1. Introduction
Osteoarthritis (OA) is the most prevalent musculoskeletal disease and a common joint degenerative disease. OA is a global health burden and is accountable for substantial health costs [
1,
2]. It is characterized by chronic pain and functional disability, and the knee is the most affected among the joints [
3]. The hallmark of the disease is the loss of a joint tissue, the cartilage [
3].
OA diagnosis often occurs late, i.e., when the destruction of articular tissues has reached a late stage. This is of importance as although OA is characterized by being a disease of “older age”, younger people are more and more being affected by this disease [
4]. Moreover, its two most prominent risk factors, age and body mass index (BMI) [
5], are also of considerable concern for the healthcare system, as there is a growing number of aging and obese people worldwide who will soon confront the system with an unsustainable draw for OA individuals. Above all, there is not yet a curative cure (in the form of disease-modifying OA drugs [DMOADs]) for this disease [
3,
6]. Currently, OA treatments only relieve symptoms.
To be able to combat the rise of this disease, there is a critical need to identify, at an early stage, individuals at risk of having a structurally progressive disease, i.e., rapid degradation of cartilage. Indeed, therapeutic strategies used early during the pathological process may permit to reduce/stop the structural progression of the disease. In turn, this would lead to an improvement of the symptoms. This is important, as in recent years there has been an issue about the safety of some of the symptom relief treatments, which were related to potential detrimental systemic impacts such as cardiovascular risks, increased risk of morbidity, and even mortality [
7,
8]. Moreover, the identification of individuals at risk of having a structural progressive disease is also of high significance for the development of DMOADs. Hence, a great part of the challenge in the development of such drugs is often the inclusion of patients in trials with advanced OA (severe cartilage loss), making it difficult to reduce or stop the degenerative process, therefore not suitable for DMOAD therapy, and impeding the power analysis of such trials.
Early identification of OA structural progressors currently depends on clinical judgment with the help of radiographic evaluation. However, it is well known that X-rays are not sensitive enough to detect early knee articular alteration [
9,
10]. Therefore, it is of great importance to develop automated and practical tools that will identify, at an early stage, OA patients for whom articular tissue alterations will progress rapidly.
A variety of fluid biomarkers has been evaluated for such discrimination. However, despite a significant body of research in this field, there is not yet a validated signature for early diagnosis or prognosis of the disease [
11]. Limitations with fluid biomarkers include, among others, the fact that there is often no direct correlation with joint structural changes, the poorly defined association with age-related changes, some being related to obesity and cannot distinguish between OA and obesity, and that the use of only one fluid biomarker cannot fully reflect the complex patterns underlying this disease.
At present, for optimal forecasting of joint structural alterations, increasing evidence points toward the use of articular structural (tissue) markers. At first, cartilage alteration was evaluated as a marker for the knee. However, when cartilage begins to show degradation as evaluated by clinical features and/or radiography it is already at a moderate stage of the disease. Recently, the change in knee bone was suggested as an accurate marker to identify early OA structural progressors; knee bone alteration was shown to precede cartilage losses and contribute to the development of the disease [
12,
13,
14,
15,
16,
17].
Over the years, many methodologies were introduced to evaluate such bony changes and included bone attrition, joint incongruity, periarticular area, shape, and curvature [
13,
14,
16,
18,
19,
20,
21,
22,
23,
24]. However, some used radiographic determination, which could lead to imprecision due to its dependence on the acquisition method and/or statistical modelling involving a component that is operator-dependent, which may introduce errors. Others used magnetic resonance imaging (MRI), and among the developed technologies, certain had shortcomings. For example, for the bone area, the assessment is subjective with inconsistent associations with knee structural progression. Machine learning (ML) techniques, coupled with MRI, have opened new possibilities for large-scale data integration to assess precise measurements of OA status in a multidimensional manner. Recently, by using these two methodologies (MRI and bone change), the measurement of the bone shape vector [
25] and the subchondral bone length (SBL) [
26] were reported. Yet, the bone shape vector was developed only for one bone, the femur, and included in its measurement the osteophytes (bony projections), which may induce inaccuracy in bone shape measurement changes, while the SBL uses 2D shape measurement. Another MRI fully automated methodology was developed and assessed the bone curvature (BC) [
20]. This BC assessment methodology in addition to being quantitative, is patient-based, and, while preserving the measured bone surface, did remove two bone alterations (peripheral osteophytes and bone marrow lesions [BML], including edema and cysts) that could interfere with the bone measurement [
20,
27]. By using this system, BC alteration was shown to precede cartilage volume loss (CVL), in addition to predicting the effectiveness of OA treatment [
20].
