Abstract
Myocardial infarction is the main driver of heart failure due to ischemia and subsequent cell death, and cell-based strategies have emerged as promising therapeutic methods to replace dead tissue in cardiovascular diseases. Research in this field has been dramatically advanced by the development of laboratory-induced pluripotent stem cells (iPSCs) that harbor the capability to become any cell type. Like other experimental strategies, stem cell therapy must meet multiple requirements before reaching the clinical trial phase, and in vivo models are indispensable for ensuring the safety of such novel therapies. Specifically, translational studies in large animal models are necessary to fully evaluate the therapeutic potential of this approach; to empirically determine the optimal combination of cell types, supplementary factors, and delivery methods to maximize efficacy; and to stringently assess safety. In the present review, we summarize the main strategies employed to generate iPSCs and differentiate them into cardiomyocytes in large animal species; the most critical differences between using small versus large animal models for cardiovascular studies; and the strategies that have been pursued regarding implanted cells’ stage of differentiation, origin, and technical application.
1. Introduction
Heart failure (HF) is the end-stage clinical syndrome for a variety of cardiovascular diseases (CVD) and is currently the leading cause of morbi-mortality worldwide [1]. HF most commonly develops after acute myocardial infarction (MI), when the injured myocardial tissue fails to recover or regenerate [2]. A significant proportion of patients develops pathological ventricular remodeling and progressive HF despite the use of evidence-based medical therapies [3,4]. Although cell-based strategies have emerged for myocardial regeneration after MI, it remains unclear which cell type is optimal for completely restoring cardiac tissue. From the arduous experimental race to find the ideal cell source, has emerged the ability to induce lineage dedifferentiation of easily accessible somatic cells into induced pluripotent stem cells (iPSCs). This has radically opened a promising therapeutic alternative for CVD, since iPSCs are capable of generating an unlimited range and quantity of clinically relevant cell types, including cardiac cell populations [5,6,7].
Like other experimental strategies, stem cell therapy must meet multiple requirements before reaching the clinical trial phase, and in vivo models are indispensable for ensuring the safety of such novel therapies. Most cardiovascular pre-clinical studies using iPSCs have been conducted in small animal models, the findings support their potential therapeutic benefits to improve cardiac function [8,9,10,11]. However, there remains a need for MI translational studies in large animal models to fully evaluate the therapeutic potential of this approach, and to empirically determine the optimal combination of cell types, supplementary factors, and delivery methods to maximize efficacy and stringently assess safety. In cardiovascular research, such studies are usually performed in swine models, but can also use sheep, dog, and macaque models [12], with each species being better suited for specific applications. In this review, we will describe both in vitro and in vivo state-of-the-art techniques for iPSC cardiac differentiation, as well as the main translational studies investigating iPSC cardiac therapy.
2. iPSC Generation and Cardiac Differentiation
2.1. Obtaining iPSCs from Large Animals
Pluripotent stem cells are defined as being able to differentiate into any cell type, regardless of their germ layer, and as being germline transmitted. Such cells could originally be obtained only by isolating embryonic stem cells (ESCs) from the inner cell mass of early-stage embryos [13,14,15]. However, in 2006, Takahashi and Yamanaka reported that the experimental induction and co-expression of Oct3/4, Klf4, Sox2, and c-Myc could force the retrograde de-differentiation of adult somatic cells into a pluripotent state [16]. To date, murine ESCs are the only cell lines proven to be germline transmitted—the most stringent criteria of pluripotency—and are thus considered true (“naïve”) pluripotent ESCs. Most other ESC/iPSC lines, including those generated from human and large animal cells, share more common characteristics with mouse epiblast-derived stem cells, which cannot form chimeras after blastocyst injection, and are not considered as flexible (“primed”) [17,18,19,20,21]. Indeed, new studies suggest that the molecular mechanisms of the naive state of pluripotency in pre-implantation embryos of other species (including rabbit or primate) may differ from those identified in rodent embryos. Specifically, not all the transcription factors that define naive pluripotency in mice are expressed in their epiblasts [22,23,24], and the signaling pathways that activate these genes and the timing of their upregulation during pre-implantation development also differ [25,26].
The first iPSCs were generated through virus-mediated (mainly lentiviruses and retroviruses) transduction of transcription factors, leading to direct integration within the host cell genome, which remains the gold standard of iPSC generation today. However, methods that circumvent genome modification, also called non-integrating techniques, are being extensively developed and evaluated [27,28,29]. Indeed, the dominant trends in reprogramming technology are designed to prevent insertional mutagenesis (by using non-integrative approaches) and contamination of donor cells with co-cultured feeder cells (by using xeno-free conditions and defined extracellular matrix components). The most widely adopted methodologies include the use of direct non-integrative vectors (e.g., single-stranded RNA viruses; double-stranded DNA Viruses and episomes) [30,31,32,33]. Here, Sendai virus (SV), a single stranded negative-sense RNA virus that replicates in the cytoplasm, is widely used in a broad range of research experiments mainly due to its high transduction efficiency, its rapid detectable transgene expression and its auto-erasable nature as a vector [34,35,36]. SV replicates independent of cell cycle, unlike other approaches where the exogenous genes are expressed only as the cell divides, producing very high copy numbers of the target gene. Notably, SV is particularly useful when reprogramming derived blood cells, such as CD34 positive cells, T-cells, and PBMCs, but also work well with fibroblasts, keratinocytes, and other cell-types [37,38]. Other non-integrative methods include integrating vectors that exhibit subsequent excision (e.g., piggyBac transposons) [39,40,41], or transient expression of small molecules (e.g., mRNA or proteins) (Figure 1) [42,43,44,45].
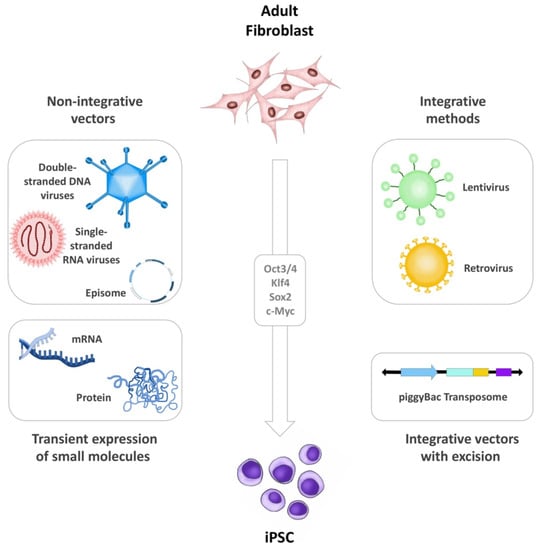
Figure 1.
iPSC generation. Illustration summarizing the different methods for iPSC generation including non-integrative vectors, integrative methods, transient expression of small molecules, and integrative vectors that exhibit subsequent excision. The figure was designed and hand-drawn by CG-M.
Diverse combinations of vectors and targeted reprogramming genes have been used to generate iPSCs from most farm animals and agriculturally important ungulates [46,47], and these studies highlight some rather interesting commonalities among protocols. Skin fibroblast isolation through biopsy seems to be the preferred cell source for starting iPSC generation, and it appears to be important that cells are obtained from animals that are as young as possible (days old instead of weeks or months). This already indicates a low robustness of the re-programming protocols, as researchers must utilize less mature cells, possibly through epigenetic marks. For instance, many of these iPSC lines exhibit inconsistent expression of typical human and murine pluripotency surface markers, such as SSEA-1, SSEA-3, SSEA-4, TRA-1-81, and TRA-1-60. Moreover, some of these markers display confounding effects, as they can also be expressed in the trophectoderm or in more differentiated stages [48]. The differences in epiblast development among these species could be thwarting reprogramming efforts, leading scientists to generate cells towards different developmental points. This could explain why iPSC generation using large animal models may require different culture conditions and yield varying marker profiles.
The swine model is emerging as the large animal of choice [49]. Literature mining in Pubmed using the search terms: ((“induced pluripotent stem cells”[MeSH Terms] OR (“induced”[All Fields] AND “pluripotent”[All Fields] AND “stem”[All Fields] AND “cells”[All Fields]) OR “induced pluripotent stem cells”[All Fields] OR “ipsc”[All Fields]) AND (“swine”[MeSH Terms] OR “swine”[All Fields] OR “swines”[All Fields])) AND ((journalarticle[Filter]) AND (english[Filter]) AND (2006:2021[pdat])), including only original articles in English language from January 2006 until October 2021, revealed over 300 studies using iPSCs and swine models, of which 103 describe the generation, maintenance, or differentiation of porcine-derived iPSCs (piPSCs), reflecting the clear clinical transfer intention [50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65]. In contrast, there were only 18 original articles involving iPSCs derived from horse [60,66,67,68,69,70,71,72,73], 15 from cattle [74,75,76,77,78,79,80], 15 from dog [81,82,83,84,85,86], and 14 from sheep/goat [87,88,89,90,91,92,93,94]. In most of these iPSC lines, pluripotency heavily depends on fibroblast growth factor and Activin/Nodal signaling (NANOG and LIN28) [80,95]. In some cases, these factors can replace KLF4 and c-Myc, which have reportedly carried oncogenic potential. Such “primed” colonies are characterized by flattened morphology and can be difficult to passage as single cells. However, by applying selective growth procedures immediately following reprogramming, it is possible to generate LIF/STAT3-dependent iPSCs (which more closely resemble naïve status) from swine cells [96].
Several reports show that additional strategies to enhance pluripotency yield and capabilities can drastically improve iPSC generation and maintenance. Wu et al. recently reported that m6A modification (the most prevalent modification in eukaryotic mRNAs) modulates the SOCS3/JAK2/STAT3 pathway, and thereby plays an important role in regulating the pluripotency of porcine-derived iPSCs [97]. Therefore, de-methylation protocols (such as those employing 5′-AZA-2′deoxycytidine) performed prior to de-differentiation efforts can have significant impacts on the success of iPSC generation [98]. Other strategies include the use of iPSC lines established from murine origin to extract nuclear and cytoplasmic factors that can be later transfected into host cells. In such methods, researchers aim to blindly add varying cellular contents that, although largely unknown, could hypothetically play a role in reprogramming, and thus could improve the de-differentiation process (a form of shotgun approach). Notably, the transcriptomic profiling of pluripotent cells highlights that pluripotency and cell expansion could be increased by the addition of specific small molecules to culture media, or the use of more refined media [99]. Bingbo et al. recently described that IRF-1 expression in the inner cell mass of a porcine early blastocyst enhances the pluripotency of piPSCs, partly through promotion of the JAK-STAT pathway [100]. The authors performed ChIP-Seq analysis, which revealed that IRF-1 activates genes related to the JAK-STAT pathway, and expression of IL7 and STAT3. Inhibition of STAT3 phosphorylation reverted the expression of primed genes in IRF-1-overexpressing cells, while addition of IL7 in culture medium resulted in no apparent changes in the cell morphology, anatomopathological staining results, or expression of pluripotency-related genes. Additionally, IRF-1 knockdown during reprogramming appeared to reduce reprogramming efficiency, whereas IRF-1 overexpression yielded the opposite effect. Such studies are uncovering a plethora of transcription factors that play key roles in optimizing pluripotency.
Most importantly, these studies show that species-specific, human, and mouse transcription factors, as well as combinations of transcription factors from different species, can be used to reprogram other animal cells.
2.2. Cardiac Lineage iPSC Differentiation in Large Animal Models
Direct iPSC delivery through transplantation or injection has become the most studied approach in regenerative medicine [101,102,103,104,105,106,107]. However, iPSC translational efforts have been dampened by concerns regarding the teratogenic potential of undifferentiated cells, as well as the lack of timely stimuli to carefully guide the in situ differentiation of these cells [108]. Therefore, increasing research attention has been focused on the in vitro differentiation of iPSCs in the laboratory setting, to obtain the desired cell type, which can then be applied to the patient. Notably, the differentiation of murine and human iPSCs into cardiomyocytes (iPSC-CMs) has been extensively described and is now standard practice in laboratories worldwide [109,110,111,112,113,114,115,116,117,118,119].
In vivo, it is thought that cardiac progenitor cells initially originate from the mesoderm following activation of the tumor growth factor-β (TGF-β) signaling cascades, and subsequent inhibition of the Wnt/β-catenin pathway (Figure 2). Most cardiac differentiation protocols attempt to replicate distinct stages of this developmental process and fall into two categories: small molecule-based or based on WNT signaling pathway inhibition. Briefly, both protocols usually involve growing purified iPSC monolayers in Matrigel (Corning Life Sciences, Glendale, CA, USA) up to 90% confluence before starting the reprogramming procedures. Next, the small molecule-based protocols usually entail the activation of Activin A and BMP4 (and sometimes the use of specific cytokines, such as VEGF and DKK1) in a timed sequence. On the other hand, WNT signaling pathway inhibition involves the use of CHIR-99021 (2 mg) and IWR-1 endo (10 mg) (Figure 2) [120,121,122,123,124,125,126,127,128]. Moreover, insulin is required to observe functional beating, but is counter-productive during initial stages, as it inhibits differentiation. Thus, almost all protocols start with insulin-free media, and involve insulin addition only after reprogramming has been established [129,130,131,132,133,134].
