Transcriptional Regulation of Abscission Zones
Abstract
1. Introduction
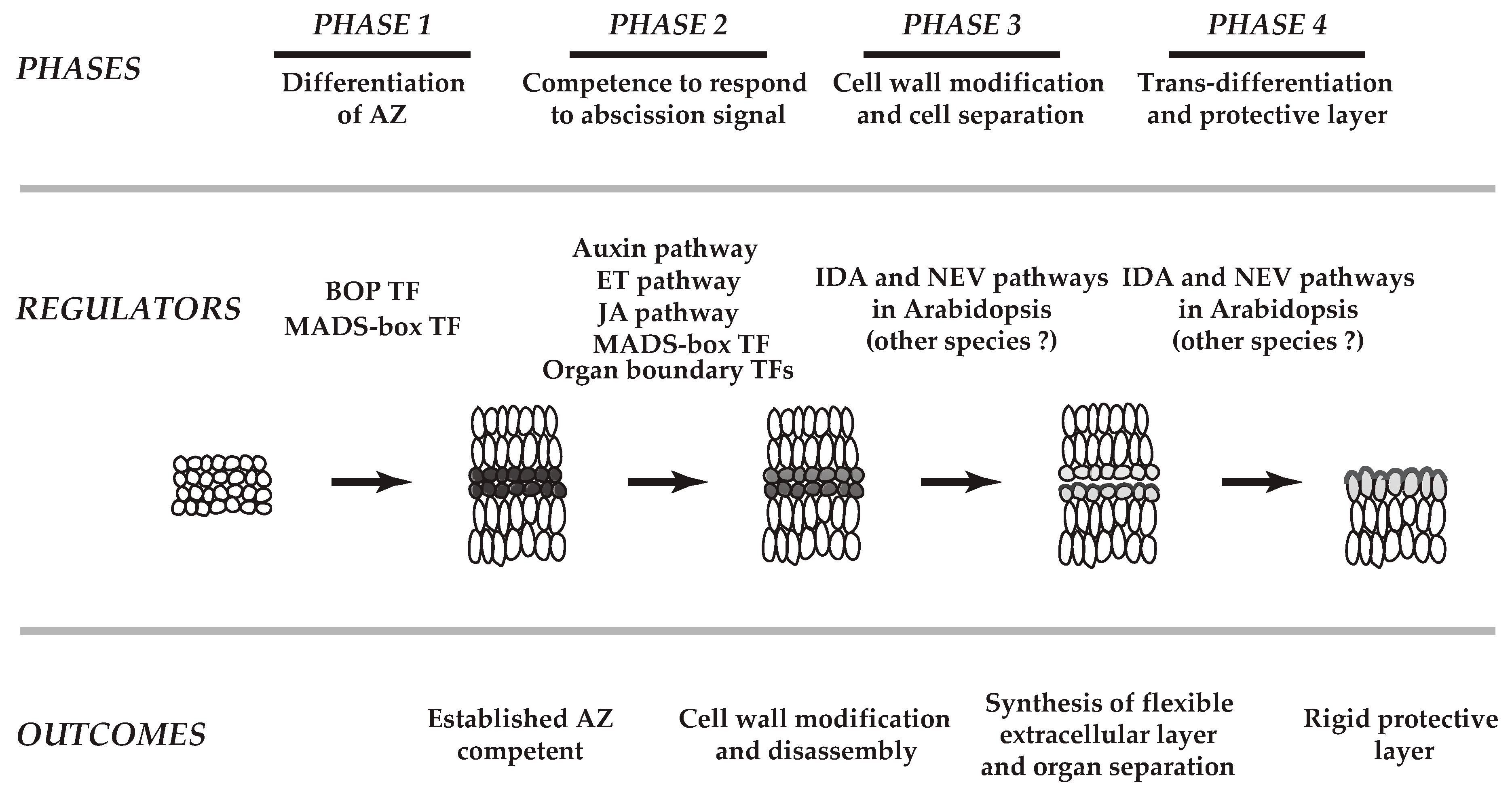
2. Cellular Changes in Various Abscission Systems
3. Comparative Analysis of Transcriptomes in Diverse Abscission Systems
3.1. Variability in Cell Wall Disassembly
3.2. Reconstruction of a Flexible Extracellular Matrix during Separation
4. Transcriptional Regulatory Networks in the Soybean AZ
4.1. Transcription Factors (TFs) in the AZ
4.2. Regulatory Modules in Abscission
4.2.1. Regulatory Module for Cell Proliferation and Differentiation
4.2.2. Regulatory Module for Integration of Other Hormone Signaling
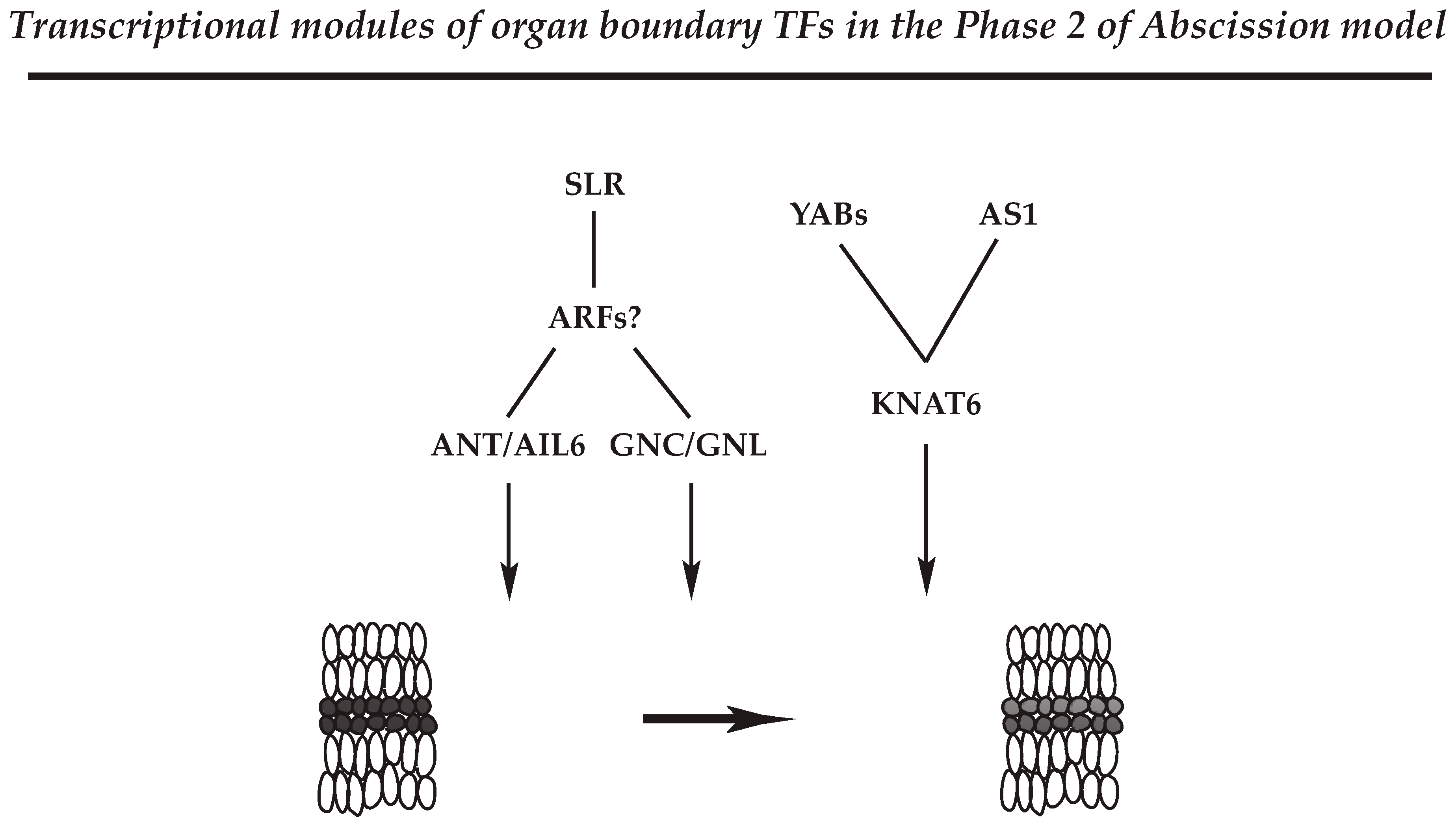
4.2.3. Regulatory Module for Organ Polarity and Separation Boundary Determination
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Addicott, F.T. Abscission; California University Press: Berkely, CA, USA, 1982. [Google Scholar]
- Patterson, S.E. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 2001, 126, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.M.; Gonzalez-Carranza, Z.H.; Azam-Ali, S.; Tang, S.; Shahid, A.A.; Roberts, J.A. The manipulation of auxin in the abscission zone cells of Arabidopsis flowers reveals that indoleacetic acid signaling is a prerequisite for organ shedding. Plant Physiol. 2013, 162, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.E.; Whitelaw, C.A. Signals in abscission. New Phytol. 2001, 151, 323–339. [Google Scholar] [CrossRef]
- Bleecker, A.B.; Patterson, S.E. Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 1997, 9, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Meir, S.; Philosoph-Hadas, S.; Sundaresan, S.; Selvaraj, K.S.; Burd, S.; Ophir, R.; Kochanek, B.; Reid, M.S.; Jiang, C.Z.; Lers, A. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 2010, 154, 1929–1956. [Google Scholar] [CrossRef]
- Kim, J. Four shades of detachment: Regulation of floral organ abscission. Plant Signal. Behav. 2014, 9, e976154. [Google Scholar] [CrossRef]
- Inman, T. The Causes that Determine the Fall of Leaves; Proc. Literary and Phiolosophical Soc.: Liverpool, UK, 1848; pp. 89–92. [Google Scholar]
- Abeles, F.B.; Morgan, P.W.; Salveit, M.E. Ethylene in Plant Biology, 2nd ed.; Academic Press: New York, NY, USA, 1992. [Google Scholar]
- Roberts, J.A.; Elliott, K.A.; Gonzalez-Carranza, Z.H. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 2002, 53, 131–158. [Google Scholar] [CrossRef]
- Sexton, R.; Roberts, J.A. Cell biology of abscission. Annu. Rev. Plant Physiol. 1982, 33, 133–162. [Google Scholar] [CrossRef]
- Tucker, M.L.; Kim, J. Abscission research: What we know and what we still need to study. Stewart Postharvest Rev. 2015, 11, 1–7. [Google Scholar] [CrossRef]
- Iwahori, S.; Tominaga, S.; Higuchi, S. Retardation of abscission of citrus leaf and fruitlet explants by brassinolide. Plant Growth Regul. 1990, 9, 119–125. [Google Scholar] [CrossRef]
