being efficient against all but two strains (98.5% in vitro efficacy, n=127), while IMR+ATM had an effectiveness rate of 79.8% (n=103). Among the 26 strains with no synergy between IMR+ATM, 7 were KPC producers, two tested positive for OXA-48-like, 5 produced NDM, and the remaining 12 were double carbapenemase producers (NDM and OXA-48 type).
Discussion
With rates of resistance surpassing 83% against all tested last-resort antimicrobials except for cefiderocol, the current study demonstrates worrisome results in terms of therapeutic options for infections produced by microorganisms similar to those isolated.
These results are even more concerning when considering the temporal trends of carbapenemase production in Romania. A decade ago, a six-month retrospective study described OXA-48-like as the most commonly encountered carbapenemase produced by CRKP strains in Romania, followed by NDM and VIM, with no double carbapenemase producers identified []. Since then, other studies revealed that the landscape has shifted, with a rise in the proportion of NDM and double NDM plus OXA-48-like producing strains, results confirmed by the current study [].
These concerning resistance results often correlate with more severe clinical presentations, representing a significant challenge both for medical professionals and the healthcare system. Moreover, an increase in resistance rates and a higher proportion of double carbapenemase-producing Gram-negative strains may occur, in parallel with the persistent overuse of antibiotics in hospitals. A previous study, including Klebsiella pneumoniae isolates from a Romanian hospital between 2019 and 2021, revealed an increase of almost 20% in carbapenem resistance rates, demonstrating the need for novel therapies to become available [].
The observed discrepancies between phenotypic and genotypic antibiotic resistance of the tested isolates, as described in the literature, may be the result of various factors, including but not limited to reduced expression of the resistance genes, a heteroresistant subpopulation, variable performance of the phenotypic or genotypic assays; further molecular characterisation may be needed [,].
The two in vitro synergistic combinations, ceftazidime/avibactam plus aztreonam and imipenem/relebactam plus aztreonam, respectively, showed encouraging in vitro results; however, it remains to be seen whether they will maintain their efficacy in clinical practice, as proposed by current guidelines [,]. As some available case series showed, these combinations represent promising real-life treatment options with good safety profiles; however, larger cohort studies are needed to assess thoroughly their feasibility in clinical use [,].
The current study was conducted in a tertiary care infectious diseases hospital; therefore, the estimated rates of resistance may differ from other clinical settings and may lead to an overestimation and larger-scale studies are needed to assess the magnitude of this issue. Another limitation is the lack of larger-scale genetic testing, which resulted in a limited number of strains genotypically tested (64/129, 49.6%).
Conclusions
The study provides a one-year overview of antimicrobial resistance rates and patterns to last-resort antimicrobials in Klebsiella pneumoniae isolates from patients admitted to a tertiary care hospital. The results suggest that carbapenemase-producing Klebsiella pneumoniae is an increasingly concerning public health issue in Romania, thus underlining the need for efficient therapeutic alternatives. The study also demonstrated the in vitro efficacy of two antimicrobial combinations, which may represent a viable option for clinical use.
Author Contributions
Conceptualisation: MOD, AR, DT; sample collection: MOD, DT; sample processing: MOD, DT, DFT; data analysis: MOD, DT, DF; genetic testing: DFT, MT; writing-original draft: MOD; writing-review and editing: MOD, AR, DT, DF; validation: AR, DT; project supervision: DT. All authors read and approved the final version of the manuscript.
Funding
The author MOD has been the beneficiary of a private scholarship offered by Johnson & Johnson Romania through Carol Davila University of Medicine and Pharmacy for the completion of this project. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author, upon reasonable request.
Ethics
The study was approved by the Institutional Review Board of National Institute for Infectious Diseases “Prof. Dr. Matei Balș” (number C9720/19.09.2024).
Acknowledgments
The authors would like to thank their laboratory colleagues for their support in the collection of samples. Additionally, they express their gratitude to Johnson & Johnson Romania for supporting young researchers at the start of their scientific careers.
Conflicts of Interest
All authors—none to declare.
