Never the Two Shall Mix: Robust Indel Markers to Ensure the Fidelity of Two Pivotal and Closely-Related Accessions of Brachypodium distachyon
Abstract
1. Introduction
2. Results
2.1. Identification of Indels or Indel Groups and Marker Development From Brachypodium distachyon Putative Orthologs of Arabidopsis thaliana Genes
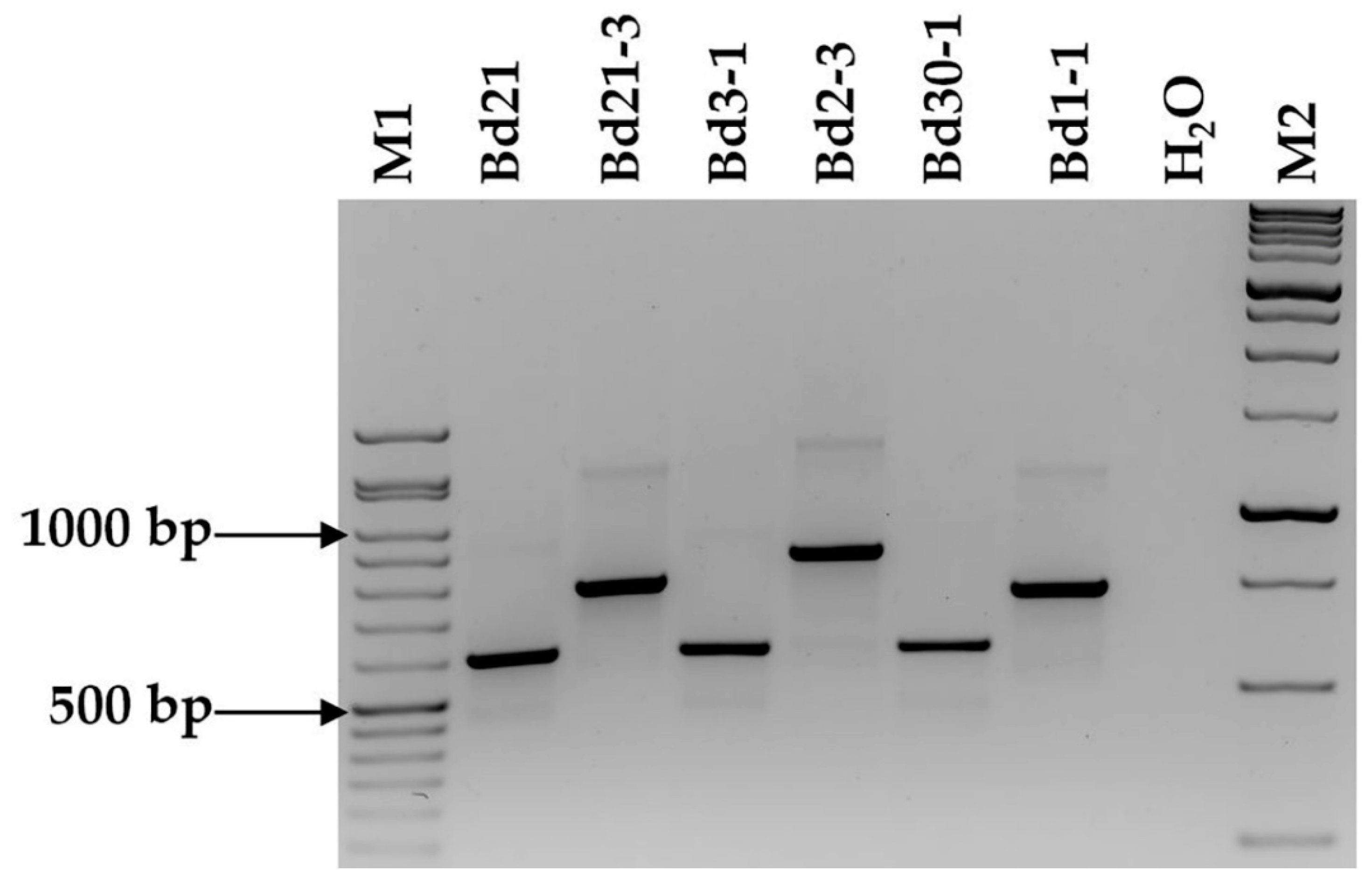
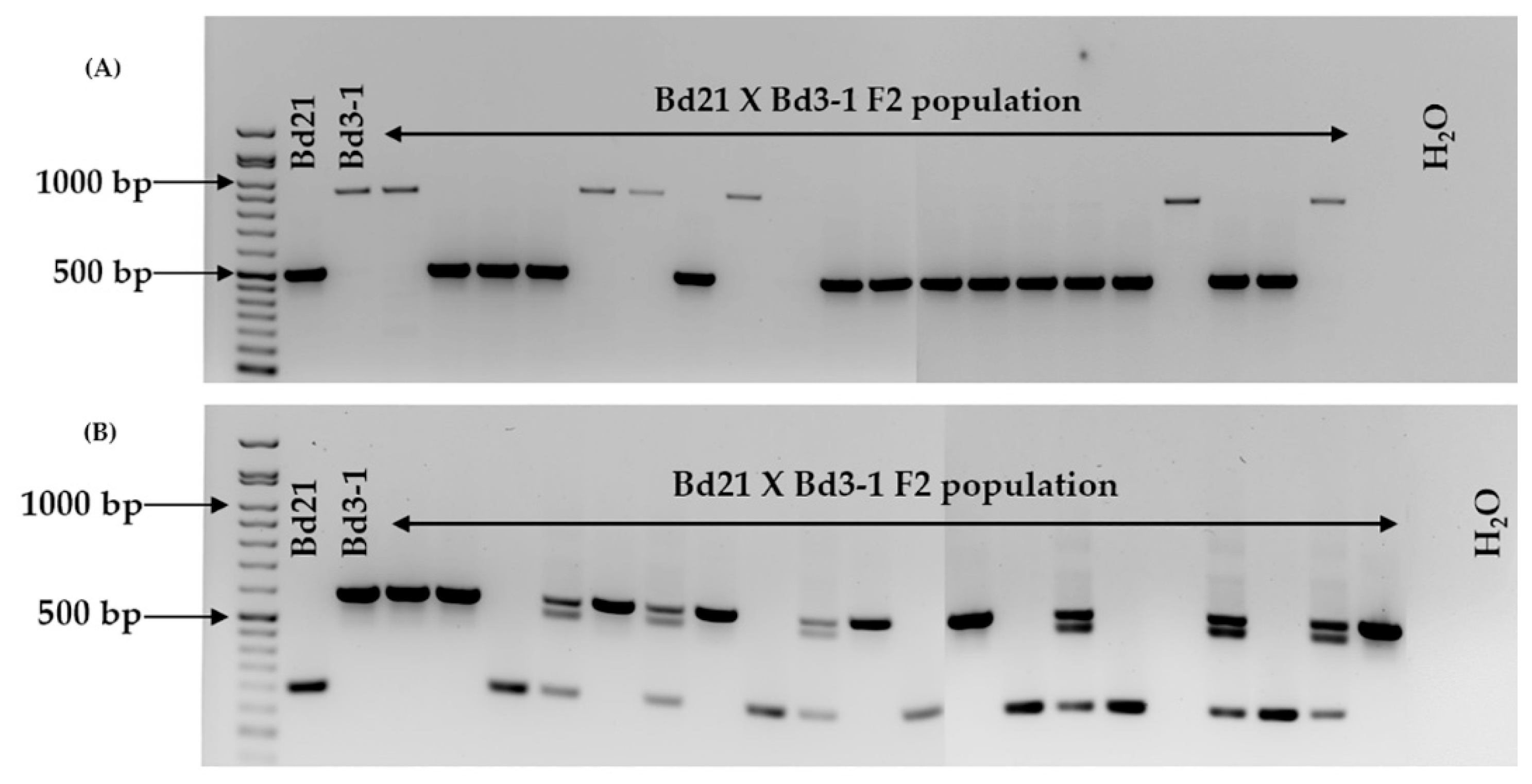
2.2. Marker Assessment on Six Brachypodium distachyon Accessions and F2 Population
3. Discussion
4. Materials and Methods
4.1. Identification of Indels or Indel Groups and Primer Design
4.2. Marker Validation on Brachypodium Distachyon Accessions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hinsley, A.; de Boer, H.J.; Fay, M.F.; Gale, S.W.; Gardiner, L.M.; Gunasekara, R.S.; Kumar, P.; Masters, S.; Metusala, D.; Roberts, D.L.; et al. A review of the trade in orchids and its implications for conservation. Bot. J. Linn. Soc. 2018, 186, 435–455. [Google Scholar] [CrossRef]
- Munafo, J.P.; Gianfagna, T.J. Chemistry and biological activity of steroidal glycosides from the Lilium genus. Nat. Prod. Rep. 2015, 32, 454–477. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.K.; Wu, X.W.; Lee, A.K.; Roh, M.S. Growth and flowering physiology, and developing new technologies to increase the flower numbers in the Genus Lilium. Hortic. Environ. Biotechnol. 2013, 54, 373–387. [Google Scholar] [CrossRef]
- Paul, J.Y.; Harding, R.; Tushemereirwe, W.; Dale, J. Banana21: From gene discovery to deregulated golden bananas. Front. Plant Sci. 2018, 9, 558. [Google Scholar] [CrossRef] [PubMed]
- Masani, M.Y.A.; Izawati, A.M.D.; Rasid, O.A.; Parveez, G.K.A. Biotechnology of oil palm: Current status of oil palm genetic transformation. Biocatal. Agric. Biotechnol. 2018, 15, 335–347. [Google Scholar] [CrossRef]
- Linder, H.P.; Lehmann, C.E.R.; Archibald, S.; Osborne, C.P.; Richardson, D.M. Global grass (Poaceae) success underpinned by traits facilitating colonization, persistence and habitat transformation. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1125–1144. [Google Scholar] [CrossRef]