- Niederhuth, C.E.; Cho, S.K.; Seitz, K.; Walker, J.C. Letting Go is Never Easy: Abscission and Receptor-Like Protein Kinases. J. Integr. Plant Biol. 2013, 55, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- McKim, S.M.; Stenvik, G.E.; Butenko, M.A.; Kristiansen, W.; Cho, S.K.; Hepworth, S.R.; Aalen, R.B.; Haughn, G.W. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 2008, 135, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Jinn, T.L.; Stone, J.M.; Walker, J.C. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000, 14, 108–117. [Google Scholar] [PubMed]
- Cho, S.K.; Larue, C.T.; Chevalier, D.; Wang, H.; Jinn, T.L.; Zhang, S.; Walker, J.C. Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 15629–15634. [Google Scholar] [CrossRef] [PubMed]
- Butenko, M.A.; Patterson, S.E.; Grini, P.E.; Stenvik, G.E.; Amundsen, S.S.; Mandal, A.; Aalen, R.B. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 2003, 15, 2296–2307. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Leslie, M.E.; Darnielle, L.; Lewis, M.W.; Taylor, S.M.; Luo, R.; Geldner, N.; Chory, J.; Randazzo, P.A.; Yanofsky, M.F.; et al. Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 2009, 136, 1909–1918. [Google Scholar] [CrossRef]
- Burr, C.A.; Leslie, M.E.; Orlowski, S.K.; Chen, I.; Wright, C.E.; Daniels, M.J.; Liljegren, S.J. CAST AWAY, a Membrane-Associated Receptor-Like Kinase, Inhibits Organ Abscission in Arabidopsis. Plant Physiol. 2011, 156, 1837–1850. [Google Scholar] [CrossRef]
- Leslie, M.E.; Lewis, M.W.; Youn, J.Y.; Daniels, M.J.; Liljegren, S.J. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development 2010, 137, 467–476. [Google Scholar] [CrossRef]
- Lewis, M.W.; Leslie, M.E.; Fulcher, E.H.; Darnielle, L.; Healy, P.N.; Youn, J.Y.; Liljegren, S.J. The SERK1 receptor-like kinase regulates organ separation in Arabidopsis flowers. Plant J. Cell Mol. Biol. 2010, 62, 817–828. [Google Scholar] [CrossRef]
- Lewis, M.W.; Leslie, M.E.; Liljegren, S.J. Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 2006, 9, 59–65. [Google Scholar] [CrossRef]
- Fang, S.C.; Fernandez, D.E. Effect of Regulated Overexpression of the MADS Domain Factor AGL15 on Flower Senescence and Fruit Maturation. Plant Physiol. 2002, 130, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, D.E.; Heck, G.R.; Perry, S.E.; Patterson, S.E.; Bleecker, A.B.; Fang, S.C. The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 2000, 12, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.J.; Lehti-Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.K.; Hsu, W.H.; Lee, P.F.; Thiruvengadam, M.; Chen, H.I.; Yang, C.H. The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J. 2011, 68, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Patterson, S.E.; Bleecker, A.B. Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol. 2004, 134, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. Arabidopsis Dehiscence Zone Polygalacturonase1 (Adpg1), Adpg2, and Quartet2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Dotson, B.; Rey, C.; Lindsey, J.; Bleecker, A.B.; Binder, B.M.; Patterson, S.E. New clothes for the jasmonic acid receptor COI1: Delayed abscission, meristem arrest and apical dominance. PLoS ONE 2013, 8, e60505. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Patterson, S.E.; Binder, B.M. Reducing jasmonic acid levels causes ein2 mutants to become ethylene responsive. FEBS Lett. 2013, 587, 226–230. [Google Scholar] [CrossRef]