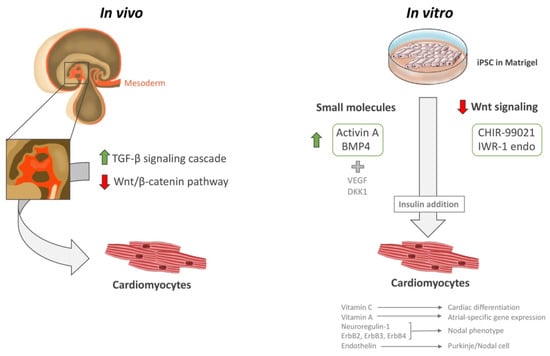
Figure 2.
iPSC cardiac differentiation. Illustration showing the in vivo and in vitro methods for iPSC cardiac differentiation, including small molecules and Wnt signaling downregulation. Insulin is a necessary step to mature CMs into a beating state. The figure was designed and hand-drawn by CG-M.
Certain vitamins and cytokines also reportedly help in cardiac differentiation and influence the efficiency and specificity of the resulting cells. For instance, ascorbic acid (vitamin C) promotes cardiac differentiation of pluripotent cells, and retinoic acid (vitamin A) promotes atrial-specific gene expression [129,135,136,137,138]. A signaling pathway involving neuroregulin-1 (NRG-1; an important regulator of cardiac development and function) and the receptor tyrosine kinases ErbB2, ErbB3, and ErbB4 orientates cells towards a nodal phenotype [139]. Additionally, endothelin, a paracrine factor secreted by endothelial cells in the arterial walls, is apparently involved in Purkinje/nodal cell differentiation [140].
While some studies show that these cells can efficiently couple to the injured myocardium and restore cardiac function, many have also reported arrhythmogenic issues. To explain this, several studies demonstrate that iPSC-CM cultures can be highly heterogeneous, comprising both ventricular- and nodal-like CMs. Nonetheless, prominent works have demonstrated regenerative capacities when non-human primates are transplanted with allogenic iPSC-CMs in a post-MI setting. In 2016, Shiba and colleagues reported that injected iPSC-CMs electrically coupled with host CMs resulted in improved cardiac contractile function at 4 and 12 weeks after transplantation [114]. However, these cells clearly elicited transient episodes of ventricular tachycardia, and the study only included five subjects transplanted with a single iPSC line. Moreover, the authors noted that the 12-week observation period after cell transplantation does not allow a definitive conclusion regarding graft survival without chronic rejection.
Unfortunately, few studies have examined the use of autologous or allogeneic iPSC-CMs in large animal models. As the end-goal in biomedicine is the clinical applicability of such therapies, human iPSC-CMs are the cell type of choice in the vast majority of investigations. Together with the ease of obtaining iPSC lines of human origin compared to from other large animal models, this has led to predominant use of human iPSCs to test stem cell therapy in other species [141,142]. In such experiments, animals must be immunosuppressed to maintain the cells after transplant. Although such practices are faster and easier to implement, the result cannot be considered to faithfully reproduce most of the diseases under investigation. Most cardiac differentiation protocols currently employed in large animal models are directly extrapolated from those effective in human and murine cells. However, as the murine protocols do not work well with human cells and vice versa, there is little scientific basis to think that either of these would optimally translate to other animal species (with the exception of human protocols that reportedly work well in other primates). Yet, few efforts have been directed towards developing better more specific protocols for use in other large animals, which impedes the development of more reliable pre-clinical models.
3. Large Animal Models for Translational Cardiovascular Studies
3.1. The Importance of Large Animal Models in Cardiac Stem Cell Therapies
When approaching the development of new treatment, to justify the huge time and economic efforts required, it is critical to carefully select what animal models should be used to determine the likelihood that a therapeutic approach will be clinically successful [143,144,145,146]. Although no animal model can ever completely substitute for human studies, large animal models have supplied critically important information about how humans might respond to specific therapeutic strategies. Most importantly, when an intervention improves the main outcomes in a well-designed trial using any experimental model, this proof of concept is a valuable piece of evidence guiding further studies. Compared to data from small species, well-designed large animal studies can better predict human trial outcomes [147,148,149].
3.2. Critical Differences between Large and Small Animal Models in Pre-Clinical Studies
From the practical standpoint, small animal species have tremendous advantages for pre-clinical research [150,151,152]. The high availability of reagents to test molecular pathways, and the relatively low cost of studying a large number of subjects, facilitate the derivation of mechanistic insights relating to the therapeutic being studied. Specifically, mice have been a species of choice for studies of stem cell biology in mammals, due to their reduced cost, high offspring generation, and ease of genetic manipulation.
Nevertheless, positive results in small animal models (mostly murine) have frequently been followed by clinical trials failing to confirm efficacy, justifying skepticism regarding the reliability of findings obtained in studies of small species [153,154,155]. These negative outcomes may be explained by the lack of disease complexity in the animal model used; and by the differences between rodents and humans in terms of size, life span, heart rate, and the innate and adaptive immune response systems. Moreover, different pathologies that can be more easily induced in small models (such as hypertension, diabetes mellitus, or hypoxia) are used as proxy to imitate some of the cellular and physiological changes in human diseases, which limits the capacity to extrapolate findings in the animal models. Many of these models fail to precisely recapitulate particular human disease phenotypes, especially in stem cell research, which has compelled investigators to examine animal species that may be more predictive of humans.
Compared to mice, large animals are often better models of human disease phenotypes [156,157,158,159,160], due their physiological factors are close to humans. Moreover, large animals also have similarities in terms of number and types of stem cells that can be reproducibly extracted and handled in sufficient amount for analysis and for different applications. Key drivers of translational applicability include advances in experimental surgery, and the ability to use equipment and techniques developed for human applications for cell delivery and animal monitoring. These features let researchers to study the safety of applications, dosages of biologics, and a delivery method that can be easily translated to humans. This is the case of the development of different approaches and techniques, such as surgical options and imaging technologies. Large animal models allow to test survival, activation and differentiation of the implanted cell by non-invasive monitoring. Clearly, it is undesirable to interpret the results of highly interventional therapies by performing experiments solely in small animal models and extrapolating the conclusions to human trials [161,162].
This is not to imply that large animal models do not also have intrinsic limitations. The most important limitation is the high cost of animal maintenance. Research in large animal species involve larger and specialized housing and surgical facilities, including higher costs associated to feed, veterinary care, and surgical costs. Additionally, their longer reproductive cycles and slow growth rates make that pre-clinical trials are slower and less economical. More specifically relevant to cell therapy, research in large animals is complicated by the relative absence of stable and well-characterized stem cell lines and protocols for their maintenance, differentiation, and cell status monitoring; and the limited availability of species-specific antibodies, expression microarrays, and other research reagents. Finally, complex diseases are difficult to fully capture with any model and when considering all previous points, studies involving large animal models can end up being poorly designed and insufficiently powered. In these studies, costs have a big impact in the design, which many times constrain the budget and forces to include too few animals in each study arm, so that false-positive outcomes can often occur by chance alone.
Nevertheless, since large animal models provide a setting that is closer to the human situation than those found in rodent models, large species studies are essential to justify the risks and costs of clinical trials [147].
3.3. Pre-Clinical Stem Cell Research for Cardiovascular Diseases
Among larger animal species, dogs, pigs, sheep, and non-human primates are suitable models for CVD-related studies. These animals have physiological parameters similar to humans (Table 1), and their size allows the use of echocardiography and cardiac magnetic resonance imaging techniques that are already widely used in the clinic, which yields information that is more rapidly transferrable and relevant.

Table 1.
Cardiac similarities and differences of large animals compared to humans.
These large species have exhibited marked improvement in cardiac function following stem cell treatments using a variety of cells, including skeletal myoblasts, bone marrow and adipose tissue-derived stem cells, cardiac stem cells, and endothelial adult stem cells [163,164,165]. Moreover, meta-analysis studies involving large animals that have received cardiac stem cells as therapy for ischemic heart disease clearly demonstrate that these models can predict clinical trial outcomes, and that such treatments are safe; however, the specific reported cardiac improvements are mixed and the gathered datasets very heterogeneous [149]. These studies can potentially address a variety of important issues before the performance of clinical trials, including determination of the optimal cell type and delivery method, time of administration, and types of clinical condition for which a treatment can be beneficial.
4. Applications of iPSC-Based Therapies in Large Animal Models of MI
All the iPSC-based therapies in large animal models are summarized in Table 2. Key published literature was searched using PubMed. We applied the search terms: (“induced pluripotent stem cells” OR “ipsc”) AND (“large animals” OR “swine” OR “pig”) and (“induced pluripotent stem cells” OR “ipsc”) AND (“large animals” OR “swine” OR “pig” AND (“myocardial infarction”) including only original articles in English language until October 2021.

Table 2.
iPSC studies in large animals. MI: myocardial infarction; HF: heart failure; hESC-CMs: human embryonic stem cell-derived cardiomyocytes; hiPSC-CMs: human induced pluripotent stem cell-derived cardiomyocytes; hiPSC-ECs: human induced pluripotent stem cell-derived endothelial cells; hiPSC-MSCs: human induced pluripotent stem cell-derived mesenchymal stem cells; hMSCs: human mesenchymal stem cells; hiPSC-SMCs: human induced pluripotent stem cell-derived smooth muscle cells; Tac: Tacrolimus; CsA: Cyclosporine; MMF: Mycophenolate mofetil; PSL: prednisolone; Methylene prednisolone: METH-PSL; NS: not specified.
4.1. Undifferentiated iPSCs
Some groups have generated piPSCs, opening up a wide range of possibilities to conduct pre-clinical testing in pigs without requiring immunosuppressive treatment [48,55,56,181]. However, good differentiation of piPSCs to CMs has not yet been achieved, and most piPSC studies performed in swine have used undifferentiated cells. Nevertheless, one major concern is the possibility of uncontrolled tumorigenesis after stem cell administration [182] and studying allogeneic transplantation in immune-competent pigs could reveal whether the immune system can control undifferentiated cells that may remain after transplantation.
In 2013, Li and colleagues published the first study assessing intramyocardial injection of allogeneic piPSCs in a pig model of MI [166,167]. First, they tested intracoronary administration of piPSCs, and found no effect on the infarct zone or in terms of cardiac perfusion, mainly due to cell washout through the blood circulation. Thus, to ensure piPSC engraftment, they performed a second study testing the direct intramyocardial injection of 2 × 107 piPSCs into infarct zones (8 sites) and border zones (12 sites). Six weeks after injection, treated animals exhibited an improved left ventricular ejection fraction (LVEF), better myocardial perfusion (likely due to vascular endothelial cell differentiation of the engrafted iPSCs), less oxidative stress, angiogenesis in the border zone, upregulated connexin 43 expression, and less ventricular tachycardia inducibility. Importantly, no tumors were detected in any animal, despite the use of undifferentiated cells. Collectively, their data demonstrated that direct intramyocardial injection of piPSCs is safe and can decrease infarct size and improve cardiac function; however, the slight reported improvements were clinically irrelevant.
In 2014, the same research team published another study using an identical piPSC cell line, dose, and delivery route in an immune-suppressed MI porcine model to alleviate the non-specific immune response and the acute inflammatory reactions [168]. At 6 weeks after treatment, the iPSC group showed less pronounced LV structural abnormality and cardiac dysfunction and accompanied again by a slightly reduced scar size. Notably, most of the transplanted cells were found in the border zone, and they had differentiated into vascular endothelial cells (ECs), which were integrated into pre-existing vessels or generating new vessels, and to a lesser extent into myoblasts. Thus, iPSC transplantation was associated with significantly increased vascular density and reduced myocardial apoptosis in the border zone. Moreover, the iPSC group exhibited significantly increased proangiogenic and antiapoptotic factors, and significantly attenuated CM hypertrophy. In conclusion, these results suggested that piPSC transplantation can result in cardiac functional recovery, mainly by promoting angiogenesis, inhibiting apoptosis, and ameliorating cardiac remodeling.
Years later, our group conducted another study that proved the safety of transplanting allogeneic piPSCs using three different engineered constructs that were implanted into an immune-competent MI swine model [169]. We tested the adipose graft transposition procedure (AGTP) [183], an acellular human pericardial scaffold (scaffold), and a combination of both (AGTP-scaffold), with or without 0.5–1 × 106 piPSCs, which was a lower cell concentration than used in the previously described works. Thirty days after implantation, histopathological analyses confirmed no presence of piPSCs within the host myocardium or biomatrices. The AGTP-scaffold group showed significantly higher vascularization, irrespective of piPSC delivery, in both the infarct and border zones. Consistent with the disappearance of the implanted cells, and unlike in other studies, these treatments did not yield functional benefit in terms of LVEF, cardiac output, ventricular volumes, or necrotic mass. On the other hand, histopathological examination of the heart at 90 days of follow-up confirmed the absence of teratoma formation in all animals. Therefore, we concluded that residual undifferentiated piPSCs should pose no safety concern when used in an allogeneic context in immune-competent recipients, at least in cardiac regenerative medicine.
Taken together, the above-mentioned studies have demonstrated that allogeneic iPSC therapy is safe in terms of teratogenesis; however, no clinical benefits are obtained due to the host immune reaction against the delivered cells.
4.2. Differentiated iPSCs
Theoretically, iPSCs have the capacity to differentiate into any other cell type present in the body, independent of the germ layer of choice. This process can be directed by using specific gene regulation methods that target highly conserved developmental molecular pathways, with each differentiation protocol being specific to the desired cell line. Undifferentiated iPSCs are more flexible at delivery but are much less effective at improving tissue-specific problems. In contrast, iPSC-derived cell lines have the potential to more effectively target distinct cellular dysfunctions and have been the central focus of iPSC-based therapeutic research for some years now.