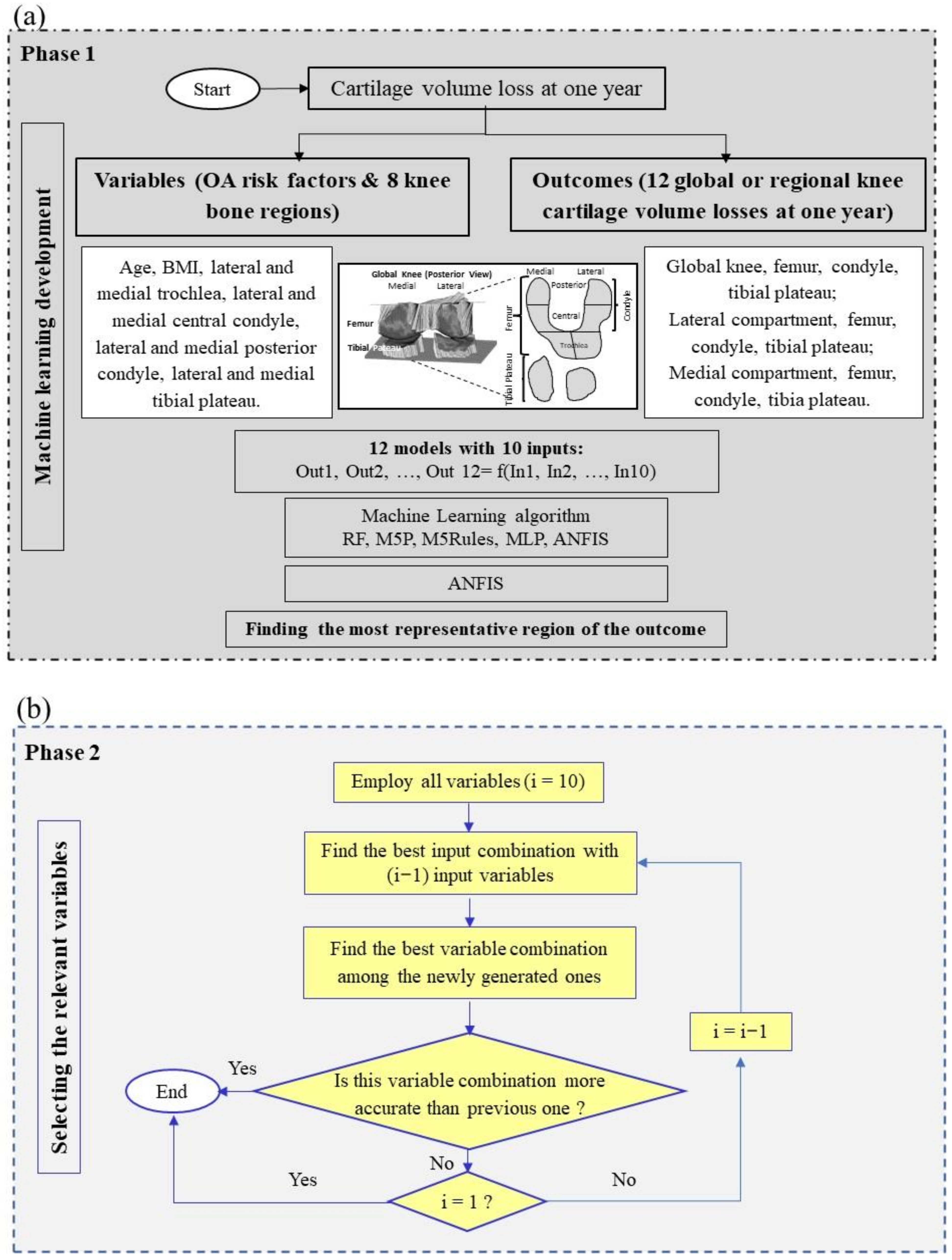
In the search for a model/tool that could offer an objective and quantitative assessment in the early forecasting of knee OA structural progressors, we hypothesized that knee BC features at baseline could predict, for an individual, CVL at one year. A primary concern was the understanding of which bone regions can play an effective role in such a prediction. Second, was the developed model able to predict CVL at one year in more than one knee subregion with the same baseline variables. Third, could the developed model be accurate for both genders, and fourth, could it be replicated and extended to another OA cohort for the prediction of outcomes? To answer these questions, we (i) applied feature selection by using ML algorithms on a fairly large sample to find the most important BC regions, (ii) developed advanced gender-based prediction models that provide high prediction performance on all the cartilage regions, and (iii) evaluated the reproducibility of the developed models by using an external cohort of OA patients from a clinical trial. Data revealed that the combination of five knee BC region values at baseline could predict CVL after one year on 12 knee regions with high accuracy and reproducibility.
