Optimizing Pneumonia Diagnosis Using RCGAN-CTL: A Strategy for Small or Limited Imaging Datasets
Abstract
1. Introduction
- (1)
- The quality of synthesized images is significantly enhanced by utilizing an enhanced RCGAN for data generation, which allows for the fusion of synthetic images belonging to the same category but possessing distinct characteristics through the input of a random vector and additional condition vectors.
- (2)
- CTL benefits from the use of the pre-training network to restrict the input transfer learning images and ensure the quality of synthetic images involved in training. Furthermore, transfer learning can update the parameters of the pre-training network to dynamically alter the conditional restrictions.
- (3)
- The RCGAN model is not simply linked to the CTL model, and the parameter update of CTL impacts the update of the random vector of RCGAN, resulting in more pliable and diverse features of the generated images in each cycle and reducing the overfitting phenomenon of the training process.
2. Theoretical Background
2.1. Conditional Generative Adversarial Networks
2.2. Conditional Transfer Learning Based on Shared Parameters
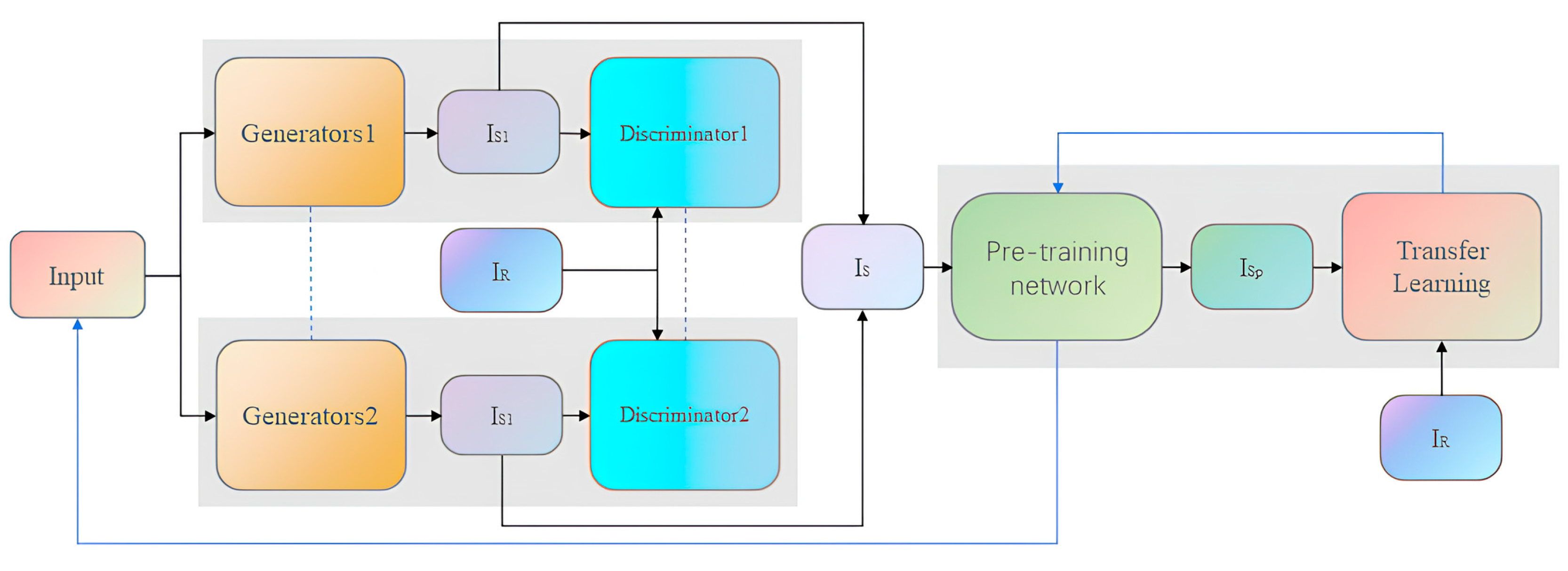
3. Proposed Method
3.1. Experimental Data
3.2. Data Pre-Processing
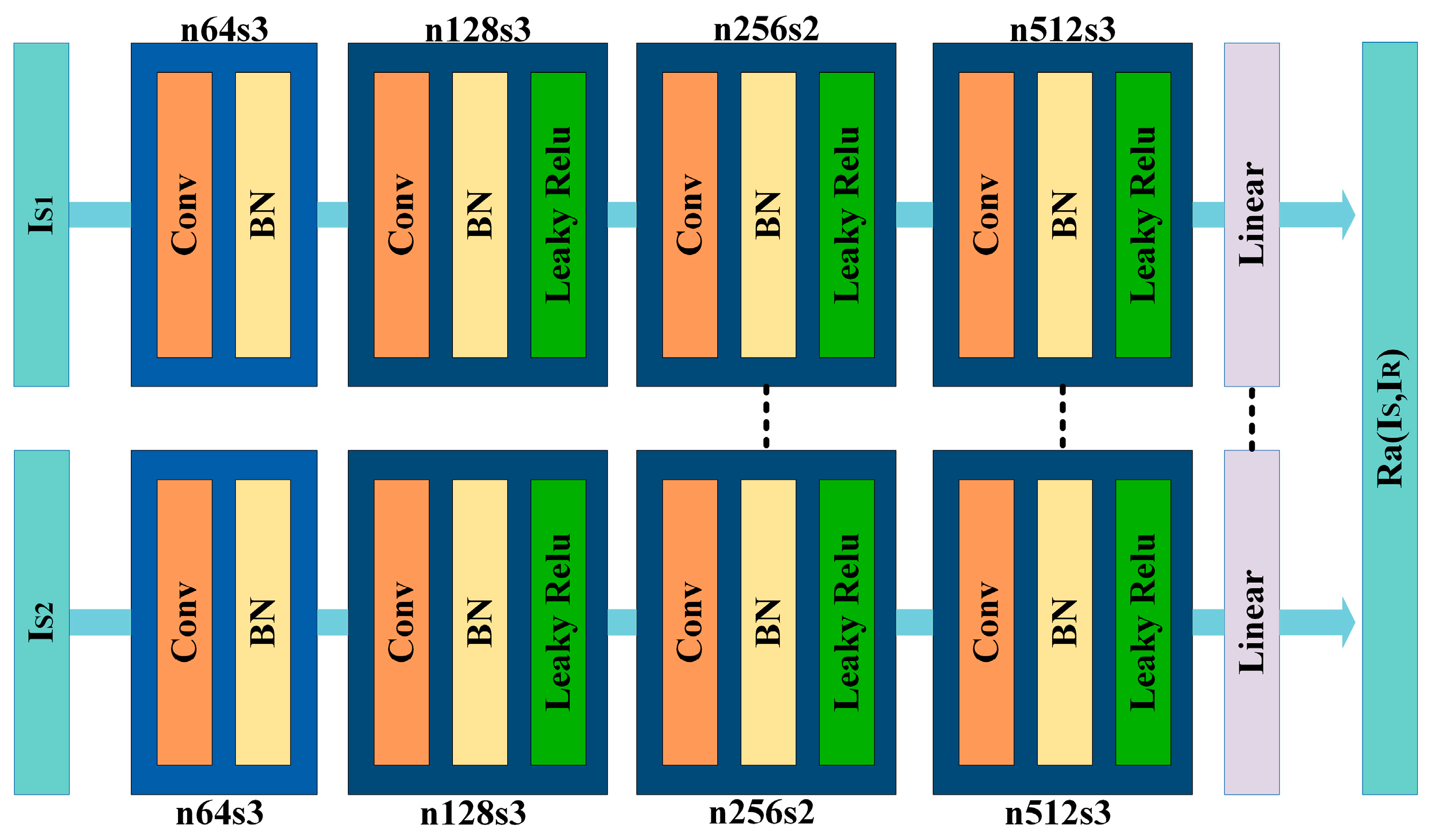
3.3. Data Generation Module
3.4. Data Classification Module
4. Experiment Analysis
4.1. Experimental Setup
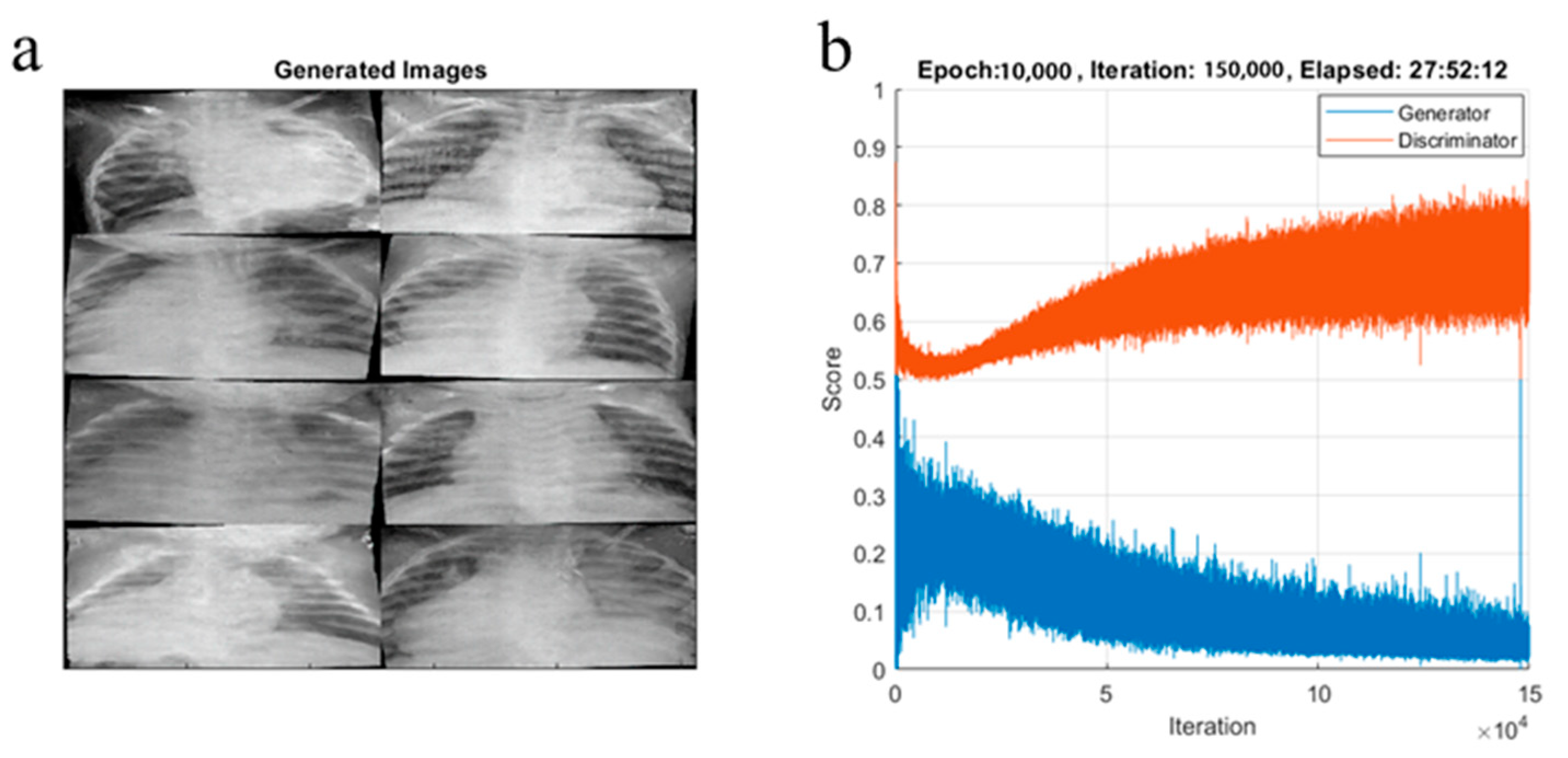
4.2. Performance Results
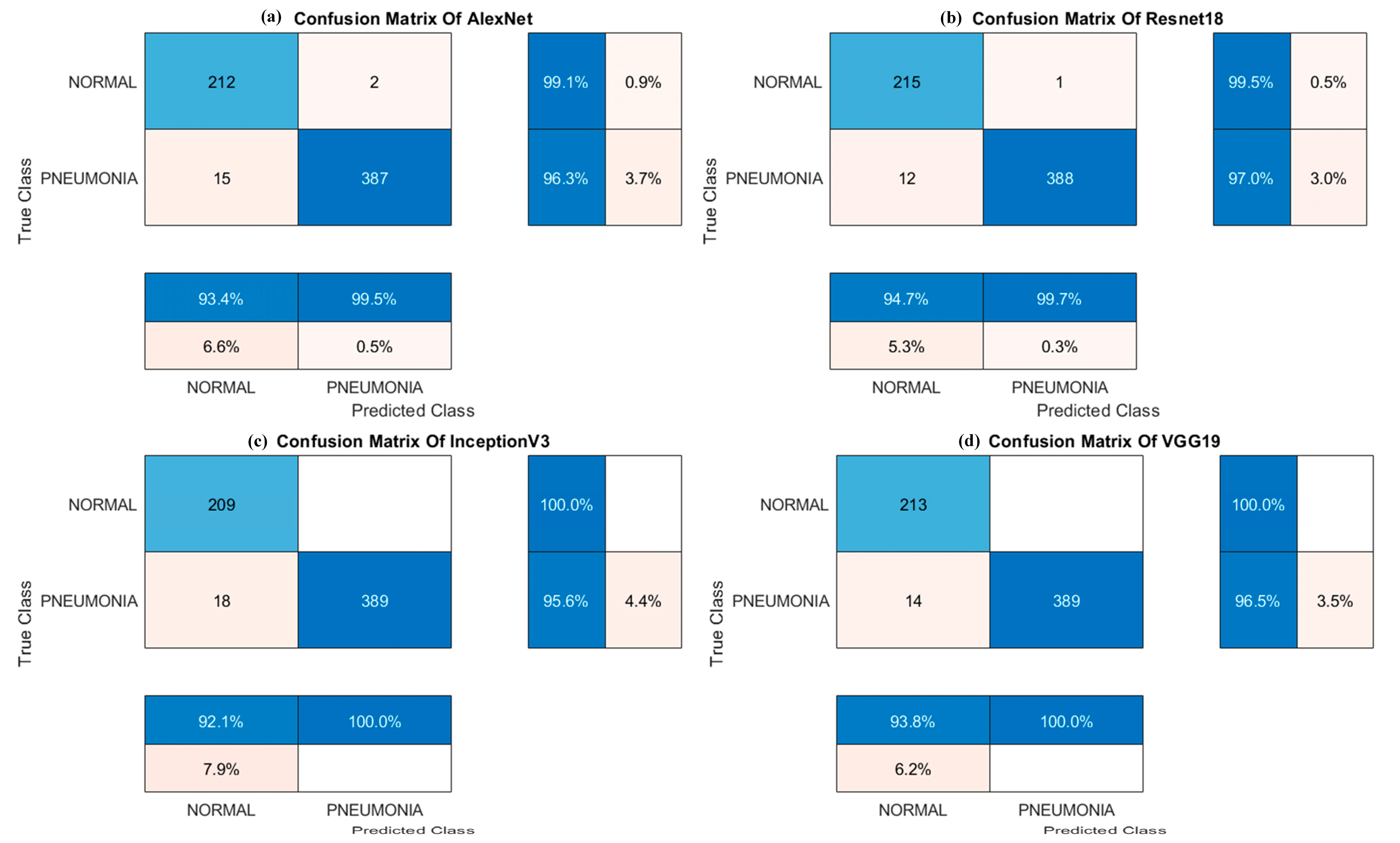
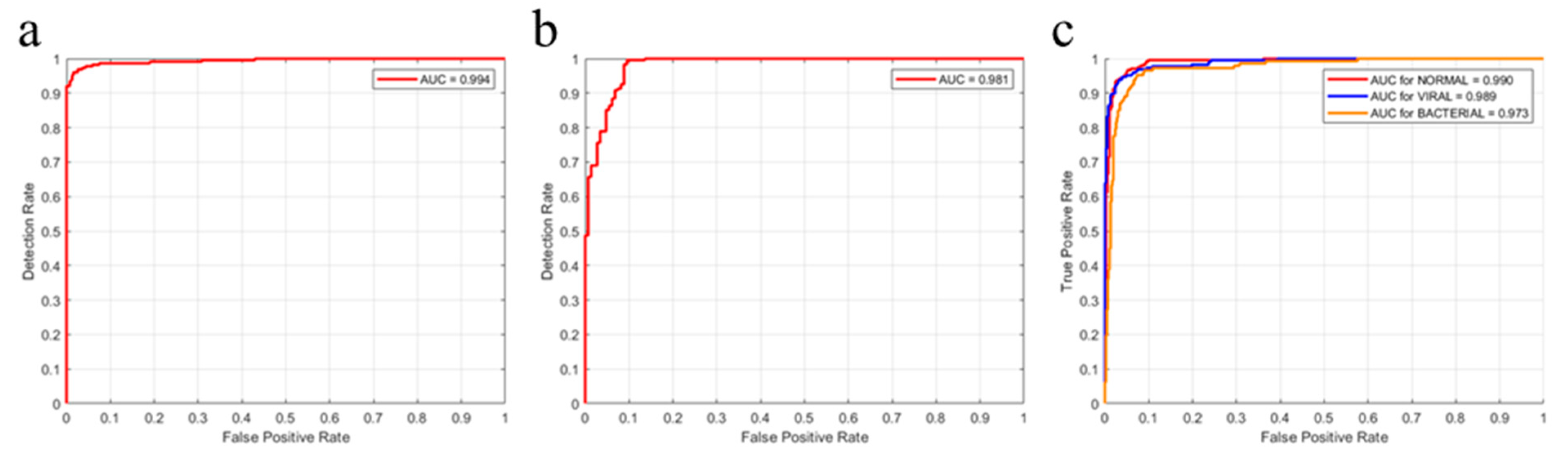
4.3. Performance Evaluation
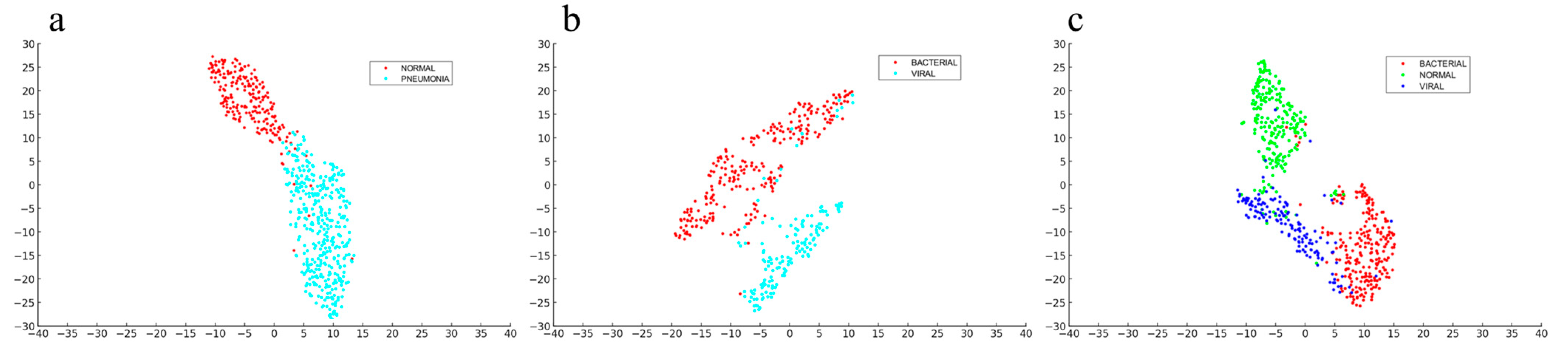
4.4. Feature Visualization
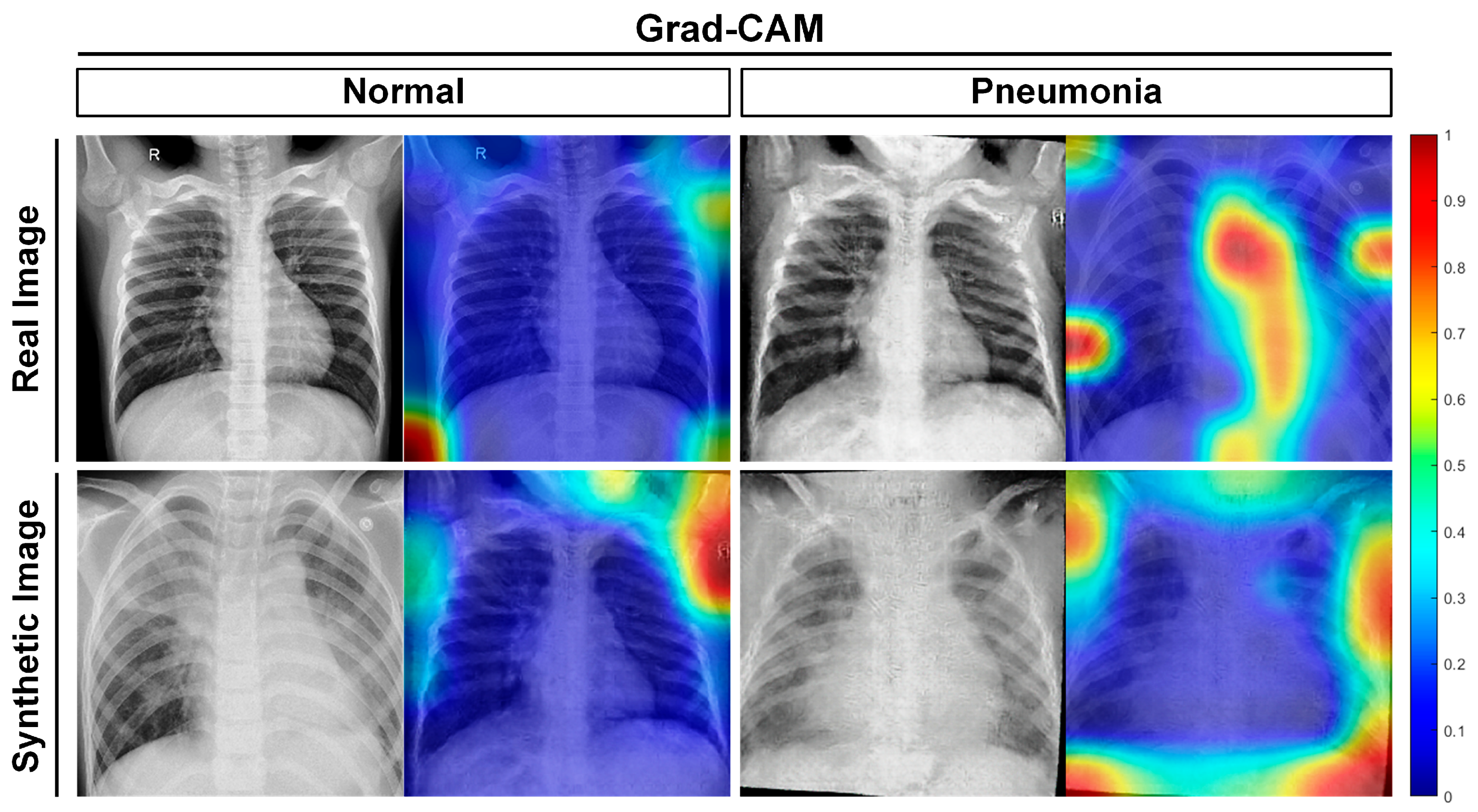
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mehta, T.; Mehendale, N. Classification of X-ray images into COVID-19, pneumonia, and TB using cGAN and fine-tuned deep transfer learning models. Res. Biomed. Eng. 2021, 37, 803–813. [Google Scholar] [CrossRef]
- Showkat, S.; Qureshi, S. Efficacy of Transfer Learning-based ResNet models in Chest X-ray image classification for detecting COVID-19 Pneumonia. Chemom. Intell. Lab. Syst. 2022, 224, 104534. [Google Scholar] [CrossRef]