References
- Wang, G.; Zhao, G.; Chao, X.; Xie, L.; Wang, H. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020, 17, 6278. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Zhu, Y.; Ouyang, J.; et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome. 2020, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Xu, Q.; Kenéz, Á.; Chen, S.; Yang, G. Klebsiella pneumoniae infection is associated with alterations in the gut microbiome and lung metabolome. Microbiol Res. 2022, 263, 127139. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Zhu, Z.C.; Zhu, J.; Chen, L.; Du, H. Antibiotic heteroresistance in Klebsiella pneumoniae: Definition, detection methods, mechanisms, and combination therapy. Microbiol Res. 2024, 283, 127701. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, K.; Ding, C.; Wang, S.; Wu, W.; Liu, X. Exploring multidrug-resistant Klebsiella pneumoniae antimicrobial resistance mechanisms through whole genome sequencing analysis. BMC Microbiol. 2023, 23, 245. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Surveillance atlas of infectious diseases. Accessed on: 3 July 2025. Available at: https://atlas.ecdc.europa.eu/public/index.aspx.
- European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2023. Stockholm: ECDC; 2024.
- European Committee on Antimicrobial Susceptibility Testing. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance eucast: Resistance mechanisms. Accessed on: 1 August 2025. Available at: https://www.eucast.org/resistance_mechanisms.
- Verschelden, G.; Noeparast, M.; Stoefs, A.; et al. Aztreonam-avibactam synergy, a validation and comparison of diagnostic tools. Front Microbiol. 2023, 14, 1322180. [Google Scholar] [CrossRef] [PubMed]
- Turcitu, M.A. Universal workflow for pathogen detection using Real Time (RT)PCR. Rom J Vet Med Pharmacol. 2024, 49, 276–287. [Google Scholar]
- Barathidasan, R.; Sharmila, F.M.; Raj, R.V.; Dhanalakshmi, G.; Anitha, G.; Dhodapkar, R. Pooled sample testing for COVID-19 diagnosis: evaluation of bi-directional matrix pooling strategies. J Virol Methods. 2022, 304, 114524. [Google Scholar] [CrossRef] [PubMed]
- Lixandru, B.E.; Cotar, A.I.; Straut, M.; et al. Carbapenemase-producing Klebsiella pneumoniae in Romania: A six-month survey. PLoS One. 2015, 10, e0143214. [Google Scholar] [CrossRef] [PubMed]
- Lazar, D.S.; Nica, M.; Dascalu, A.; et al. Carbapenem-resistant NDM and OXA-48-like producing K. pneumoniae: from menacing superbug to a mundane bacteria; A retrospective study in a romanian tertiary hospital. Antibiotics (Basel). 2024, 13, 435. [Google Scholar] [CrossRef] [PubMed]
- Cireșă, A.; Tălăpan, D.; Vasile, C.-C.; Popescu, C.; Popescu, G.-A. Evolution of antimicrobial resistance in Klebsiella pneumoniae over 3 years (2019-2021) in a tertiary hospital in Bucharest, Romania. Antibiotics (Basel). 2024, 13, 431. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, A.; Chasan, R.; Nowak, M.D.; et al. Correlation between identification of β-lactamase resistance genes and antimicrobial susceptibility profiles in Gram-negative bacteria: A laboratory data analysis. Microbiol Spectr. 2022, 10, e0148521. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Patel, R. Molecular diagnostics for genotypic detection of antibiotic resistance: current landscape and future directions. JAC Antimicrob Resist. 2023, 5, dlad018. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Heil, E.L.; Justo, J.A.; Mathers, A.J.; Satlin, M.J.; Bonomo, R.A. Infectious Diseases Society of America 2024 guidance on the treatment of antimicrobial-resistant Gram-negative infections. Clin Infect Dis. 2024, ciae403. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Carrara, E.; Retamar, P.; et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines for the treatment of infections caused by multidrug-resistant Gram-negative bacilli (endorsed by European Society of Intensive Care Medicine). Clin Microbiol Infect. 2022, 28, 521–547. [Google Scholar] [CrossRef] [PubMed]
- Leanza, C.; Mascellino, M.T.; Volpicelli, L.; et al. Real-world use of imipenem/cilastatin/relebactam for the treatment of KPC-producing Klebsiella pneumoniae complex and difficult-to-treat resistance (DTR) Pseudomonas aeruginosa infections: a single-center preliminary experience. Front Microbiol. 2024, 15, 1432296. [Google Scholar] [CrossRef] [PubMed]
- Cismaru, I.M.; Văcăroiu, M.C.; Soium, E.; et al. Synergistic combination of aztreonam and ceftazidime-avibactam-a promising defense strategy against OXA-48 + NDM Klebsiella pneumoniae in Romania. Antibiotics (Basel). 2024, 13, 550. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2025.