- Meng, X.; Zhou, J.; Tang, J.; Li, B.; de Oliveira, M.V.; Chai, J.; He, P.; Shan, L. Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 2016, 14, 1330–1338. [Google Scholar] [CrossRef]
- Mao, L.; Begum, D.; Chuang, H.W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. Jointless is a MADS-box gene controlling tomato flower abscission zone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J. Organ abscission: Exit strategies require signals and moving traffic. Curr. Opin. Plant Biol. 2012, 15, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Estornell, L.H.; Agusti, J.; Merelo, P.; Talon, M.; Tadeo, F.R. Elucidating mechanisms underlying organ abscission. Plant Sci. 2013, 199–200, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Meir, S.; Philosoph-Hadas, S.; Riov, J.; Tucker, M.L.; Patterson, S.E.; Roberts, J.A. Re-evaluation of the ethylene-dependent and-independent pathways in the regulation of floral and organ abscission. J. Exp. Bot. 2019, 70, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, D.; Li, A.; Sun, X.; Zhang, R.; Wu, L.; Liang, Y.; Mao, L. Transcriptome analysis of tomato flower pedicel tissues reveals abscission zone-specific modulation of key meristem activity genes. PLoS ONE 2013, 8, e55238. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Lashbrook, C.C. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: Enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis Zinc Finger Protein2. Plant Physiol. 2008, 146, 1305–1321. [Google Scholar] [CrossRef]
- Nakano, T.; Fujisawa, M.; Shima, Y.; Ito, Y. Expression profiling of tomato pre-abscission pedicels provides insights into abscission zone properties including competence to respond to abscission signals. BMC Plant Biol. 2013, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, M.; Aït Barka, E.; Clément, C.; Vaillant-Gaveau, N.; Jacquard, C. Cross-talk between environmental stresses and plant metabolism during reproductive organ abscission. J. Exp. Bot. 2015, 66, 1707–1719. [Google Scholar] [CrossRef]
- Zhang, X.-L.; Qi, M.-L.; Xu, T.; Lu, X.-L.; Li, T.-L. Proteomics profiling of ethylene-induced tomato flower pedicel abscission. J. Proteom. 2015, 121, 67–87. [Google Scholar] [CrossRef]
- Kim, J.; Sundaresan, S.; Philosoph-Hadas, S.; Yang, R.; Meir, S.; Tucker, M.L. Examination of the abscission-associated transcriptomes for soybean, tomato, and Arabidopsis highlights the conserved biosynthesis of an extensible extracellular matrix and boundary layer. Front. Plant Sci. 2015, 6, 1109. [Google Scholar] [CrossRef]