- Chakraborty, S.; Paul, S.; Hasan, K.M.A. A Transfer Learning-Based Approach with Deep CNN for COVID-19- and Pneumonia-Affected Chest X-ray Image Classification. SN Comput. Sci. 2022, 3, 17. [Google Scholar] [CrossRef]
- Avola, D.; Bacciu, A.; Cinque, L.; Fagioli, A.; Marini, M.R.; Taiello, R. Study on transfer learning capabilities for pneumonia classification in chest-X-rays images. Comput. Methods Programs Biomed. 2022, 221, 106833. [Google Scholar] [CrossRef]
- Manickam, A.; Jiang, J.; Zhou, Y.; Sagar, A.; Soundrapandiyan, R.; Dinesh Jackson Samuel, R. Automated pneumonia detection on chest X-ray images: A deep learning approach with different optimizers and transfer learning architectures. Measurement 2021, 184, 109953. [Google Scholar] [CrossRef]
- Mamalakis, M.; Swift, A.J.; Vorselaars, B.; Ray, S.; Weeks, S.; Ding, W.; Clayton, R.H.; Mackenzie, L.S.; Banerjee, A. DenResCov-19: A deep transfer learning network for robust automatic classification of COVID-19, pneumonia, and tuberculosis from X-rays. Comput. Med. Imaging Graph. 2021, 94, 102008. [Google Scholar] [CrossRef]