4.2.1. Allogeneic iPSC-CMs: Non-Primate Models
Autologous iPSC transplantation therapy avoids the need for immunosuppression and related problems, such as malignancy and infection. However, the clinical application of this approach is limited by high costs, safety concerns, and challenges related to manufacturing and regulation. To overcome these limitations, an iPSC bank has been developed to store iPSC lines with established safety, with the aim of transplanting iPSC derivatives in an allogeneic manner. However, as previously stated, a potential disadvantage of allogeneic transplantation lies in the immune reaction response, which is responsible for graft rejection [184]. The major histocompatibility complex (MHC) plays an essential role in the post-transplant immune response [185,186]. Therefore, donor/recipient MHC matching can decrease the rejection rate following organ and cell transplantation [187]. To that end, the establishment of iPSC lines from healthy donors with homozygous MHC alleles could be useful for minimizing the number of banked iPSC lines [188,189].
To that end, Kawamura and co-workers generated iPSC-CMs with a homozygous MHC HT1 haplotype line from the cynomolgus macaque, which has an MHC structure identical to that of humans [170]. They transplanted 3.3 × 106 iPSC-CM sheets at the subcutaneous level and by intramyocardial injection in MHC-matched macaques (with heterozygous MHC haplotypes) and MCH-mismatched macaques (without identical MHC alleles) in conjunction with immune suppression treatment. Compared to the MHC-mismatched group, the MHC-matched group displayed a higher engraftment rate and less infiltration of immune cells (CD3+ and CD4+ T cells). However, MHC-matched transplantation with single or no immune-suppressive drugs still induced a substantial host immune response to the graft. Thus, although MHC-matched transplantation reduced the immunogenicity of allogeneic iPSC-CMs, successful engraftment still required appropriate immune suppression.
Additionally, in 2016, another study examined allogeneic iPSC intramyocardial transplantation in a non-human primate model [114]. Two weeks after MI, 4 × 108 iPSC-CMs were injected into the infarct and border zones of MHC-matched and MHC-mismatched monkeys under immunosuppressive treatment. In the MHC-mismatched group, implanted iPSC-CMs were thoroughly rejected due to severe infiltration of T lymphocytes at 4 weeks after transplantation. However, in the MHC-matched group, the grafted iPSC-CMs survived for 12 weeks with no evidence of immune rejection and exhibited electrical coupling with host CMs. Despite evidence of improved cardiac contractile function at 4 and 12 weeks after iPSC-CM transplantation, the incidence of ventricular tachycardia was significantly increased compared to vehicle-treated controls. No animal showed tumor formation. Collectively, their data demonstrated that allogeneic iPSC-CM transplantation regenerates the infarcted non-human primate heart. However, there remains a need for further research on controlling post-transplant arrhythmias.
Most recently, Sawa’s group tested the effects of iPSC-derived cardiac sheet transplantation in both MHC-matched and MHC-mismatched immunosuppressed macaques. In line with previous findings, the treated animals showed significantly improved cardiac function with less fibrosis and higher vascular density, compared to sham animals, and homozygous MHC haplotypes were preferred to avoid immune rejection. Unfortunately, no arrhythmic inducibility analysis was reported [171].
4.2.2. Xenogeneic hiPSC-CMs
Recent studies have reported methods for the highly efficient differentiation of CMs from hiPSCs, which show typical electrophysiological function and pharmacological responsiveness [190,191]. Transplantation of hiPSC-CMs would mechanically contribute to directly improving cardiac function, among other benefits. However, low retention of the transplanted cells remains a primary factor limiting the effectiveness of this cell therapy [173,192,193,194].
In the onerous fight to identify the ideal approach to ensure hiPSC-CM engraftment, three different techniques have been tested: cell sheets, intracoronary infusion, and intramyocardial injections. Direct intramyocardial or intracoronary injections of dissociated single cells yields an engraftment rate of <10% immediately after transplantation [195,196]. Therefore, some studies have focused on developing innovative new injection techniques to improve cellular retention, such as co-transplantation with human MSCs that release antiapoptotic factors [197]. Moreover, in recent years, various new tissue engineering approaches, including cell sheets, have been developed to enhance cell delivery in myocardial regeneration therapy. In contrast to the needle injection technique, a cell sheet can drive a large number of cells to damaged tissue without transplanted cell loss or injury to the host myocardium. Furthermore, the arrangement of hiPSC-CMs in three-dimensional patches promotes their continuous maturation [174,198,199].
Dr. Sawa’s lab has acquired expertise in generating hiPSC-CM sheets. In their first study in a porcine MI model, cell sheets generated from 2.5 × 107 hiPSC-CMs yielded improved cardiac function (LVEF) and myocardial perfusion, attenuating LV remodeling. Furthermore, treated animals consistently exhibited significantly less accumulation of interstitial fibrosis in border zones, and increased myocardial vascular density, mainly through paracrine effects. The lack of teratoma formation in animals that received hiPSC-CM sheets confirmed the safety of this treatment [176]. However, the authors reported poor engraftment of the transplanted cells, affecting the long-term effectiveness of the treatment. This low cell retention was attributed to ischemia caused by poor vascularization and inflammation in the transplanted sites, as also suggested in other studies [200,201,202]. To overcome this issue, Sawa’s group has focused on a new approach using an omentum flap [177,178]. The omentum is a vascular-rich organ that contains abundant angiogenic factors and has anti-inflammatory effects and was thus expected to help to sustain a blood supply for the cell sheets [203]. The group examined the survival of hiPSC-CMs, enriched with commercial human mesenchymal stem cells (hMSCs), with or without an omentum pedicle, after transplantation into healthy mini-pigs [177] and a mini-pig MI model [178]. They observed significantly improved hiPSC-CM survival in mini-pigs with the omentum than without omentum. Over three months, both groups showed a steady decrease of cell survival, but the proportion of the decrease was significantly less in the omentum group. Moreover, the omentum group exhibited increased capillary density, and upregulated VEGF, SDF-1, and bFGF expression in the transplanted area. Therefore, the omentum pedicle yielded enhanced hiPSC-CM survival in vivo, and produced longer therapeutic effects after MI.
Most recently, studies have tested the addition of different proteins to ensure hiPSC-CM retention and proliferation in swine. Notably, Ye’s lab reported that thymosin β4 (Tb4) improves the engraftment of hiPSC-CM fibrin scaffolds in a porcine model of sub-acute MI [101]. They demonstrated that co-treatment with Tb4 significantly enhanced hiPSC-CM engraftment, induced vasculogenesis, promoted proliferation of CMs and ECs, improved LV systolic function, and reduced infarct size. Moreover, hiPSC-CM engraftment was not correlated with incidence of ventricular arrhythmia, and no tumorigenesis was detected in the immunosuppressed animals. Correspondingly, Zhao et al. have shown that hiPSC-CMs overexpressing cyclin D2 were associated with improved LV function; reduced infarct size; less fibrosis, ventricular hypertrophy, and CM apoptosis; and increased vessel density [179].
Preclinical and clinical studies of stem cell-based cardiac regenerative therapy have widely applied intracoronary infusion and intramyocardial injection (either via direct open-chest or transendocardial catheter-based injections) [204]. Intracoronary cell delivery into a recanalized infarct-related or target artery is safe and practical (e.g., it can be performed using standard balloon catheters); however, its effectiveness is compromised by the low cellular retention in the damaged myocardium. Compared to the intracoronary route, direct intramyocardial injection often results in slightly better retention [205]. Several strategies have been investigated for improving transplanted cell engraftment and viability—including cell aggregation, biomaterials or scaffolds, and pro-survival factors [122,206,207].
Fukuda’s lab has generated spheroids of purified hiPSC-CMs and used gelatin hydrogel as a biomatrix to enhance engraftment [180,206] and developed a new injection device for optimal 3D distribution of these materials in the myocardial layer [208]. They have demonstrated that aggregated CMs (spheroids) are less likely to be cleared from the host heart by normal circulation compared to non-aggregated CMs [206], and that gelatin hydrogel enhances CM engraftment [180]. Their new device comprises six needles, each with six elliptical holes facing in various directions, and with a domed tip to minimize damage to the myocardium. Compared to conventional needle injection procedures, direct epicardial injection of spheroids resulted in better distribution and retention in a layer within the myocardium, and no detrimental effects on cell viability, spheroid shape, or size were detected in healthy pigs. Interestingly, cell injections in gelatin hydrogel increased the retention of beads by approximately 20-fold compared to the retention rate using saline. Furthermore, they verified that their injection device provided a more equal three-dimensional distribution of transplanted cells in all layers of the host myocardium compared to conventional procedures [208].
Most recently, Kawaguchi et al. transplanted intramyocardially hiPSC-derived cardiac spheroids in pigs after cryoinjury. In line with previous works, treated animals exhibited improved cardiac function and reduced infarct size. However, the engraftment of CMs was unusual, and the treated pigs suffered ventricular arrhythmic events correlated with tachycardia [103].
4.2.3. Combination of Multiple hiPSC-Cardiovascular Lineage Cell Populations: Cardiomyocytes, Endothelial, and Smooth Muscle Cells
Both cardiac muscles and vessels are excessively damaged following MI. Therefore, therapeutic strategies should be focused on comprehensively repairing both together, to achieve true cardiac repair. In vitro data strongly suggest that CMs exhibit better survival and resistance to hypoxic injury when co-cultured with ECs and smooth muscle cells (SMCs) than when cultured alone [172,209,210]. Thus, co-administration with ECs and SMCs could enhance the engraftment of transplanted CMs, as well as improve LV myocardial perfusion, cardiac metabolism, and contractile activity through release of signaling molecules [211,212,213]. Consistent with this hypothesis, some groups have tested the differentiation of iPSCs to ECs and SMCs, and their combination with iPSC-CMs and growth factors to treat MI.
In 2014, Ye et al. transplanted hiPSC-CMs, -ECs, and -SMCs into injured hearts of a porcine MI model, with comparison of two delivery methods: through direct intramyocardial injections or through a fibrin patch loaded with insulin growth factor 1 (IGF-1)-microspheres [172]. Four weeks after transplantation, the cell + patch + IGF animals showed the best engraftment rate, ~20-fold greater than achieved with any other delivery method used in the porcine MI model [214]. Their results also demonstrated that co-administration of multiple hiPSC-cardiovascular linage cell transplantation significantly improved LV function, vascular density, and cardiac metabolism; and yielded reductions of infarct size, ventricular wall stress, and apoptosis, without inducing ventricular arrhythmias. The increased angiogenesis was related to paracrine effects, as suggested by other studies [209,211]. Together, these findings reveal that co-administration of trilineage cardiac cell populations, conjugated by microspheres of IGF-1, improves engraftment and graft effectiveness.
Most recently, clinical-sized cardiac tissue sheets (L-CTSs) comprising hiPSC-CMs and hiPSC-derived vascular cells (ECs and vascular mural cells) were also evaluated in a porcine MI model [173]. Transplantation of L-CTSs restored wall motion of the transplanted region, improved cardiac function in terms of higher LV systolic function and LVEF, and yielded reduced fibrosis and greater capillary density in the border region. L-CTS transplantation induced neovascularization, prompting CM hypertrophy attenuation and reducing global LV remodeling, as reported in previous works [215,216]. Once again, the authors stated that the therapeutic mechanism was mainly mediated by paracrine mechanisms, considering that the small engrafted cells cannot be responsible for the reinforcement of mechanical ventricular contraction.
In the same year, Gao et al. developed human cardiac muscle patches (hCMPs) by suspending 4 × 106 iPSC-CMs, 2 × 106 iPSC-SMCs, and 2 × 106 iPSC-ECs in a fibrin scaffold that covers the acute ischemic area in pigs [174]. The cell engraftment rate was 11% at 4 weeks after transplantation, and the hCMP treatment was associated with significantly improved LV function, reduction on infarct size, myocardial wall stress, and myocardial hypertrophy, and less apoptosis in the border area. Notably, the exosomes released from the hCMP exhibited cytoprotective properties that improved CM survival and neovascularization activity, and protected myocytes of the border from apoptosis. These paracrine mechanisms, as well as the microenvironment of the patch itself, likely contributed to the relatively high engraftment rate, which is consistent with the results from studies in rodents [11,217,218].
4.2.4. iPSC-Derived Mesenchymal Stem Cells
Besides iPSCs, mesenchymal stem cells (MSCs)—which can be obtained from several tissues, including bone marrow, fat, placenta, Wharton’s jelly, etc.—have been exhaustively examined as another promising cell type for HF treatment. The differentiation of MSCs from iPSCs (hiPSC-MSCs) has also been successfully tested, and some groups have demonstrated that hiPSC-MSCs exhibit better proliferative capacity, survival, and therapeutic efficacy for tissue repair than bone marrow-derived MSCs [219,220,221]. In parallel, Liao et al. compared the safety and efficacy of human embryonic stem cell-derived CMs (hESC-CMs) vs. hiPSC-MSCs following transplantation in a porcine model of MI-induced HF [175]. At 8 weeks post-transplantation, both cell groups exhibited significantly improved LV function. However, the percentage of infarcted area normalized to body weight did not significantly differ in the hiPSC-MSC group or hESC-CM group compared with in the MI group, suggesting that cell transplantation did not lead to cardiac regeneration. Notably, only the hiPSC-MSC group exhibited markedly increased vessel density and expressions of TGF-α and VEGF-A. These results show that hiPSC-MSCs can stimulate angiogenesis by upregulating the myocardial expression of angiogenic cytokines after transplantation. Moreover, histological assays confirmed the immunomodulatory potential of the hiPSC-MSCs, which activated regulatory T cells and reduced inflammatory cells in the myocardium.