- Kim, J.; Yang, J.; Yang, R.; Sicher, R.C.; Chang, C.; Tucker, M.L. Transcriptome analysis of soybean leaf abscission identifies transcriptional regulators of organ polarity and cell fate. Front. Plant Sci. 2016, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Domingos, S.; Fino, J.; Cardoso, V.; Sánchez, C.; Ramalho, J.C.; Larcher, R.; Paulo, O.S.; Oliveira, C.M.; Goulao, L.F. Shared and divergent pathways for flower abscission are triggered by gibberellic acid and carbon starvation in seedless Vitis vinifera L. BMC Plant Biol. 2016, 16, 38. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.K.; Reid, P.D. Role of polygalacturonase in bean leaf abscission. Plant Physiol. 1979, 63, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.; Goren, R.; Riov, J. The role of cellulase and polygalacturonase in abscission of young and mature Shamouti orange fruits. Physiol. Plant. 1975, 34, 1–7. [Google Scholar] [CrossRef]
- Huberman, M.; Goren, R. Exo- and endo-cellular cellulase and polygalacturonase in abscission zones of developing orange fruits. Physiol. Plant. 1979, 45, 189–196. [Google Scholar] [CrossRef]
- Morre, D.J. Cell wall dissolution and enzyme secretion during leaf abscission. Plant Physiol. 1968, 43, 1545–1559. [Google Scholar]
- Riov, J. A polygalacturonase from citrus leaf explants: Role in abscission. Plant Physiol. 1974, 53, 312–316. [Google Scholar] [CrossRef]
- Moline, H.E.; LaMotte, C.E.; Gochnauer, C.; McNamer, A. Further comparative studies of pectin esterase in relation to leaf and flower abscission. Plant Physiol. 1972, 50, 655–659. [Google Scholar] [CrossRef][Green Version]
- Sexton, R.; Redshaw, A. The role of cell expansion in the abscission of Impatiens sultani leaves. Ann. Bot. 1981, 48, 745–756. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Wall structure and wall loosening. A look backwards and forwards. Plant Physiol. 2001, 125, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Bornman, C.; Addicott, F.; Spurr, A. Auxin and gibberellin effects on cell growth and starch during abscission in cotton. Plant Physiol. 1966, 41, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Keegstra, K. Plant cell walls. Plant Physiol. 2010, 154, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.S. Ethylene and abscission. Hortscience 1985, 20, 45–50. [Google Scholar]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Yoon, T.H.; Lee, J.; Jeon, S.Y.; Lee, J.H.; Lee, M.K.; Chen, H.; Yun, J.; Oh, S.Y.; Wen, X. A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 2018, 173, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Lanahan, M.B.; Yen, H.C.; Giovannoni, J.J.; Klee, H.J. The never ripe mutation blocks ethylene perception in tomato. Plant Cell 1994, 6, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.L.; Yang, R. IDA-like gene expression in soybean and tomato leaf abscission and requirement for a diffusible stelar abscission signal. AoB Plants 2012, 2012, pls035. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Carranza, Z.H.; Elliott, K.A.; Roberts, J.A. Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3719–3730. [Google Scholar] [CrossRef]
- Kim, J.; Shiu, S.-H.; Thoma, S.; Li, W.-H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006, 7, R87. [Google Scholar] [CrossRef]