- Loey, M.; Manogaran, G.; Khalifa, N.E.M. A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural. Comput. Appl. 2020. [Google Scholar] [CrossRef]
- Kaderuppan, S.S.; Wong, W.L.E.; Sharma, A.; Woo, W.L. O-Net: A Fast and Precise Deep-Learning Architecture for Computational Super-Resolved Phase-Modulated Optical Microscopy. Microsc. Microanal. 2022, 28, 1584–1598. [Google Scholar] [CrossRef]
- Ayan, E.; Unver, H.M. Diagnosis of Pneumonia from Chest X-ray Images Using Deep Learning. In Proceedings of the 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), Istanbul, Turkey, 24–26 April 2019. [Google Scholar]
- Hammoudi, K.; Benhabiles, H.; Melkemi, M.; Dornaika, F.; Arganda-Carreras, I.; Collard, D.; Scherpereel, A. Deep Learning on Chest X-ray Images to Detect and Evaluate Pneumonia Cases at the Era of COVID-19. J. Med. Syst. 2021, 45, 75. [Google Scholar] [CrossRef]
- Luján-García, J.; Yáñez-Márquez, C.; Villuendas-Rey, Y.; Camacho-Nieto, O. A Transfer Learning Method for Pneumonia Classification and Visualization. Appl. Sci. 2020, 10, 2908. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Li, Q.; Lu, L.; Li, Z.; Wu, W.; Liu, Z.; Jeon, G.; Yang, X. Coupled GAN With Relativistic Discriminators for Infrared and Visible Images Fusion. IEEE Sens. J. 2021, 21, 7458–7467. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Qian, N. On the momentum term in gradient descent learning algorithms. Neural Netw. 1999, 12, 145–151. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. arXiv 2015, arXiv:1502.01852. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2016, arXiv:1512.03385. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Loffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference On Computer Vision and Pattern Recognition (Cvpr), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef]
- Mahmud, T.; Rahman, M.A.; Fattah, S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020, 122, 103869. [Google Scholar] [CrossRef]
- Rajaraman, S.; Candemir, S.; Kim, I.; Thoma, G.; Antani, S. Visualization and Interpretation of Convolutional Neural Network Predictions in Detecting Pneumonia in Pediatric Chest Radiographs. Appl. Sci. 2018, 8, 1715. [Google Scholar] [CrossRef]
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, D.; Tiwari, P.; Moreira, C.; Damaševičius, R.; de Albuquerque, V.H.C. A Novel Transfer Learning Based Approach for Pneumonia Detection in Chest X-ray Images. Appl. Sci. 2020, 10, 559. [Google Scholar] [CrossRef]
- Cai, T.T.; Ma, R. Theoretical Foundations of t-SNE for Visualizing High-Dimensional Clustered Data. arXiv 2021, arXiv:2105.07536. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (Iccv), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Bharati, S.; Podder, P.; Mondal, M.; Prasath, V.B. Medical imaging with deep learning for COVID-19 diagnosis: A comprehensive review. arXiv preprint 2021, arXiv:2107.09602. [Google Scholar]
- Ain, Q.U.; Akbar, S.; Hassan, S.A.; Naaqvi, Z. Diagnosis of Leukemia Disease through Deep Learning using Microscopic Images. In Proceedings of the 2022 2nd International Conference on Digital Futures and Transformative Technologies (ICoDT2), Rawalpindi, Pakistan, 24–26 May 2022; pp. 1–6. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2019; pp. 178–190. [Google Scholar]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.