5. Conclusions
Preclinical studies in large animal models of MI indicate that iPSC therapy might be a promising approach for treating CVDs. However, cell therapies are notably complex, and studies in large animal models could be performed to fine-tune several factors to further improve the clinical outcomes resulting from such advanced treatments. Specific aspects requiring further investigation include the optimal delivery method (cell sheets vs. intramyocardial injections), and type of cell administered (undifferentiated vs. iPSC-CMs vs. co-administration of multiple iPSC-derived cardiac cell types).
The desirable stem cell for regenerative medicine should be immunologically compatible, easily accessible, with high in vitro expansion, long-term survival, and able to integrate into the host target tissue. Currently, the MSC are the most extensively cell source tested in preclinical and clinical trials, due its easy isolation, high plasticity, and immunomodulation potential [222]. However, results show that the benefits are modest and mainly due to their paracrine effect, not to their nesting and differentiation in the host tissue. Contrary, iPSC, despite their capacity of teratoma formation (that can be evaded by a previous differentiation into committed lineages), are able to migrate, nest and integrate with the host myocardium, acting as new functional cardiac cells. Nevertheless, iPSC therapy not only still requires high immunosuppression to ensure its safety and effectiveness, but also their arrhythmic potential has to be solved before reaching the clinical setting. Considering the results of the preclinical studies with MSC and iPSC, it is difficult to conclude which of the two cell sources offers more notable positive outcomes. In this sense, MSCs, due to their immunomodulatory potential, may offer greater benefit in the context of acute MI when the inflammatory plethora of cytokines is active and on which the extension of the definitive myocardial scar (tissue at risk) depends. On the other hand, iPSC-CM could be a promising therapy not only for acute but also for chronic MI, where cardiac regeneration is feasible. Taken together, one possibility to consider in the search for the best therapeutic results lies in the joint administration of MSC and iPSC to simultaneously resolve post-MI inflammation, iPSC rejection, as well as cardiac regeneration, converging in a cardiac functional improvement.
The available data show that co-administration of hiPSC-cardiovascular lineage cell populations conjugated with certain biological molecules yield the most robust response in terms of promoting myocardial repair, as this method comprehensively approaches all injured tissues of the heart (rather than only CMs) to achieve true cardiac repair. Additionally, cell-sheet delivery systems are probably the most effective for delivering large numbers of cells without cell loss due to post-transplantation physical strain, hypoxia, or cell washout through the vascular or lymphatic system. However, a primary factor limiting the efficacy of this approach is the low proportion of transplanted cells that survive in the recipient heart just a few weeks following graft administration. Many research groups have begun examining this problem and are providing innovative solutions to improve the cell engraftment rate—such as new injection techniques, combination of the cell sheets with the omentum flap, or adding specific proteins. Additional research is clearly still needed to overcome engraftment issues, as well as to study more intricate cellular interactions, such as innervation (critical to the electric coupling of the graft and recipient tissue), vascularization, and final integration, it has been less than 15 years since the discovery of iPSCs, let alone since their application as stem cell therapy. If the progression of future research resembles the success over the past decade—as indeed seems to be the case—there will still be plenty of opportunities to marvel at the solutions that have yet to come and that will render stem cell approaches routine clinical practice.
Author Contributions
Conceptualization: C.G.-M.; manuscript writing: D.M.-F., O.I.-E., and C.G.-M.; manuscript final revision: C.G.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the Instituto de Salud Carlos III (PI17/01487, PI18/00256, PIC18/00014, ICI19/00039, PI19/01788, and ICI20/00135) funds, Red de Terapia Celular-TerCel (RD16/00111/0006), CIBER Cardiovascular (CB16/11/00403) projects, as part of the Plan Nacional de I + D + I, and co-funded by ISCIII-Subdirección General de Evaluación y el Fondo Europeo de Desarrollo Regional (FEDER) by the Generalitat de Catalunya [SGR2017 00483], and the CERCA Programme/Generalitat de Catalunya, Sociedad Española de Cardiología, and Societat Catalana de Cardiologia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Khan, M.A.; Hashim, M.J.; Mustafa, H.; Baniyas, M.Y.; Al Suwaidi, S.K.B.M.; AlKatheeri, R.; Alblooshi, F.M.K.; Almatrooshi, M.E.A.H.; Alzaabi, M.E.H.; Al Darmaki, R.S.; et al. Global Epidemiology of Ischemic Heart Disease: Results from the Global Burden of Disease Study. Cureus 2020, 12, e9349. [Google Scholar] [CrossRef]
- McMurray, J.; Pfeffer, M. Heart failure. Lancet 2005, 365, 1877–1889. [Google Scholar] [CrossRef]
- Koitabashi, N.; Kass, D. Reverse remodeling in heart failure: Mechanisms and therapeutic opportunities. Nat. Rev. Cardio 2011, 9, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Towbin, J.A.; Bowles, N.E. The failing heart. Nature 2002, 415, 227–233. [Google Scholar] [CrossRef]
- Yamashita, J.; Itoh, H.; Hirashima, M.; Ogawa, M.; Nishikawa, S.; Yurugi, T.; Naito, M.; Nakao, K.; Nishikawa, S. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature 2000, 408, 92–96. [Google Scholar] [CrossRef]
- Yamashita, J.K.; Takano, M.; Hiraoka-Kanie, M.; Shimazu, C.; Peishi, Y.; Yanagi, K.; Nakano, A.; Inoue, E.; Kita, F.; Nishikawa, S.-I. Prospective identification of cardiac progenitors by a novel single cell-based cardiomyocyte induction. FASEB J. 2005, 19, 1534–1536. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, H.; Ikuno, T.; Takeda, M.; Fukushima, H.; Marui, A.; Katayama, S.; Shimizu, T.; Ikeda, T.; Okano, T.; Sakata, R.; et al. Human iPS cell-engineered cardiac tissue sheets with cardiomyocytes and vascular cells for cardiac regeneration. Sci. Rep. 2014, 4, 6716. [Google Scholar] [CrossRef]
- Yeung, E.; Fukunishi, T.; Bai, Y.; Bedja, D.; Pitaktong, I.; Mattson, G.; Jeyaram, A.; Lui, C.; Ong, C.S.; Inoue, T.; et al. Cardiac regeneration using human-induced pluripotent stem cell-derived biomaterial-free 3D-bioprinted cardiac patch in vivo. J. Tissue Eng. Regen. Med. 2019, 13, 2031–2039. [Google Scholar] [CrossRef]
- Wendel, J.S.; Ye, L.; Tao, R.; Zhang, J.; Zhang, J.; Kamp, T.J.; Tranquillo, R.T. Functional Effects of a Tissue-Engineered Cardiac Patch From Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes in a Rat Infarct Model. Stem. Cells Transl. Med. 2015, 4, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Wang, Y.; Tang, Y.; Ali, A.; Hu, H.; Maienschein-Cline, M.; Ashraf, M. Cardiac Progenitors Induced from Human Induced Pluripotent Stem Cells with Cardiogenic Small Molecule Effectively Regenerate Infarcted Hearts and Attenuate Fibrosis. Shock 2018, 50, 627–639. [Google Scholar] [CrossRef]
- Gao, L.; Kupfer, M.; Jung, J.; Yang, L.; Zhang, P.; da Sie, Y.; Tran, Q.; Ajeti, V.; Freeman, B.; Fast, V.; et al. Myocardial Tissue Engineering With Cells Derived From Human-Induced Pluripotent Stem Cells and a Native-Like, High-Resolution, 3-Dimensionally Printed Scaffold. Circ. Res. 2017, 120, 1318–1325. [Google Scholar] [CrossRef]
- Tsang, H.G.; Rashdan, N.A.; Whitelaw, C.B.A.; Corcoran, B.M.; Summers, K.M.; MacRae, V.E. Large animal models of cardiovascular disease. Cell Biochem. Funct. 2016, 34, 113–132. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Hutchins, R. Embryonic stem cells. Stem Cells Dev. 2007, 16, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Vazin, T.; Freed, W.J. Human embryonic stem cells: Derivation, culture, and differentiation: A review. Restor. Neurol. Neurosci. 2010, 28, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Pourquie, O. Human embryonic stem cells get organized. Nature 2018, 558, 35–36. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Ware, C.B. Concise Review: Lessons from Naïve Human Pluripotent Cells. Stem Cells 2017, 35, 35–41. [Google Scholar] [CrossRef]
- Stirparo, G.G.; Boroviak, T.; Guo, G.; Nichols, J.; Smith, A.; Bertone, P. Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development 2018, 145, dev169672. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef]
- Rossant, J. Mouse and human blastocyst-derived stem cells: Vive les differences. Development 2015, 142, 9–12. [Google Scholar] [CrossRef]
- Samanta, M.; Kalantry, S. Generating primed pluripotent epiblast stem cells: A methodology chapter. Curr. Top. Dev. Biol. 2020, 138, 139–174. [Google Scholar]
- Nakamura, T.; Yabuta, Y.; Okamoto, I.; Sasaki, K.; Iwatani, C.; Tsuchiya, H.; Saitou, M. Single-cell transcriptome of early embryos and cultured embryonic stem cells of cynomolgus monkeys. Sci. Data 2017, 4, 170067. [Google Scholar] [CrossRef] [PubMed]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Reyes, A.P.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef]
- Boroviak, T.; Nichols, J. Primate embryogenesis predicts the hallmarks of human naïve pluripotency. Development 2017, 144, 175–186. [Google Scholar] [CrossRef]
- Boroviak, T.; Loos, R.; Lombard, P.; Okahara, J.; Behr, R.; Sasaki, E.; Nichols, J.; Smith, A.; Bertone, P. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev. Cell 2015, 35, 366–382. [Google Scholar] [CrossRef]
- Piliszek, A.; Madeja, Z.E. Pre-implantation Development of Domestic Animals. Curr. Top. Dev. Biol. 2018, 128, 267–294. [Google Scholar]
- Haridhasapavalan, K.K.; Borgohain, M.P.; Dey, C.; Saha, B.; Narayan, G.; Kumar, S.; Thummer, R.P. An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene 2019, 686, 146–159. [Google Scholar] [CrossRef]
- Cherkashova, E.A.; Leonov, G.E.; Namestnikova, D.D.; Solov’eva, A.A.; Gubskii, I.L.; Bukharova, T.B.; Gubskii, L.V.; Goldstein, D.V.; Yarygin, K.N. Methods of Generation of Induced Pluripotent Stem Cells and Their Application for the Therapy of Central Nervous System Diseases. Bull. Exp. Biol. Med. 2020, 168, 566–573. [Google Scholar] [CrossRef]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.L.; Loh, C.Y.Y. Episomal Induced Pluripotent Stem Cells: Functional and Potential Therapeutic Applica-tions. Cell Transplant 2019, 28, 112S–131S. [Google Scholar] [CrossRef]
- Skowron, K.; Tomsia, M.; Czekaj, P. An experimental approach to the generation of human embryonic stem cells equivalents. Mol. Biotechnol. 2014, 56, 12–37. [Google Scholar] [CrossRef]
- Mansouri, M.; Bellon-Echeverria, I.; Rizk, A.; Ehsaei, Z.; Cianciolo-Cosentino, C.; Silva, C.S.; Xie, Y.; Boyce, F.M.; Davis, M.W.; Neuhauss, S.C.; et al. Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat. Commun. 2016, 7, 11529. [Google Scholar] [CrossRef]
- Ye, H.; Wang, Q. Efficient Generation of Non-Integration and Feeder-Free Induced Pluripotent Stem Cells from Human Peripheral Blood Cells by Sendai Virus. Cell Physiol. Biochem. 2018, 50, 1318–1331. [Google Scholar] [CrossRef] [PubMed]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef]