- Allwright, M.R.; Taylors, G. Molecular breeding for improved second generation bioenergy crops. Trends Plant Sci. 2016, 21, 43–54. [Google Scholar] [CrossRef]
- Yang, J.D.; Udvardi, M. Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. J. Exp. Bot. 2018, 69, 855–865. [Google Scholar] [CrossRef]
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef]
- Bolser, D.M.; Kerhornou, A.; Walts, B.; Kersey, P. Triticeae resources in ensembl plants. Plant Cell Physiol. 2015, 56, e3. [Google Scholar] [CrossRef]
- Mochida, K.; Shinozaki, K. Unlocking triticeae genomics to sustainably feed the future. Plant Cell Physiol. 2013, 54, 1931–1950. [Google Scholar] [CrossRef] [PubMed]
- Gurel, F.; Ozturk, Z.N.; Ucarli, C.; Rosellini, D. Barley genes as tools to confer abiotic stress tolerance in crops. Front. Plant Sci. 2016, 7, 1137. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Garcia-Pereira, M.J.; Gracia, M.P.; Igartua, E.; Casas, A.M.; Contreras-Moreira, B. Large differences in gene expression responses to drought and heat stress between elite barley cultivar Scarlett and a Spanish landrace. Front. Plant Sci. 2017, 8, 647. [Google Scholar] [CrossRef] [PubMed]
- Brew-Appiah, R.A.T.; York, Z.B.; Krishnan, V.; Roalson, E.H.; Sanguinet, K.A. Genome-wide identification and analysis of the ALTERNATIVE OXIDASE gene family in diploid and hexaploid wheat. PLoS ONE 2018, 13, e0201439. [Google Scholar] [CrossRef] [PubMed]
- Brew-Appiah, R.A.T.; Sanguinet, K.A. Considerations of AOX functionality revealed by critical motifs and unique domains. Int. J. Mol. Sci. 2018, 19, 2972. [Google Scholar] [CrossRef] [PubMed]
- Alptekin, B.; Langridge, P.; Budak, H. Abiotic stress miRNomes in the Triticeae. Funct. Integr. Genom. 2017, 17, 145–170. [Google Scholar] [CrossRef] [PubMed]
- Galvez, S.; Merida-Garcia, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramirez-Gonzalez, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019, 19, 295–309. [Google Scholar] [CrossRef]
- Tack, J.; Barkley, A.; Nalley, L.L. Effect of warming temperatures on US wheat yields. Proc. Natl. Acad. Sci. USA 2015, 112, 6931–6936. [Google Scholar] [CrossRef]