- Kim, J.; Patterson, S.E. Expression divergence and functional redundancy of polygalacturonases in floral organ abscission. Plant Signal. Behavior 2006, 1, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Valliyodan, B.; Prince, S.; Wan, J.; Nguyen, H. Characterization of the XTH Gene Family: New Insight to the Roles in Soybean Flooding Tolerance. Int. J. Mol. Sci. 2018, 19, 2705. [Google Scholar]
- Niederhuth, C.E.; Patharkar, O.R.; Walker, J.C. Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-seq. BMC Genom. 2013, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Mauch, F. The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 2005, 8, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Voigt, C.A. Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci. 2014, 5, 168. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.L.; Sexton, R.; del Campillo, E.; Lewis, L.N. Bean abscission cellulase: Characterization of a cDNA clone and regulation of gene expression by ethylene and auxin. Plant Physiol. 1988, 88, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Kalaitzis, P.; Solomos, T.; Tucker, M.L. Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 1997, 113, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Lashbrook, C.C.; Cai, S. Cell wall remodeling in Arabidopsis stamen abscission zones: Temporal aspects of control inferred from transcriptional profiling. Plant Signal. Behavior 2008, 3, 733–736. [Google Scholar] [CrossRef]
- Del Campillo, E.; Lewis, L.N. Identification and kinetics of accumulation of proteins induced by ethylene in bean abscission zones. Plant Physiol. 1992, 98, 955–961. [Google Scholar] [CrossRef]
- Herbers, K.; Monke, G.; Badur, R.; Sonnewald, U. A simplified procedure for the subtractive cDNA cloning of photoassimilate-responding genes: Isolation of cDNAs encoding a new class of pathogenesis-related proteins. Plant Mol. Biol. 1995, 29, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Thoma, S.; Hecht, U.; Kippers, A.; Botella, J.; De Vries, S.; Somerville, C. Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 1994, 105, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.X.; Malitsky, S.; De Oliveira, S.; Branigan, C.; Franke, R.B.; Schreiber, L.; Aharoni, A. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 2011, 7, e1001388. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K. The formation and function of plant cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 2012, 3, 77. [Google Scholar] [CrossRef]
- Hepworth, S.R.; Pautot, V.A. Beyond the divide: Boundaries for patterning and stem cell regulation in plants. Front. Plant Sci. 2015, 6, 1052. [Google Scholar] [CrossRef]
- Lelli, K.M.; Slattery, M.; Mann, R.S. Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 2012, 46, 43–68. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar]
- Eshed, Y.; Baum, S.F.; Bowman, J.L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 1999, 99, 199–209. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Perea, J.V.; Bowman, J.L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef]