- Ban, H.; Nishishita, N.; Fusaki, N.; Tabata, T.; Saeki, K.; Shikamura, M.; Takada, N.; Inoue, M.; Hasegawa, M.; Kawamata, S.; et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA 2011, 108, 14234–14239. [Google Scholar] [CrossRef]
- Okumura, T.; Horie, Y.; Lai, C.Y.; Lin, H.T.; Shoda, H.; Natsumoto, B.; Fujio, K.; Kumaki, E.; Okano, T.; Ono, S.; et al. Robust and highly efficient hiPSC generation from patient non-mobilized peripheral blood-derived CD34+ cells using the auto-erasable Sendai virus vector. Stem Cell Res. Ther. 2019, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Seki, T.; Yuasa, S.; Oda, M.; Egashira, T.; Yae, K.; Kusumoto, D.; Nakata, H.; Tohyama, S.; Hashimoto, H.; Kodaira, M.; et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010, 7, 11–14. [Google Scholar] [CrossRef]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. PiggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Brightwell, S.; Kaji, K. PiggyBac Transposon Mediated Reprogramming and Flow Cytometry Analysis of CD44 and ICAM1 Cell-Surface Marker Changes. Methods Mol. Biol. 2016, 1357, 285–923. [Google Scholar]
- Behringer, R.; Gertsenstein, M.; Nagy, K.V.; Nagy, A. Reprogramming Mouse Fibroblasts with piggyBac Transposons. Cold Spring Harb. Protoc. 2017, 2017, pdb.prot092627. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.A. RNA-based tools for nuclear reprogramming and lineage-conversion: Towards clinical applications. J. Cardiovasc. Transl. Res. 2013, 6, 956–968. [Google Scholar] [CrossRef]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef]
- Warren, L.; Lin, C. mRNA-Based Genetic Reprogramming. Mol. Ther. 2019, 27, 729–734. [Google Scholar] [CrossRef]
- Seo, B.J.; Hong, Y.J.; Do, J.T. Cellular Reprogramming Using Protein and Cell-Penetrating Peptides. Int. J. Mol. Sci. 2017, 18, 552. [Google Scholar] [CrossRef]
- Ezashi, T.; Telugu, B.P.; Roberts, R.M. Induced pluripotent stem cells from pigs and other ungulate species: An alternative to embryonic stem cells? Reprod. Domest. Anim. 2012, 47, 92–97. [Google Scholar] [CrossRef]
- Su, Y.; Zhu, J.; Salman, S.; Tang, Y. Induced pluripotent stem cells from farm animals. J. Anim. Sci. 2020, 98, skaa343. [Google Scholar] [CrossRef] [PubMed]
- Ezashi, T.; Telugu, B.P.; Alexenko, A.P.; Sachdev, S.; Sinha, S.; Roberts, R.M. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 10993–10998. [Google Scholar] [CrossRef] [PubMed]
- Iborra-Egea, O.; Martínez-Falguera, D.; Roura, S.; Bayes-Genis, A.; Raya, Á.; Gálvez-Montón, C. Porcine iPSC generation: Testing different protocols to a successful application. Methods Mol. Biol. 2021. [Google Scholar] [CrossRef]
- Gao, X.; Nowak-Imialek, M.; Chen, X.; Chen, D.; Herrmann, D.; Ruan, D.; Chen, A.C.H.; Eckersley-Maslin, M.A.; Ahmad, S.; Lee, Y.L.; et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 2019, 21, 687–699. [Google Scholar] [CrossRef]
- Xu, J.; Yu, L.; Guo, J.; Xiang, J.; Zheng, Z.; Gao, D.; Shi, B.; Hao, H.; Jiao, D.; Zhong, L.; et al. Generation of pig induced pluripotent stem cells using an extended pluripotent stem cell culture system. Stem Cell Res. Ther. 2019, 10, 193. [Google Scholar] [CrossRef]
- Zhong, C.; Wu, J.; Izpisua-Belmonte, J.C. Pig Chimeric Model with Human Pluripotent Stem Cells. Methods Mol. Biol. 2019, 2005, 101–124. [Google Scholar]
- Ezashi, T.; Yuan, Y.; Roberts, R.M. Pluripotent Stem Cells from Domesticated Mammals. Annu. Rev. Anim. Biosci. 2016, 4, 223–253. [Google Scholar] [CrossRef]
- Guo, R.; Morimatsu, M.; Feng, T.; Lan, F.; Chang, D.; Wan, F.; Ling, Y. Stem cell-derived cell sheet transplantation for heart tissue repair in myocardial infarction. Stem Cell Res. Ther. 2020, 11, 19. [Google Scholar] [CrossRef]
- Esteban, M.A.; Xu, J.; Yang, J.; Peng, M.; Qin, D.; Li, W.; Jiang, Z.; Chen, J.; Deng, K.; Zhong, M.; et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J. Biol. Chem. 2009, 284, 17634–17640. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chen, J.; Ren, J.; Bao, L.; Liao, J.; Cui, C.; Rao, L.; Li, H.; Gu, Y.; Dai, H.; et al. Generation of pig induced pluripotent stem cells with a drug-inducible system. J. Mol. Cell Biol. 2009, 1, 46–54. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, C.; Zhang, P.; Li, X.; Liu, T.; Pu, Y.; Li, Y.; Cao, Z.; Cao, H.; Liu, Y.; et al. Efficient reprogramming of naïve-like induced pluripotent stem cells from porcine adipose-derived stem cells with a feeder-independent and serum-free system. PLoS ONE 2014, 9, e85089. [Google Scholar]
- Ruan, W.; Han, J.; Li, P.; Cao, S.; An, Y.; Lim, B.; Li, N. A novel strategy to derive iPS cells from porcine fibroblasts. Sci. China Life Sci. 2011, 54, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Guo, Y.; Li, Z.; Liu, Y.; Gao, X.; Gao, Y.; Cheng, X.; Hu, J.; Wang, H. Porcine induced pluripotent stem cells require LIF and maintain their developmental potential in early stage of embryos. PLoS ONE 2012, 7, e51778. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Song, G.; An, X.; Fan, A.; Tan, W.; Tang, B.; Zhang, X.; Li, Z. miRNAs promote generation of porcine-induced pluripotent stem cells. Mol. Cell Biochem. 2014, 389, 209–218. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Y.; Cui, Y.; Liu, Y.; Yu, T.; Wang, H. Generation of intermediate porcine iPS cells under culture condition favorable for mesenchymal to epithelial transition. Stem. Cell Rev. Rep. 2015, 11, 24–38. [Google Scholar] [CrossRef]
- Kues, W.A.; Herrmann, D.; Barg-Kues, B.; Haridoss, S.; Nowak-Imialek, M.; Buchholz, T.; Streeck, M.; Grebe, A.; Grabundzija, I.; Merkert, S.; et al. Derivation and characterization of sleeping beauty transposon-mediated porcine induced pluripotent stem cells. Stem Cells Dev. 2013, 22, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Allegrucci, C.; Alberio, R. Modulation of pluripotency in the porcine embryo and iPS cells. PLoS ONE 2012, 7, e49079. [Google Scholar] [CrossRef]
- Chakritbudsabong, W.; Sariya, L.; Pamonsupornvichit, S.; Pronarkngver, R.; Chaiwattanarungruengpaisan, S.; Ferreira, J.N.; Setthawong, P.; Phakdeedindan, P.; Techakumphu, M.; Tharasanit, T.; et al. Generation of a pig induced pluripotent stem cell (piPSC) line from embryonic fibroblasts by incorporating LIN28 to the four transcriptional factor-mediated reprogramming: VSMUi001-D. Stem Cell Res. 2017, 24, 21–24. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, Q.; Deng, W.; Wang, H.; Liu, K.; Fu, H.; Zhao, Q.; Wang, X.; Liu, L. Epigenetic Modifiers Facilitate Induction and Pluripotency of Porcine iPSCs. Stem Cell Rep. 2017, 8, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Nagy, K.; Sung, H.K.; Zhang, P.; Laflamme, S.; Vincent, P.; Agha-Mohammadi, S.; Woltjen, K.; Monetti, C.; Michael, I.P.; Smith, L.C. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. Rep. 2011, 7, 693–702. [Google Scholar] [CrossRef]
- Khodadadi, K.; Sumer, H.; Pashaiasl, M.; Lim, S.; Williamson, M.; Verma, P.J. Induction of pluripotency in adult equine fibroblasts without c-MYC. Stem Cells Int. 2012, 2012, 29160. [Google Scholar] [CrossRef]
- Breton, A.; Sharma, R.; Diaz, A.C.; Parham, A.G.; Graham, A.; Neil, C.; Whitelaw, C.B.; Milne, E.; Donadeu, F.X. Derivation and characterization of induced pluripotentstem cells from equine fibroblasts. Stem Cells Dev. 2013, 22, 611–621. [Google Scholar] [CrossRef]
- Whitworth, D.J.; Ovchinnikov, D.A.; Sun, J.; Fortuna, P.R.; Wolvetang, E.J. Generation and characterization of leukemia inhibitory factor-dependent equineinduced pluripotent stem cells from adult dermal fibroblasts. Stem Cells Dev. 2014, 23, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.M.; Kim, A.Y.; Lee, E.J.; Park, J.K.; Park, S.I.; Cho, S.G.; Kim, H.K.; Kim, S.Y.; Jeong, K.S. Generation of Equine-Induced Pluripotent Stem Cellsand Analysis of Their Therapeutic Potential for Muscle Injuries. Cell Transpl. 2016, 25, 2003–2016. [Google Scholar] [CrossRef]
- Quattrocelli, M.; Giacomazzi, G.; Broeckx, S.Y.; Ceelen, L.; Bolca, S.; Spaas, J.H.; Sampaolesi, M. Equine-Induced Pluripotent Stem Cells Retain Lineage Commitment Tward Myogenic and Chondrogenic Fates. Stem Cell Rep. 2016, 6, 55–63. [Google Scholar] [CrossRef]
- Moro, L.N.; Amin, G.; Furmento, V.; Waisman, A.; Garate, X.; Neiman, G.; La Greca, A.; Santín-Velazque, N.L.; Luzzani, C.; Sevlever, G.E.; et al. MicroRNA characterization in equine induced pluripotent stem cells. PLoS ONE 2018, 13, e0207074. [Google Scholar] [CrossRef] [PubMed]
- Pessôa, L.V.F.; Pires, P.R.L.; Del Collado, M.; Pieri, N.C.G.; Recchia, K.; Souza, A.F.; Perecin, F.; da Silveira, J.C.; de Andrade, A.F.C.; Ambrosio, C.E.; et al. Generation and miRNA Characterization of Equine Induced Pluripotent Stem Cells Derived from Fetal and Adult Multipotent Tissues. Stem Cells Int. 2019, 2019, 1393791. [Google Scholar] [CrossRef]
- Han, X.; Han, J.; Ding, F.; Cao, S.; Lim, S.S.; Dai, Y.; Zhang, R.; Zhang, Y.; Lim, B.; Li, N. Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 2011, 21, 1509–1512. [Google Scholar] [CrossRef]
- Sumer, H.; Liu, J.; Malaver-Ortega, L.F.; Lim, M.L.; Khodadadi, K.; Verma, P.J. NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J. Anim. Sci. 2011, 89, 708–716. [Google Scholar] [CrossRef]
- Cao, H.; Yang, P.; Pu, Y.; Sun, X.; Yin, H.; Zhang, Y.; Zhang, Y.; Li, Y.; Liu, Y.; Fang, F.; et al. Characterization of bovine induced plu-ripotent stem cells by lentiviral transduction of reprogramming factor fusion proteins. Int. J. Biol. Sci. 2012, 8, 498–511. [Google Scholar] [CrossRef]
- Talluri, T.R.; Kumar, D.; Glage, S.; Garrels, W.; Ivics, Z.; Debowski, K.; Behr, R.; Niemann, H.; Kues, W.A. Derivation and characterization of bovine induced plu-ripotent stem cells by transposon-mediated reprogramming. Cell Reprogram. 2015, 17, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Malaver-Ortega, L.F.; Sumer, H.; Liu, J.; Verma, P.J. Inhibition of JAK-STAT ERK/MAPK and Glycogen Synthase Kinase-3 Induces a Change in Gene Expression Profile of Bovine Induced Pluripotent Stem Cells. Stem Cells Int. 2016, 2016, 5127984. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Z.; Zhang, J.; Yang, J.; Gao, X.; Wu, B.; Zhao, G.; Bao, S.; Hu, S.; Liu, P.; et al. Characterization of the single-cell derived bovine induced pluripotent stem cells. Tissue Cell 2017, 49, 521–527. [Google Scholar] [CrossRef]
- Pillai, V.V.; Kei, T.G.; Reddy, S.E.; Das, M.; Abratte, C.; Cheong, S.H.; Selvaraj, V. Induced pluripotent stem cell generation from bovine somatic cells indicates unmet needs for pluripotency sustenance. Anim. Sci. J. 2019, 90, 1149–1160. [Google Scholar] [CrossRef]
- Shimada, H.; Nakada, A.; Hashimoto, Y.; Shigeno, K.; Shionoya, Y.; Nakamura, T. Generation of canine induced pluripotent stem cells by retroviral transduc-tion and chemical inhibitors. Mol. Reprod. Dev. 2010, 77, 2. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Suhr, S.T.; Chang, E.A.; Wang, K.; Ross, P.J.; Nelson, L.L.; Venta, P.J.; Knott, J.G.; Cibelli, J.B. Generation of leukemia inhibitory factor and basic fi-broblast growth factor-dependent induced pluripotent stem cells from canine adult somatic cells. Stem Cells Dev. 2011, 20, 1669–1678. [Google Scholar] [CrossRef]
- Lee, A.S.; Xu, D.; Plews, J.R.; Nguyen, P.K.; Nag, D.; Lyons, J.K.; Han, L.; Hu, S.; Lan, F.; Liu, J.; et al. Preclinical derivation and imaging of autologously transplanted canine induced pluripotent stem cells. J. Biol. Chem. 2011, 286, 32697–32704. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.; Thomas, R.; Tsai, S.; Bischoff, S.; Lim, J.H.; Breen, M.; Olby, N.J.; Piedrahita, J.A. Growth requirements and chromosomal instability of induced pluripotent stem cells generated from adult canine fibroblasts. Stem Cells Dev. 2013, 22, 951–963. [Google Scholar] [CrossRef]