4. Discussion
At present, we cannot discriminate, early during the OA process, patients for whom cartilage will degrade rapidly from those for whom the progression will be slow. Such discrimination would not only assist to modify the disease trajectory with a personalized clinical treatment plan but would represent a unique opportunity to intervene before cartilage degradation becomes too severe. Moreover, it would also enable patient screening for clinical trials for the development of DMOADs. Indeed, such drug trials have not yet achieved significant results, which appears to be mainly due to OA recruitment, in that patients have, for the most part, moderate to severe cartilage damage. Consequently, the effect of a DMOAD could not be observed with enough statistical power. This study was undertaken to fulfill these needs.
To achieve CVL forecasting, evidence points toward the use of joint tissue markers and, more recently, BC was suggested for the knee. We developed a gender-based model in which five BC regions at baseline (lateral tibial plateau, medial central condyle, lateral posterior condyle, and lateral and medial trochlea) enable the prediction of 12 global and regional CVL at one year with very good accuracy for both genders: OAI, R ≥ 0.79 (testing stage) and Naproxen (validation) R ≥ 0.78, except for the medial tibial plateau for women.
As we aimed to detect CVL for multiple (12 global/regional) outcomes, a two-phase ML-based methodology was performed. In Phase 1, after comparing the accuracy and benefits of five ML algorithms, ANFIS was found to be the most reliable for prediction. The selection of ANFIS was not surprising as it has the advantage over other ML methodologies of capturing the nonlinear structure of a problem, an adaptive capability and a rapid learning capacity as it combines a neural network with fuzzy logic, in addition to a significant potential for predicting systems with high uncertainty and in a dynamic nature.