- Eshed, Y.; Izhaki, A.; Baum, S.F.; Floyd, S.K.; Bowman, J.L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef] [PubMed]
- Sawa, S.; Watanabe, K.; Goto, K.; Kanaya, E.; Morita, E.H.; Okada, K. Filamentous Flower, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Stahle, M.I.; Kuehlich, J.; Staron, L.; von Arnim, A.G.; Golz, J.F. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 2009, 21, 3105–3118. [Google Scholar] [CrossRef]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowman, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef]
- Gómez-Mena, C.; Sablowski, R. Arabidopsis Thaliana Homeobox Gene1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 2008, 20, 2059–2072. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Mutwil, M. Towards revealing the functions of all genes in plants. Trends Plant Sci. 2013. [Google Scholar] [CrossRef]
- Sorrells, T.R.; Johnson, A.D. Making sense of transcription networks. Cell 2015, 161, 714–723. [Google Scholar] [CrossRef]
- Lee, T.I.; Rinaldi, N.J.; Robert, F.; Odom, D.T.; Bar-Joseph, Z.; Gerber, G.K.; Hannett, N.M.; Harbison, C.T.; Thompson, C.M.; Simon, I. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 2002, 298, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Shen-Orr, S.; Itzkovitz, S.; Kashtan, N.; Chklovskii, D.; Alon, U. Network motifs: Simple building blocks of complex networks. Science 2002, 298, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Shen-Orr, S.S.; Milo, R.; Mangan, S.; Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002, 31, 64. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Araya, C.L.; Brdlik, C.; Cayting, P.; Cheng, C.; Cheng, Y.; Gardner, K.; Hillier, L.W.; Janette, J.; Jiang, L. Comparative analysis of regulatory information and circuits across distant species. Nature 2014, 512, 453. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.; Gao, G. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef]
- Nole-Wilson, S.; Tranby, T.L.; Krizek, B.A. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol. Biol. 2005, 57, 613–628. [Google Scholar] [CrossRef]
- Prasad, K.; Grigg, S.P.; Barkoulas, M.; Yadav, R.K.; Sanchez-Perez, G.F.; Pinon, V.; Blilou, I.; Hofhuis, H.; Dhonukshe, P.; Galinha, C. Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr. Biol. 2011, 21, 1123–1128. [Google Scholar] [CrossRef]
- Krizek, B. Aintegumenta and Aintegumenta-Like6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 2009, 150, 1916–1929. [Google Scholar] [CrossRef]
- Louie, D.S.; Addicott, F.T. Applied auxin gradients and abscission in explants. Plant Physiol. 1970, 45, 654–657. [Google Scholar] [CrossRef]
- Bi, Y.M.; Zhang, Y.; Signorelli, T.; Zhao, R.; Zhu, T.; Rothstein, S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005, 44, 680–692. [Google Scholar] [CrossRef]
- Naito, T.; Kiba, T.; Koizumi, N.; Yamashino, T.; Mizuno, T. Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1557–1560. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.; Behringer, C.; Zourelidou, M.; Schwechheimer, C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2013, 110, 13192–13197. [Google Scholar] [CrossRef] [PubMed]