- Whitworth, D.J.; Ovchinnikov, D.A.; Wolvetang, E.J. Generation and characterization of LIF-dependent canine induced pluripotent stem cells from adult dermal fibroblasts. Stem Cells Dev. 2012, 21, 2288–2297. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Hatoya, S.; Kanegi, R.; Sugiura, K.; Wijewardana, V.; Kuwamura, M.; Tanaka, M.; Yamate, J.; Izawa, T.; Takahashi, M.; et al. Generation of functional platelets from canine induced pluripotent stem cells. Stem Cells Dev. 2013, 22, 2026–2035. [Google Scholar] [CrossRef]
- Bao, L.; He, L.; Chen, J.; Wu, Z.; Liao, J.; Rao, L.; Ren, J.; Li, H.; Zhu, H.; Qian, L.; et al. Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 2011, 21, 600–608. [Google Scholar] [CrossRef]
- Li, Y.; Cang, M.; Lee, A.S.; Zhang, K.; Liu, D. Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS ONE 2011, 6, e15947. [Google Scholar] [CrossRef]
- Liu, J.; Balehosur, D.; Murray, B.; Kelly, J.M.; Sumer, H.; Verma, P.J. Generation and characterization of reprogrammed sheep induced pluripo-tent stem cells. Theriogenology 2012, 77, 338–346.e1. [Google Scholar] [CrossRef]
- Shi, H.; Fu, Q.; Li, G.; Ren, Y.; Hu, S.; Ni, W.; Guo, F.; Shi, M.; Meng, L.; Zhang, H.; et al. Roles of p53 and ASF1A in the Repro-gramming of Sheep Kidney Cells to Pluripotent Cells. Cell Reprogram. 2015, 17, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.; Liu, P.; Gao, J.; Jin, M.; Xu, T.; Zuo, Y.; Liang, H.; Liu, D. Generation of Arbas Cashmere Goat Induced Pluripotent Stem Cells Through Fibroblast Reprogramming. Cell Reprogram. 2015, 17, 297–305. [Google Scholar] [CrossRef]
- Song, H.; Li, H.; Huang, M.; Xu, D.; Gu, C.; Wang, Z.; Dong, F.; Wang, F. Induced pluripotent stem cells from goat fibroblasts. Mol. Reprod Dev. 2013, 80, 1009–1017. [Google Scholar] [CrossRef]
- Sandmaier, S.E.; Nandal, A.; Powell, A.; Garrett, W.; Blomberg, L.; Donovan, D.M.; Talbot, N.; Telugu, B.P. Generation of induced pluripotent stem cells from domestic goats. Mol. Reprod. Dev. 2015, 82, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zuo, Q.; Wang, Y.; Song, J.; Yang, H.; Zhang, Y.; Li, B. Inducing goat pluripotent stem cells with four transcription factor mRNAs that activate endogenous promoters. BMC Biotechnol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Chung, M.J.; Park, S.; Son, J.Y.; Lee, J.Y.; Yun, H.H.; Lee, E.J.; Lee, E.M.; Cho, G.J.; Lee, S.; Park, H.S.; et al. Differentiation of equine induced pluripotent stem cells into mesenchymal lineage for therapeutic use. Cell Cycle 2019, 18, 2954–2971. [Google Scholar] [CrossRef] [PubMed]
- Telugu, B.P.; Ezashi, T.; Roberts, R.M. Porcine induced pluripotent stem cells analogous to naïve and primed embryonic stem cells of the mouse. Int. J. Dev. Biol. 2010, 54, 1703–1711. [Google Scholar] [CrossRef]
- Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Wang, F.; Wang, Y.; Wang, X. m 6 A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 2019, 10, 171. [Google Scholar] [CrossRef]
- Wang, G.; Weng, R.; Lan, Y.; Guo, X.; Liu, Q.; Liu, X.; Lu, C.; Kang, J. Synergetic effects of DNA methylation and histone modification during mouse induced pluripotent stem cell generation. Sci. Rep. 2017, 7, 39527. [Google Scholar] [CrossRef] [PubMed]
- Habekost, M.; Jørgensen, A.L.; Qvist, P.; Denham, M. Transcriptomic profiling of porcine pluripotency identifies species-specific reprogramming requirements for culturing iPSCs. Stem Cell Res. 2019, 41, 101645. [Google Scholar] [CrossRef]
- Shi, B.; Gao, D.; Zhong, L.; Zhi, M.; Weng, X.; Xu, J.; Li, J.; Du, X.; Xin, Y.; Gao, J.; et al. IRF-1 expressed in the inner cell mass of the porcine early blastocyst enhances the pluripotency of induced pluripotent stem cells. Stem Cell Res. Ther. 2020, 11, 505. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Loo, S.J.; Gao, Y.; Tao, Z.H.; Su, L.P.; Wang, C.X.; Zhang, S.L.; Mu, Y.H.; Cui, Y.H.; Abdurrachim, D.; et al. Thymosin β4 increases cardiac cell proliferation, cell engraftment, and the reparative potency of human induced-pluripotent stem cell-derived cardiomyocytes in a porcine model of acute myocardial infarction. Theranostics 2021, 11, 7879–7895. [Google Scholar] [CrossRef] [PubMed]
- Biagi, D.; Fantozzi, E.T.; Campos-Oliveira, J.C.; Naghetini, M.V.; Ribeiro, A.F., Jr.; Rodrigues, S.; Ogusuku, I.; Vanderlinde, R.; Christie, M.L.A.; Mello, D.B.; et al. In Situ Maturated Early-Stage Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Improve Cardiac Function by Enhancing Segmental Contraction in Infarcted Rats. J. Pers Med. 2021, 11, 374. [Google Scholar] [CrossRef]
- Kawaguchi, S.; Soma, Y.; Nakajima, K.; Kanazawa, H.; Tohyama, S.; Tabei, R.; Hirano, A.; Handa, N.; Yamada, Y.; Okuda, S.; et al. Intramyocardial Transplantation of Human iPS Cell-Derived Cardiac Spheroids Improves Cardiac Function in Heart Failure Animals. JACC Basic Transl. Sci. 2021, 6, 239–254. [Google Scholar] [CrossRef]
- Nishida, M.; Tanaka, Y.; Tanaka, Y.; Amaya, S.; Tanaka, N.; Uyama, H.; Masuda, T.; Onishi, A.; Sho, J.; Yokota, S.; et al. Human iPS cell derived RPE strips for secure delivery of graft cells at a target place with minimal surgical invasion. Sci. Rep. 2021, 11, 21421. [Google Scholar] [CrossRef]
- Duarri, A.; Rodríguez-Bocanegra, E.; Martínez-Navarrete, G.; Biarnés, M.; García, M.; Ferraro, L.L.; Kuebler, B.; Aran, B.; Izquierdo, E.; Aguilera-Xiol, E.; et al. Transplantation of Human Induced Pluripotent Stem Cell-Derived Retinal Pigment Epithelium in a Swine Model of Ge-ographic Atrophy. Int. J. Mol. Sci. 2021, 22, 10497. [Google Scholar] [CrossRef] [PubMed]
- Thavapalachandran, S.; Le, T.Y.L.; Romanazzo, S.; Rashid, F.N.; Ogawa, M.; Kilian, K.A.; Brown, P.; Pouliopoulos, J.; Barry, A.M.; Fahmy, P.; et al. Pluripotent stem cell-derived mesenchymal stromal cells improve cardiac function and vascularity after myocardial infarction. Cytotherapy 2021, 23, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Lingam, S.; Liu, Z.; Yang, B.; Wong, W.; Parikh, B.H.; Ong, J.Y.; Goh, D.; Wong, D.S.L.; Tan, Q.S.W.; Tan, G.S.W.; et al. cGMP-grade human iPSC-derived retinal photoreceptor precursor cells rescue cone photoreceptor damage in non-human primates. Stem Cell Res. Ther. 2021, 12, 464. [Google Scholar] [CrossRef]
- Volarevic, V.; Markovic, B.S.; Gazdic, M.; Volarevic, A.; Jovicic, N.; Arsenijevic, N.; Armstrong, L.; Djonov, V.; Lako, M.; Stojkovic, M. Ethical and Safety Issues of Stem Cell-Based Therapy. Int. J. Med. Sci. 2018, 15, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Tavakol, D.N.; Fleischer, S.; Vunjak-Novakovic, G. Harnessing organs-on-a-chip to model tissue regeneration. Cell Stem Cell 2021, 28, 993–1015. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K.; et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927. [Google Scholar] [CrossRef]
- Tang, B.L. Maturing iPSC-Derived Cardiomyocytes. Cells 2020, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Breckwoldt, K.; Letuffe-Brenière, D.; Mannhardt, I.; Schulze, T.; Ulmer, B.; Werner, T.; Benzin, A.; Klampe, B.; Reinsch, M.C.; Laufer, S.; et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017, 12, 1177–1197. [Google Scholar] [CrossRef] [PubMed]
- Shiba, Y.; Gomibuchi, T.; Seto, T.; Wada, Y.; Ichimura, H.; Tanaka, Y.; Ogasawara, T.; Okada, K.; Shiba, N.; Sakamoto, K.; et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016, 538, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Bizy, A.; Klos, M. Optimizing the Use of iPSC-CMs for Cardiac Regeneration in Animal Models. Animals 2020, 10, 1561. [Google Scholar] [CrossRef]
- Zhang, J.; Wilson, G.F.; Soerens, A.G.; Koonce, C.H.; Yu, J.; Palecek, S.P.; Thomson, J.A.; Kamp, T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009, 104, e30–e41. [Google Scholar] [CrossRef]
- Cyganek, L.; Tiburcy, M.; Sekeres, K.; Gerstenberg, K.; Bohnenberger, H.; Lenz, C.; Henze, S.; Stauske, M.; Salinas, G.; Zimmermann, W.H.; et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight 2018, 3, e99941. [Google Scholar] [CrossRef] [PubMed]
- Fujita, J. Development of Cardiac Regenerative Medicine Using Human iPS Cell-derived Cardiomyocytes. Keio J. Med. 2021, 70, 53–59. [Google Scholar] [CrossRef]
- Masumoto, H.; Yamashita, J.K. Human iPS Cell-Derived Cardiac Tissue Sheets: A Platform for Cardiac Regeneration. Curr. Treat Options Cardiovasc. Med. 2016, 18, 65. [Google Scholar] [CrossRef]
- van den Berg, C.W.; Elliott, D.A.; Braam, S.R.; Mummery, C.L.; Davis, R.P. Differentiation of Human Pluripotent Stem Cells to Cardiomyocytes under Defined Conditions. Methods Mol. Biol. 2016, 1353, 163–180. [Google Scholar] [PubMed]
- Paige, S.L.; Osugi, T.; Afanasiev, O.K.; Pabon, L.; Reinecke, H.; Murry, C.E. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS ONE 2010, 5, e11134. [Google Scholar] [CrossRef]
- Laflamme, M.A.; Chen, K.Y.; Naumova, A.V.; Muskheli, V.; Fugate, J.A.; Dupras, S.K.; Reinecke, H.; Xu, C.; Hassanipour, M.; Police, S.; et al. Cardiomyocytes derived from human embryonic stem cells in pro-survivalfactors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007, 25, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Q.; Zweigerdt, R.; Soo, S.Y.; Ngoh, Z.X.; Tham, S.C.; Wang, S.T.; Graichen, R.; Davidson, B.; Colman, A.; Sun, W. Highly enriched cardiomyocytes from human embryonic stem cells. Cytotherapy 2008, 10, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Kempf, H.; Kropp, C.; Olmer, R.; Martin, U.; Zweigerdt, R. Cardiac differentiation of human pluripotent stem cells in scalable suspension culture. Nat. Protoc. 2015, 10, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- Laco, F.; Woo, T.L.; Zhong, Q.; Szmyd, R.; Ting, S.; Khan, F.J.; Chai, C.L.L.; Reuveny, S.; Chen, A.; Oh, S. Unraveling the Inconsistencies of Cardiac Differentiation Efficiency Induced by the GSK3β Inhibitor CHIR99021 in Human Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1851–1866. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Zhang, J.; Azarin, S.M.; Zhu, K.; Hazeltine, L.B.; Bao, X.; Hsiao, C.; Kamp, T.J.; Palecek, S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Prot. 2013, 8, 162–175. [Google Scholar] [CrossRef]
- Mummery, C.; Zhang, J.; Ng, E.S.; Elliott, D.; Elefanty, A.; Kamp, T. Differentiation of Human Embryonic Stem Cells and Induced Pluripotent Stem Cells to Cardiomyocytes. Circ. Res. 2012, 111, 344–358. [Google Scholar] [CrossRef]
- Burridge, P.W.; Thompson, S.; Millrod, M.A.; Weinberg, S.; Yuan, X.; Peters, A.; Mahairaki, V.; Koliatsos, V.E.; Tung, L.; Zambidis, E.T. A Universal System for Highly Efficient Cardiac Differentiation of Human Induced Pluripotent Stem Cells That Eliminates Interline Variability. PLoS ONE 2011, 6, e18293. [Google Scholar] [CrossRef]
- Freund, C.; Oostwaard, D.W.; Monshouwer-Kloots, J.; Brink, S.V.D.; van Rooijen, M.; Xu, X.; Zweigerdt, R.; Mummery, C.; Passier, R. Insulin Redirects Differentiation from Cardiogenic Mesoderm and Endoderm to Neuroectoderm in Differentiating Human Embryonic Stem Cells. Stem Cells 2008, 26, 724–733. [Google Scholar] [CrossRef]
- Huang, C.Y.; Maia-Joca, R.P.M.; Ong, C.S.; Wilson, I.; DiSilvestre, D.; Tomaselli, G.F.; Reich, D.H. Enhancement of human iPSC-derived cardiomyocyte maturation by chemical conditioning in a 3D environment. J. Mol. Cell. Cardiol. 2020, 138, 1–11. [Google Scholar] [CrossRef]