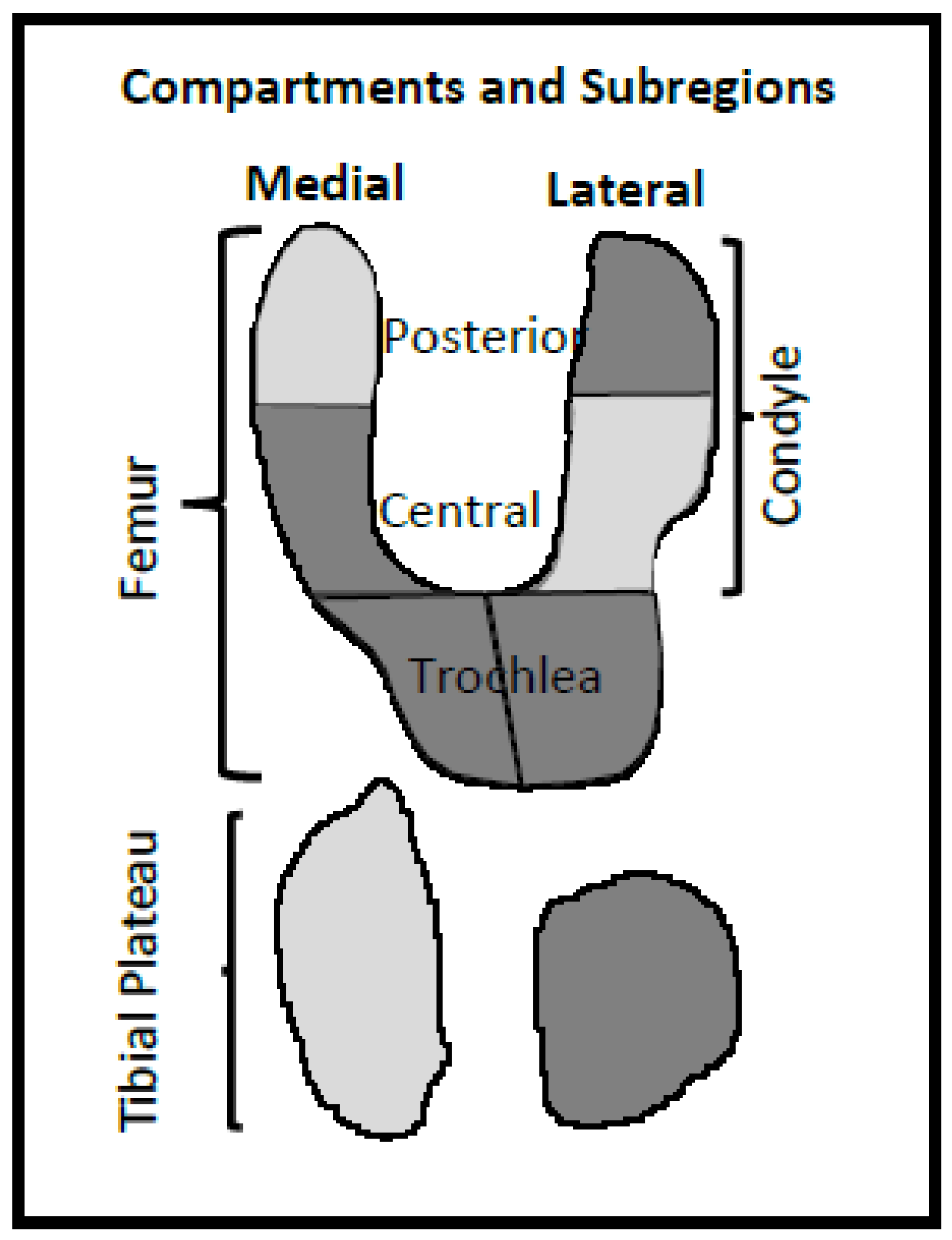
Next, data showed that the most representative region of CVL (outcome) was the medial condyle. Such a finding could reflect that the medial tibiofemoral compartment of the knee, more specifically the medial condyle, displays a higher rate of cartilage change with greater sensitivity than the other regions [
42,
43,
44,
45], as well as being highly related to OA progression and total knee replacement [
46,
47,
48].
In Phase 2, the relevant variables were selected. Reducing the number of variables for ML development and application saves resources. Moreover, having fewer misleading features not only improves the accuracy of a ML model but also removes multicollinearity, thus reducing the possibility of overfitting. To this end, we employed a systematic controllability variable reduction (removing the lowest cost features among all input variables) to identify the relevant ones.
In this study, of the five selected BC variables, the lateral tibial plateau and medial central condyle demonstrated the highest impact in prediction forecasting. This finding contrasts with a previous one in which two other BC regions, namely, medial posterior condyle and lateral central condyle, were found to be the best regions to predict CVL at two years [
20]. In the current study, the weight of these two regions appeared to be somewhat important as they were eliminated only when eight variables were examined (M30). Removing these two variables resulted in a decrease of 10.5% in R, and an increase of about 27% in RMSE and MAE, compared to model M1 (all ten variables). It should also be taken into consideration that the period examined between the two studies, as well as the methodology varied, which could be responsible for the change in the selected variables.