-W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN per-formance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Li, Y.; Liu, S.; Yang, J.; Yang, M.H. Generative face completion. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3911–3919. [Google Scholar]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–292. [Google Scholar] [CrossRef]


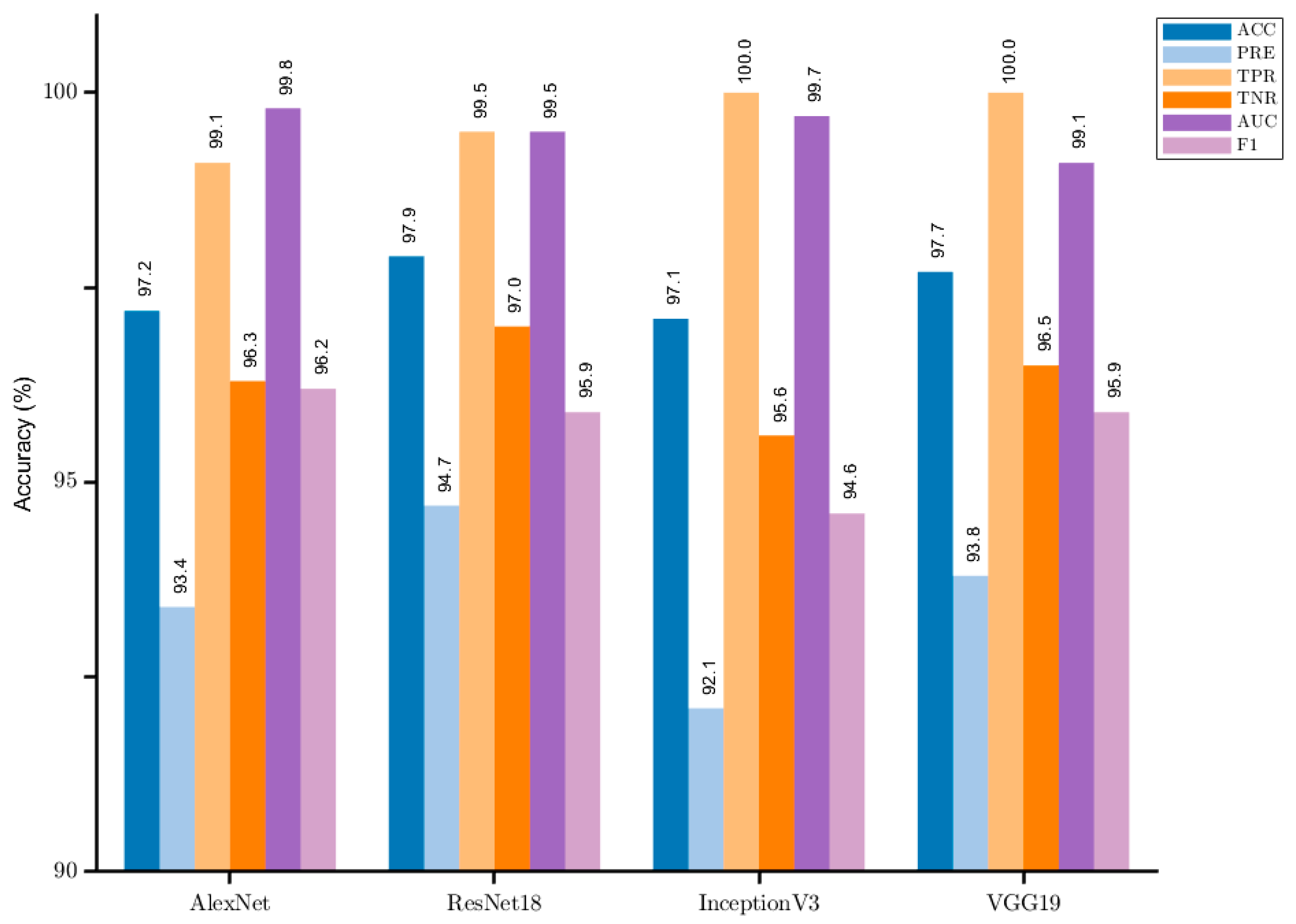

| Missions | Methods | Accuracy (%) | Precision (%) | Sensitivity (%) | Specificity (%) | F1-Score (%) | AUC (%) |
|---|---|---|---|---|---|---|---|
| Normal/pneumonia | Proposed | 97.7 | 93.8 | 100 | 96.5 | 99.1 | 95.9 |
| VGG-19 | 91.5 | 91.7 | 87.4 | 91.7 | 80.2 | 88.2 | |
| [24] | 98.1 | 98 | 98.5 | 97.9 | 98.3 | 99.4 | |
| [25] | 95.7 | 95.1 | 98.3 | 91.5 | 96.7 | 99.0 | |
| [16] | 92.8 | - | 93.3 | 90.1 | - | 96.8 | |
| [26] | 96.4 | 93.3 | 99.6 | - | - | 99.3 | |
| Bacterial pneumonia/viral pneumonia | Proposed | 94.6 | 99.6 | 92.3 | 99.2 | 95.8 | 98.1 |
| VGG-19 | 85.1 | 83.8 | 91.5 | 70.5 | 87.7 | 89.3 | |
| [24] | 95.1 | 94.4 | 96.1 | 94.3 | 95.5 | 97.6 | |
| [25] | 93.6 | 92.0 | 98.4 | 71.7 | 95.1 | 96.2 | |
| [16] | 90.7 | - | 88.6 | 90.9 | - | 94.0 | |