- Li, P.; Cavallero, S.; Gu, Y.; Chen, T.H.P.; Hughes, J.; Hassan, A.B.; Brüning, J.C.; Pashmforoush, M.; Sucov, H.M. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development 2011, 138, 1795–1805. [Google Scholar] [CrossRef]
- Lian, X.; Zhang, J.; Zhu, K.; Kamp, T.J.; Palecek, S.P. Insulin Inhibits Cardiac Mesoderm, Not Mesendoderm, Formation During Cardiac Differentiation of Human Pluripotent Stem Cells and Modulation of Canonical Wnt Signaling Can Rescue This Inhibition. Stem Cells 2012, 31, 447–457. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, T.C.; Laflamme, M.; Murry, C.E. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J. Mol. Cell. Cardiol. 2005, 39, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Kaichi, S.; Hasegawa, K.; Takaya, T.; Yokoo, N.; Mima, T.; Kawamura, T.; Morimoto, T.; Ono, K.; Baba, S.; Doi, H.; et al. Cell line-dependent differentiation of induced pluripotent stem cells into cardiomyocytes in mice. Cardiovasc. Res. 2010, 88, 314–323. [Google Scholar] [CrossRef][Green Version]
- Cao, N.; Liu, Z.; Chen, Z.; Wang, J.; Chen, T.; Zhao, X.; Ma, Y.; Qin, L.; Kang, J.; Wei, B.; et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012, 22, 219–236. [Google Scholar] [CrossRef] [PubMed]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M.; et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef]
- Xavier-Neto, J.; Neville, C.M.; Shapiro, M.D.; Houghton, L.; Wang, G.F.; Nikovits, W., Jr.; Stockdale, F.E.; Rosenthal, N. A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development 1999, 126, 2677–2687. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.Z.; Xie, Y.; Moyes, K.W.; Gold, J.D.; Askari, B.; Laflamme, M.A. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ. Res. 2010, 107, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Gassanov, N.; Er, F.; Zagidullin, N.; Hoppe, U.C. Endothelin induces differentiation of ANP-EGFP expressing embryonic stem cells towards a pacemaker phenotype. FASEB J. 2004, 18, 1710–1712. [Google Scholar] [CrossRef]
- Yoshida, Y.; Yamanaka, S. Induced Pluripotent Stem Cells 10 Years Later: For Cardiac Applications. Circ. Res. 2017, 120, 1958–1968. [Google Scholar] [CrossRef]
- Yang, X.; Pabon, L.; Murry, C.E. Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014, 114, 511–523. [Google Scholar] [CrossRef]
- Zhang, Z.; Bédard, E.; Luo, Y.; Wang, H.; Deng, S.; Kelvin, D.; Zhong, R. Animal models in xenotransplantation. Expert Opin. Investig. Drugs 2000, 9, 2051–2068. [Google Scholar] [CrossRef]
- Milani-Nejad, N.; Janssen, P.M. Small and large animal models in cardiac contraction research: Advantages and disadvantages. Pharmacol. Ther. 2014, 141, 235–249. [Google Scholar] [CrossRef]
- Fliegner, D.; Gerdes, C.; Meding, J.; Stasch, J.P. Translational In Vivo Models for Cardiovascular Diseases. Handb. Exp. Pharmacol. 2016, 232, 223–234. [Google Scholar]
- Recchia, F.A.; Lionetti, V. Animal models of dilated cardiomyopathy for translational research. Vet. Res. Commun. 2007, 31, 35–41. [Google Scholar] [CrossRef]
- Bolli, R.; Ghafghazi, S. Cell Therapy Needs Rigorous Translational Studies in Large Animal Models. J. Am. Coll. Cardiol. 2015, 66, 2000–2004. [Google Scholar] [CrossRef]
- Harding, J.; Roberts, R.M.; Mirochnitchenko, O. Large animal models for stem cell therapy. Stem Cell Res. Ther. 2013, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- van der Spoel, T.I.; Jansen of Lorkeers, S.J.; Agostoni, P.; van Belle, E.; Gyöngyösi, M.; Sluijter, J.P.; Cramer, M.J.; Doevendans, P.A.; Chamuleau, S.A. Human relevance of pre-clinical studies in stem cell therapy: Systematic review and meta-analysis of large animal models of ischaemic heart disease. Cardiovasc Res. 2011, 91, 649–658. [Google Scholar] [CrossRef]
- Wang, H. Small animal models of xenotransplantation. Methods Mol. Biol. 2012, 885, 125–153. [Google Scholar] [PubMed]
- da Silva Morais, A.; Oliveira, J.M.; Reis, R.L. Small Animal Models. Adv. Exp. Med. Biol. 2018, 1059, 423–439. [Google Scholar] [PubMed]
- Russell, J.C.; Proctor, S.D. Small animal models of cardiovascular disease: Tools for the study of the roles of metabolic syn-drome, dyslipidemia, and atherosclerosis. Cardiovasc. Pathol. 2006, 15, 318–330. [Google Scholar] [CrossRef]
- Bracken, M.B. Why animal studies are often poor predictors of human reactions to exposure. J. R. Soc. Med. 2009, 102, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Martić-Kehl, M.I.; Schibli, R.; Schubiger, P.A. Can animal data predict human outcome? Problems and pitfalls of translational animal research. Eur. J. Nucl. Med. Mol. Imaging 2012, 39, 1492–1496. [Google Scholar] [CrossRef]
- Veening-Griffioen, D.H.; Ferreira, G.S.; van Meer, P.J.K.; Boon, W.P.C.; Gispen-de Wied, C.C.; Moors, E.H.M.; Schellekens, H. Are some animal models more equal than others? A case study on the translational value of animal models of efficacy for Alzheimer’s disease. Eur. J. Pharmacol. 2019, 859, 172524. [Google Scholar] [CrossRef]
- Li, X.J.; Li, S. Large Animal Models of Huntington’s Disease. Curr. Top. Behav. Neurosci. 2015, 22, 149–160. [Google Scholar]
- Shim, J.; Al-Mashhadi, R.H.; Sørensen, C.B.; Bentzon, J.F. Large animal models of atherosclerosis—New tools for persistent problems in car-diovascular medicine. J. Pathol. 2016, 238, 257–266. [Google Scholar] [CrossRef]
- Hotham, W.E.; Henson, F.M.D. The use of large animals to facilitate the process of MSC going from laboratory to pa-tient-‘bench to bedside’. Cell Biol. Toxicol. 2020, 36, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Van der Velden, J.; Snibson, K.J. Airway disease: The use of large animal models for drug discovery. Pulm. Pharmacol. Ther. 2011, 24, 525–532. [Google Scholar] [CrossRef]
- Plews, J.R.; Gu, M.; Longaker, M.T.; Wu, J.C. Large animal induced pluripotent stem cells as pre-clinical models for studying human disease. J. Cell. Mol. Med. 2012, 16, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Alstrup, K.O.; Winterdahl, M. Imaging techniques in large animals. Scand. J. Lab. Anim. Sci. 2009, 36, 55–66. [Google Scholar] [CrossRef]
- Yang, P.C. Is Reliable In Vivo Detection of Stem Cell Viability Possible in a Large Animal Model of Myocardial Injury? Circulation 2012, 126, 388–390. [Google Scholar] [CrossRef][Green Version]
- Gandolfi, F.; Vanelli, A.; Pennarossa, G.; Rahaman, M.; Acocella, F.; Brevini, T. Large animal models for cardiac stem cell therapies. Theriogenology 2011, 75, 1416–1425. [Google Scholar] [CrossRef] [PubMed]
- Amado, L.C.; Schuleri, K.H.; Saliaris, A.P.; Boyle, A.J.; Helm, R.; Oskouei, B.; Centola, M.; Eneboe, V.; Young, R.; Lima, J.A.; et al. Multimodality Noninvasive Imaging Demonstrates In Vivo Cardiac Regeneration After Mesenchymal Stem Cell Therapy. J. Am. Coll. Cardiol. 2006, 48, 2116–2124. [Google Scholar] [CrossRef]
- Mazhari, R.; Hare, J.M. Translational Findings from Cardiovascular Stem Cell Research. Trends Cardiovasc. Med. 2012, 22, 1–6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, X.; Zhang, F.; Song, G.; Gu, W.; Chen, M.; Yang, B.; Li, D.; Wang, D.; Cao, K. Intramyocardial Injection of Pig Pluripotent Stem Cells Improves Left Ventricular Function and Perfusion: A Study in a Porcine Model of Acute Myocardial Infarction. PLoS ONE 2013, 8, e66688. [Google Scholar] [CrossRef]
- Zhang, F.; Song, G.; Li, X.; Gu, W.; Shen, Y.; Chen, M.; Yang, B.; Qian, L.; Cao, K. Transplantation of iPSc Ameliorates Neural Remodeling and Reduces Ventricular Arrhythmias in a Post-Infarcted Swine Model. J. Cell. Biochem. 2014, 115, 531–539. [Google Scholar] [CrossRef]
- Song, G.; Lingmei, Q.; Shen, Y.; Qian, L.; Kong, X.; Chen, M.; Cao, K.; Zhang, F. Transplantation of iPSc Restores Cardiac Function by Promoting Angiogenesis and Ameliorating Cardiac Remodeling in a Post-infarcted Swine Model. Cell Biophys. 2014, 71, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Gálvez-Montón, C.; Soler-Botija, C.; Iborra-Egea, O.; Díaz-Güemes, I.; Martí, M.; Iglesias-García, O.; Prat-Vidal, C.; Crisóstomo, V.; Llucià-Valldeperas, A.; Perea-Gil, I.; et al. Preclinical Safety Evaluation of Allogeneic Induced Pluripotent Stem Cell-Based Therapy in a Swine Model of Myocardial Infarction. Tissue Eng. Part C Methods 2017, 23, 736–744. [Google Scholar] [CrossRef]
- Kawamura, T.; Miyagawa, S.; Fukushima, S.; Maeda, A.; Kashiyama, N.; Kawamura, A.; Miki, K.; Okita, K.; Yoshida, Y.; Shiina, T.; et al. Cardiomyocytes Derived from MHC-Homozygous Induced Pluripotent Stem Cells Exhibit Reduced Allogeneic Immunogenicity in MHC-Matched Non-human Primates. Stem Cell Rep. 2016, 6, 312–320. [Google Scholar] [CrossRef]
- Kashiyama, N.; Miyagawa, S.; Fukushima, S.; Kawamura, T.; Kawamura, A.; Yoshida, S.; Eiraku, S.; Harada, A.; Matsunaga, K.; Watabe, T.; et al. MHC-mismatched Allotransplantation of Induced Pluripotent Stem Cell-derived Cardiomyocyte Sheets to Improve Cardiac Function in a Primate Ischemic Cardiomyopathy Model. Transplantation 2019, 103, 1582–1590. [Google Scholar] [CrossRef]
- Ye, L.; Chang, Y.-H.; Xiong, Q.; Zhang, P.; Zhang, L.; Somasundaram, P.; Lepley, M.; Swingen, C.; Su, L.; Wendel, J.S.; et al. Cardiac Repair in a Porcine Model of Acute Myocardial Infarction with Human Induced Pluripotent Stem Cell-Derived Cardiovascular Cells. Cell Stem Cell 2014, 15, 750–761. [Google Scholar] [CrossRef]
- Ishigami, M.; Masumoto, H.; Ikuno, T.; Aoki, T.; Kawatou, M.; Minakata, K.; Ikeda, T.; Sakata, R.; Yamashita, J.K.; Minatoya, K. Human iPS cell-derived cardiac tissue sheets for functional restoration of infarcted porcine hearts. PLoS ONE 2018, 13, e0201650. [Google Scholar] [CrossRef]
- Gao, L.; Gregorich, Z.R.; Zhu, W.; Mattapally, S.; Oduk, Y.; Lou, X.; Kannappan, R.; Borovjagin, A.V.; Walcott, G.P.; Pollard, A.E.; et al. Large Cardiac Muscle Patches Engineered From Human Induced-Pluripotent Stem Cell–Derived Cardiac Cells Improve Recovery From Myocardial Infarction in Swine. Circulation 2018, 137, 1712–1730. [Google Scholar] [CrossRef]
- Liao, S.; Zhang, Y.; Ting, S.; Zhen, Z.; Luo, F.; Zhu, Z.; Jiang, Y.; Sun, S.; Lai, W.-H.; Lian, Q.; et al. Potent immunomodulation and angiogenic effects of mesenchymal stem cells versus cardiomyocytes derived from pluripotent stem cells for treatment of heart failure. Stem Cell Res. Ther. 2019, 10, 78. [Google Scholar] [CrossRef]
- Kawamura, M.; Miyagawa, S.; Miki, K.; Saito, A.; Fukushima, S.; Higuchi, T.; Kuratani, T.; Daimon, T.; Shimizu, T.; Okano, T.; et al. Feasibility, Safety, and Therapeutic Efficacy of Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Sheets in a Porcine Ischemic Cardiomyopathy Model. Circulation 2012, 126, S29–S37. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, M.; Miyagawa, S.; Fukushima, S.; Saito, A.; Miki, K.; Ito, E.; Sougawa, N.; Kawamura, T.; Daimon, T.; Shimizu, T.; et al. Enhanced Survival of Transplanted Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes by the Combination of Cell Sheets With the Pedicled Omental Flap Technique in a Porcine Heart. Circulation 2013, 128, S87–S94. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, M.; Miyagawa, S.; Fukushima, S.; Saito, A.; Miki, K.; Funakoshi, S.; Yoshida, Y.; Yamanaka, S.; Shimizu, T.; Okano, T.; et al. Enhanced Therapeutic Effects of Human iPS Cell Derived-Cardiomyocyte by Combined Cell-Sheets with Omental Flap Technique in Porcine Ischemic Cardiomyopathy Model. Sci. Rep. 2017, 7, 8824. [Google Scholar] [CrossRef]