Here, the selection of the lateral tibial plateau and medial central condyle was not unexpected as they both showed a high level of bony remodeling during OA. Indeed, the tibial plateau demonstrated expansion and increased depression during the OA process [
49,
50,
51,
52], and bony changes in the lateral tibial plateau were associated with the presence of radiographic OA [
53]. Moreover, uneven lateral support of the tibial plateau has been reported to be a key factor that leads to the non-uniform settlement of the knee and a shift of the mechanical axis to the medial compartment, more specifically, on the medial central condyle [
54]. The stresses engendered could be responsible for the reported flattening of the medial central condyle bone during OA [
14,
23]. Bone remodeling in the medial central condyle could also be due to the presence of a high level of BMLs in the OA knee in this region [
55]. Although BML was removed from our BC segmentation [
27], such subchondral bone changes are suggested to increase the levels of contact stresses, thus, bone remodeling [
56].
Even though all the 12 studied global and regional cartilage regions could be predicted with high accuracy with the OAI participants, validation using OA patients from a clinical trial (Naproxen) showed that generalization was attained in all cartilage regions, except in the medial tibial plateau for women. The lower accuracy in this region in women could reflect the fact that (i) compared to the OAI, participants from the Naproxen cohort displayed more disease severity, as ascertained by the clinical parameters, (ii) during the OA process, there was a high level of cartilage thinning/loss as well as inter-subject variability in this region [
33,
43,
57], in addition to (iii) the cartilage volume of women being smaller than in men [
58].
The finding that BMI was not included in the model with five variables was rather surprising as a link between BMI and knee bone remodeling has been previously reported [
59]. However, this is still under debate as other studies have not shown such an association [
53]. Of note, the weight of this variable was, to some extent, important as it was included when six variables were investigated.
Some challenges and limitations of this study should be acknowledged. First, ANFIS was selected as the best ML algorithm for model development. Because of the use of ten input variables, a limitation of this method could have been the high computational expense due to the high number of iterations needed to achieve high accuracy. However, care was given to the selection of the appropriate number and shape of membership function in the ANFIS model as they impact the accuracy of the final results and computational complexity of the ANFIS-based model, and although a challenging task, we were able to define the appropriate membership function, i.e., Gaussian (
Table S1) for this study.
Second, in practice, for a given ML problem, multiple equivalent solutions in variable selections can exist [
60]. A shortcoming of some variable selection methods is that they injudiciously identify only a single solution, minimizing a loss function like mean squared error, classification error, etc. Yet, a single solution is not proper when variable selections can be considered both for building a predictive model with high accuracy and for knowledge discovery. In this study, we opted to employ the systematic controllability variable reduction as, instead of giving only one solution, it deals with achieving the highest accuracy by removing the lowest cost variables. In addition, with this step-by-step variable reduction, not only the sensitivity but also the synergy between two variables for the estimation of the outcome could be evaluated.
Third, we could have used the cartilage volume as the outcome. We favored this tissue volume loss as to whether baseline cartilage volume predicts future cartilage loss is questionable.
Fourth, another challenge was the CVL period to be analyzed. We chose one year to ensure both a reliable assessment of cartilage change sensitivity and high patient retention for its use in clinical practice. However, a longer period was not evaluated, and the five input variables found could differ. The next step will be to explore, for a longer observation period, whether the developed ML model using the same BC variables could also predict with high accuracy CVL for all the studied regions. Although the purpose of this study was to evaluate BC as prognosis for CVL over time, assessing a longer period of cartilage loss (e.g., two to four years) could educate us on the collinearity between these two structures. In addition, validating the ML model for longer periods converts it into an application that can be of broader use in clinical practice.
This study has several strengths. The use of MRI to assess BC and cartilage volume at baseline permits for automation of these two knee structures (thus, avoiding human error) and quantitative segmentation/measurement in the same knee [
27,
32]. In addition, the 3D nature of the MRI data over radiographs for knee tissue measurement avoids difficulties in interpreting findings that may be related to positioning during image acquisition and to projection effects. Moreover, when studying BC in OA, care should be taken not to confuse the osteophytes and BMLs with true differences in the bone. This putative problem was circumvented by the exclusion of these two tissues in the measurement methodology used [
27]. Finally, special emphasis needs to be given to the validation (reproducibility) of data using an external clinical trial cohort which, in addition to mimicking patients seen in clinical routine, adds to the robustness and generalization of the developed ML model in that the accuracy persisted.