- Behringer, C.; Schwechheimer, C. B-GATA transcription factors–insights into their structure, regulation, and role in plant development. Front. Plant Sci. 2015, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. Auxin Response Factor1 and Auxin Response Factor2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Mitina, I.; Quach, H.L.; Theologis, A. AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant J. 2005, 43, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Kerstetter, R.A.; Bollman, K.; Taylor, R.A.; Bomblies, K.; Poethig, R.S. KANADI regulates organ polarity in Arabidopsis. Nature 2001, 411, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- McAbee, J.M.; Hill, T.A.; Skinner, D.J.; Izhaki, A.; Hauser, B.A.; Meister, R.J.; Venugopala Reddy, G.; Meyerowitz, E.M.; Bowman, J.L.; Gasser, C.S. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 2006, 46, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Bonaccorso, O.; Lee, J.E.; Puah, L.; Scutt, C.P.; Golz, J.F. FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 2012, 12, 176. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, M.K.; Bowman, J.L.; Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 2002, 14, 2761–2770. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.E.; Barley, R.; Curtis, M.; Arroyo, J.M.; Dunham, M.; Hudson, A.; Martienssen, R.A. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 2000, 408, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Hazen, S.P.; Borevitz, J.O.; Harmon, F.G.; Pruneda-Paz, J.L.; Schultz, T.F.; Yanovsky, M.J.; Liljegren, S.J.; Ecker, J.R.; Kay, S.A. Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 2005, 138, 990–997. [Google Scholar] [CrossRef][Green Version]
- Gubert, C.M.; Christy, M.E.; Ward, D.L.; Groner, W.D.; Liljegren, S.J. ASYMMETRIC LEAVES1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 2014, 14, 195. [Google Scholar] [CrossRef] [PubMed]
- Ori, N.; Eshed, Y.; Chuck, G.; Bowman, J.L.; Hake, S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 2000, 127, 5523–5532. [Google Scholar] [PubMed]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef]
- Xu, B.; Li, Z.; Zhu, Y.; Wang, H.; Ma, H.; Dong, A.; Huang, H. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 2008, 146, 566–575. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Chun, J.-P.; Tucker, M.L. Transcriptional Regulation of Abscission Zones. Plants 2019, 8, 154. https://doi.org/10.3390/plants8060154
Kim J, Chun J-P, Tucker ML. Transcriptional Regulation of Abscission Zones. Plants. 2019; 8(6):154. https://doi.org/10.3390/plants8060154
Chicago/Turabian StyleKim, Joonyup, Jong-Pil Chun, and Mark L. Tucker. 2019. "Transcriptional Regulation of Abscission Zones" Plants 8, no. 6: 154. https://doi.org/10.3390/plants8060154
APA StyleKim, J., Chun, J.-P., & Tucker, M. L. (2019). Transcriptional Regulation of Abscission Zones. Plants, 8(6), 154. https://doi.org/10.3390/plants8060154