- Zhao, M.; Nakada, Y.; Wei, Y.; Bian, W.; Chu, Y.; Borovjagin, A.V.; Xie, M.; Zhu, W.; Nguyen, T.; Zhou, Y.; et al. Cyclin D2 Overexpression Enhances the Efficacy of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes for Myocardial Repair in a Swine Model of Myocardial Infarction. Circulation 2021, 144, 210–228. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Fujita, J.; Matsui, M.; Tohyama, S.; Tamura, N.; Kanazawa, H.; Seki, T.; Kishino, Y.; Hirano, A.; Okada, M.; et al. Gelatin Hydrogel Enhances the Engraftment of Transplanted Cardiomyocytes and Angiogenesis to Ameliorate Cardiac Function after Myocardial Infarction. PLoS ONE 2015, 10, e0133308. [Google Scholar] [CrossRef]
- Montserrat, N.; Bahima, E.G.; Batlle, L.; Häfner, S.; Rodrigues, A.; González, F.; Belmonte, J.C.I. Generation of Pig iPS Cells: A Model for Cell Therapy. J. Cardiovasc. Transl. Res. 2011, 4, 121–130. [Google Scholar] [CrossRef]
- Nelson, T.J.; Martinez-Fernandez, A.; Terzic, A. Induced pluripotent stem cells: Developmental biology to regenerative medicine. Nat. Rev. Cardiol. 2010, 7, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Gálvez-Montón, C.; Prat-Vidal, C.; Roura, S.; Farré, J.; Soler-Botija, C.; Llucià-Valldeperas, A.; Díaz-Güemes, I.; Sánchez-Margallo, F.M.; Arís, A.; Bayes-Genis, A. Transposition of a pericardial-derived vascular adipose flap for myocardial salvage after infarct. Cardiovasc. Res. 2011, 91, 659–667. [Google Scholar] [CrossRef][Green Version]
- Petersdorf, E.W. The major histocompatibility complex: A model for understanding graft-versus-host disease. Blood 2013, 122, 1863–1872. [Google Scholar] [CrossRef] [PubMed]
- Bach, F.H.; Bach, M.L.; Sondel, P.M. Differential function of major histocompatibility complex antigens in T-lymphocyte activation. Nat. Cell Biol. 1976, 259, 273–281. [Google Scholar] [CrossRef]
- Janeway, C.A.; Travers, P.; Walport, M.; Shlomchik, M. Immunobiology: The Immune System in Health and Disease; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Flomenberg, N.; Baxter-Lowe, L.A.; Confer, D.; Fernandez-Vina, M.; Filipovich, A.; Horowitz, M.; Hurley, C.; Kollman, C.; Anasetti, C.; Noreen, H.; et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood 2004, 104, 1923–1930. [Google Scholar] [CrossRef]
- Nakatsuji, N.; Nakajima, F.; Tokunaga, K. HLA-haplotype banking and iPS cells. Nat. Biotechnol. 2008, 26, 739–740. [Google Scholar] [CrossRef]
- Taylor, C.J.; Peacock, S.; Chaudhry, A.N.; Bradley, J.A.; Bolton, E. Generating an iPSC Bank for HLA-Matched Tissue Transplantation Based on Known Donor and Recipient HLA Types. Cell Stem Cell 2012, 11, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Germanguz, I.; Sedan, O.; Zeevi-Levin, N.; Shtrichman, R.; Barak, E.; Ziskind, A.; Eliyahu, S.; Meiry, G.; Amit, M.; Itskovitz-Eldor, J.; et al. Molecular characterization and functional properties of cardiomyocytes derived from human inducible pluripotent stem cells. J. Cell. Mol. Med. 2009, 15, 38–51. [Google Scholar] [CrossRef]
- Ren, Y.; Lee, M.Y.; Schliffke, S.; Paavola, J.; Amos, P.J.; Ge, X.; Ye, M.; Zhu, S.; Senyei, G.; Lum, L.; et al. Small molecule Wnt inhibitors enhance the efficiency of BMP-4-directed cardiac differentiation of human pluripotent stem cells. J. Mol. Cell. Cardiol. 2011, 51, 280–287. [Google Scholar] [CrossRef]
- Beauchamp, J.; Morgan, J.E.; Pagel, C.; Partridge, T.A. Dynamics of Myoblast Transplantation Reveal a Discrete Minority of Precursors with Stem Cell–like Properties as the Myogenic Source. J. Cell Biol. 1999, 144, 1113–1122. [Google Scholar] [CrossRef]
- Qu, Z.; Balkir, L.; Van Deutekom, J.C.; Robbins, P.D.; Pruchnic, R.; Huard, J. Development of Approaches to Improve Cell Survival in Myoblast Transfer Therapy. J. Cell Biol. 1998, 142, 1257–1267. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.-L.; Rokosh, G.; Sanganalmath, S.K.; Yuan, F.; Sato, H.; Mu, J.; Dai, S.; Li, C.; Chen, N.; Peng, Y.; et al. Intracoronary Administration of Cardiac Progenitor Cells Alleviates Left Ventricular Dysfunction in Rats With a 30-Day-Old Infarction. Circulation 2010, 121, 293–305. [Google Scholar] [CrossRef]
- Menasche, P. Cardiac cell therapy: Lessons from clinical trials. J. Mol. Cell. Cardiol. 2011, 50, 258–265. [Google Scholar] [CrossRef]
- Hsiao, L.-C.; Carr, C.; Chang, K.-C.; Lin, S.-Z.; Clarke, K. Stem Cell-Based Therapy for Ischemic Heart Disease. Cell Transplant. 2013, 22, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Templin, C.; Zweigerdt, R.; Schwanke, K.; Olmer, R.; Ghadri, J.-R.; Emmert, M.Y.; Müller, E.; Küest, S.M.; Cohrs, S.; Schibli, R.; et al. Transplantation and Tracking of Human-Induced Pluripotent Stem Cells in a Pig Model of Myocardial Infarction. Circulation 2012, 126, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Nunes, S.S. Bioengineering Approaches to Mature Human Pluripotent Stem Cell-Derived Cardiomyocytes. Front. Cell Dev. Biol. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Shadrin, I.; Lam, J.; Xian, H.-Q.; Snodgrass, H.R.; Bursac, N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 2013, 34, 5813–5820. [Google Scholar] [CrossRef]
- Nguyen, P.K.; Rhee, J.-W.; Wu, J.C. Adult stem cell therapy and heart failure, 2000 to 2016: A systematic review. JAMA Cardiol. 2016, 1, 831–841. [Google Scholar] [CrossRef]
- Poulin, M.-F.; Deka, A.; Mohamedali, B.; Schaer, G.L. Clinical Benefits of Stem Cells for Chronic Symptomatic Systolic Heart Failure: A Systematic Review of the Existing Data and Ongoing Trials. Cell Transplant. 2016, 25, 1911–1923. [Google Scholar] [CrossRef]
- Ishida, M.; Miyagawa, S.; Saito, A.; Fukushima, S.; Harada, A.; Ito, E.; Ohashi, F.; Watabe, T.; Hatazawa, J.; Matsuura, K.; et al. Transplantation of Human-induced Pluripotent Stem Cell-derived Cardiomyocytes Is Superior to Somatic Stem Cell Therapy for Restoring Cardiac Function and Oxygen Consumption in a Porcine Model of Myocardial Infarction. Transplantation 2019, 103, 291–298. [Google Scholar] [CrossRef]
- Chandra, A.; Srivastava, R.K.; Kashyap, M.P.; Kumar, R.; Srivastava, R.N.; Pant, A.B. The Anti-Inflammatory and Antibacterial Basis of Human Omental Defense: Selective Expression of Cytokines and Antimicrobial Peptides. PLoS ONE 2011, 6, e20446. [Google Scholar] [CrossRef]
- Beeres, S.L.M.A.; Atsma, D.E.; Van Ramshorst, J.; Schalij, M.J.; Bax, J.J. Cell therapy for ischaemic heart disease. Heart 2008, 94, 1214–1226. [Google Scholar] [CrossRef]
- Hou, D.; Youssef, E.A.-S.; Brinton, T.J.; Zhang, P.; Rogers, P.; Price, E.T.; Yeung, A.C.; Johnstone, B.H.; Yock, P.G.; March, K.L. Radiolabeled Cell Distribution After Intramyocardial, Intracoronary, and Interstitial Retrograde Coronary Venous Delivery. Circulation 2005, 112, I150–I156. [Google Scholar] [CrossRef]
- Hattori, F.; Chen, H.; Yamashita, H.; Tohyama, S.; Satoh, Y.-S.; Yuasa, S.; Li, W.; Yamakawa, H.; Tanaka, T.; Onitsuka, T.; et al. Nongenetic method for purifying stem cell–derived cardiomyocytes. Nat. Methods 2009, 7, 61–66. [Google Scholar] [CrossRef]
- Rane, A.A.; Christman, K.L. Biomaterials for the Treatment of Myocardial Infarction: A 5-Year Update. J. Am. Coll. Cardiol. 2011, 58, 2615–2629. [Google Scholar] [CrossRef]
- Tabei, R.; Kawaguchi, S.; Kanazawa, H.; Tohyama, S.; Hirano, A.; Handa, N.; Hishikawa, S.; Teratani, T.; Kunita, S.; Fukuda, J.; et al. Development of a transplant injection device for optimal distribution and retention of human induced pluripotent stem cell—derived cardiomyocytes. J. Heart Lung Transplant. 2019, 38, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.-C.E.; Wu, C.-S.; Chiou, M.-H.; Hsieh, P.-C.; Yu, H.-S. Low-Energy Helium-Neon Laser Induces Locomotion of the Immature Melanoblasts and Promotes Melanogenesis of the More Differentiated Melanoblasts: Recapitulation of Vitiligo Repigmentation In Vitro. J. Investig. Dermatol. 2006, 126, 2119–2126. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Ye, L.; Zhang, P.; Lepley, M.; Swingen, C.; Zhang, L.; Kaufman, D.; Zhang, J. Bioenergetic and Functional Consequences of Cellular Therapy: Activation of Endogenous Cardiovascular Progenitor Cells. Circ. Res. 2012, 111, 455–468. [Google Scholar] [CrossRef]
- Hsieh, P.C.; Davis, M.E.; Lisowski, L.K.; Lee, R.T. Endothelial-cardiomyocyte interactions in cardiac development and repair. Annu. Rev. Physiol. 2006, 68, 51–66. [Google Scholar] [CrossRef]
- Brutsaert, D.L. Cardiac Endothelial-Myocardial Signaling: Its Role in Cardiac Growth, Contractile Performance, and Rhythmicity. Physiol. Rev. 2003, 83, 59–115. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Hsieh, P.C.; Takahashi, T.; Song, Q.; Zhang, S.; Kamm, R.D.; Grodzinsky, A.J.; Anversa, P.; Lee, R.T. Local myocardial insulin-like growth factor 1 (IGF-1) delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc. Natl. Acad. Sci. USA 2006, 103, 8155–8160. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Hu, Q.; Wang, X.; Mansoor, A.; Lee, J.; Feygin, J.; Zhang, G.; Suntharalingam, P.; Boozer, S.; Mhashilkar, A.; et al. Bioenergetic and Functional Consequences of Bone Marrow–Derived Multipotent Progenitor Cell Transplantation in Hearts With Postinfarction Left Ventricular Remodeling. Circulation 2007, 115, 1866–1875. [Google Scholar] [CrossRef]
- Masumoto, H.; Matsuo, T.; Yamamizu, K.; Uosaki, H.; Narazaki, G.; Katayama, S.; Marui, A.; Shimizu, T.; Ikeda, T.; Okano, T.; et al. Pluripotent Stem Cell-Engineered Cell Sheets Reassembled with Defined Cardiovascular Populations Ameliorate Reduction in Infarct Heart Function Through Cardiomyocyte-Mediated Neovascularization. Stem Cells 2012, 30, 1196–1205. [Google Scholar] [CrossRef]
- Olivetti, G.; Capasso, J.M.; Meggs, L.G.; Sonnenblick, E.H.; Anversa, P. Cellular basis of chronic ventricular remodeling after myocardial infarction in rats. Circ. Res. 1991, 68, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Riegler, J.; Tiburcy, M.; Ebert, A.D.; Tzatzalos, E.; Raaz, U.; Abilez, O.J.; Shen, Q.; Kooreman, N.G.; Neofytou, E.; Chen, V.C.; et al. Human Engineered Heart Muscles Engraft and Survive Long Term in a Rodent Myocardial Infarction Model. Circ. Res. 2015, 117, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, F.; Breckwoldt, K.; Pecha, S.; Kelly, A.; Geertz, B.; Starbatty, J.; Yorgan, T.; Cheng, K.-H.; Lessmann, K.; Stolen, T.; et al. Cardiac repair in guinea pigs with human engineered heart tissue from induced pluripotent stem cells. Sci. Transl. Med. 2016, 8, 363ra148. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Liao, J.; Chang, C.; Liu, Y.; Padhiar, A.A.; Zhou, Y.; Zhou, G. Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells Hold Lower Heterogeneity and Great Promise in Biological Research and Clinical Applications. Front. Cell Dev. Biol. 2021, 9, 716907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, Z.; Jiang, D.; Liang, X.; Liao, S.; Zhang, Z.; Yue, W.; Li, X.; Chiu, S.-M.; Chai, Y.-H.; et al. iPSC-MSCs with High Intrinsic MIRO1 and Sensitivity to TNF-α Yield Efficacious Mitochondrial Transfer to Rescue Anthracycline-Induced Cardiomyopathy. Stem Cell Rep. 2016, 7, 749–763. [Google Scholar] [CrossRef]
- Lian, Q.; Zhang, Y.; Zhang, J.; Zhang, H.K.; Wu, X.; Zhang, Y.; Lam, F.F.-Y.; Kang, S.; Xia, J.C.; Lai, W.-H.; et al. Functional Mesenchymal Stem Cells Derived From Human Induced Pluripotent Stem Cells Attenuate Limb Ischemia in Mice. Circulation 2010, 121, 1113–1123. [Google Scholar] [CrossRef]
- Ambrosio, C.; Zomer, H.; Vidane, A.; Gonçalves, N. Mesenchymal and induced pluripotent stem cells: General insights and clinical perspectives. Stem Cells Cloning Adv. Appl. 2015, 8, 125–134. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).