| Normal/bacterial pneumonia/viral pneumonia | Proposed | 96.1 | 92.0 | 91.5 | 95.2 | 91.7 | 98.6 |
| VGG-19 | 78.6 | 75.6 | 83.2 | 83.1 | 79.9 | 85.6 | |
| [24] | 91.7 | 92.9 | 92.1 | 93.6 | 92.6 | 94.1 | |
| [25] | 91.7 | 91.7 | 90.5 | 95.8 | 91.1 | 93.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, K.; He, S.; Yu, Y. Optimizing Pneumonia Diagnosis Using RCGAN-CTL: A Strategy for Small or Limited Imaging Datasets. Processes 2024, 12, 548. https://doi.org/10.3390/pr12030548
Han K, He S, Yu Y. Optimizing Pneumonia Diagnosis Using RCGAN-CTL: A Strategy for Small or Limited Imaging Datasets. Processes. 2024; 12(3):548. https://doi.org/10.3390/pr12030548
Chicago/Turabian StyleHan, Ke, Shuai He, and Yue Yu. 2024. "Optimizing Pneumonia Diagnosis Using RCGAN-CTL: A Strategy for Small or Limited Imaging Datasets" Processes 12, no. 3: 548. https://doi.org/10.3390/pr12030548
APA StyleHan, K., He, S., & Yu, Y. (2024). Optimizing Pneumonia Diagnosis Using RCGAN-CTL: A Strategy for Small or Limited Imaging Datasets. Processes, 12(3), 548. https://doi.org/10.3390/pr12030548