- Leonelli, S.; Ankeny, R.A. What makes a model organism? Endeavour 2013, 37, 209–212. [Google Scholar] [CrossRef]
- Koornneef, M.; Meinke, D. The development of Arabidopsis as a model plant. Plant J. 2010, 61, 909–921. [Google Scholar] [CrossRef]
- Provart, N.J.; Alonso, J.; Assmann, S.M.; Bergmann, D.; Brady, S.M.; Brkljacic, J.; Browse, J.; Chapple, C.; Colot, V.; Cutler, S.; et al. 50 years of Arabidopsis research: Highlights and future directions. New Phytol. 2016, 209, 921–944. [Google Scholar] [CrossRef] [PubMed]
- Brutnell, T.P.; Bennetzen, J.L.; Vogel, J.P. Brachypodium distachyon and Setaria viridis: Model genetic systems for the grasses. Annu. Rev. Plant Biol. 2015, 66, 465–485. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Coupland, G. The molecular basis of diversity in the photoperiodic flowering responses of arabidopsis and rice. Plant Physiol. 2004, 135, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J.L.; Freeling, M. Grasses as a single genetic system: Genome composition, collinearity and compatibility. Trends Genet. 1993, 9, 259–261. [Google Scholar] [CrossRef]
- Freeling, M. Grasses as a single genetic system. Reassessment 2001. Plant Physiol. 2001, 125, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, K.; Kyozuka, J. Rice as a model for comparative genomics of plants. Annu. Rev. Plant Biol. 2002, 53, 399–419. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.L.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Hu, S.N.; Wang, J.; Wong, G.K.S.; Li, S.G.; Liu, B.; Deng, Y.J.; Dai, L.; Zhou, Y.; Zhang, X.Q.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Han, B.; Zhang, Q.F. Rice genome research: Current status and future perspectives. Plant Genome 2008, 1, 71–76. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.S.; Pasternak, S.; Liang, C.Z.; Zhang, J.W.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Beier, S.; Himmelbach, A.; Colmsee, C.; Zhang, X.Q.; Barrero, R.A.; Zhang, Q.S.; Li, L.; Bayer, M.; Bolser, D.; Taudien, S.; et al. Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Sci. Data 2017, 4, 170044. [Google Scholar] [CrossRef] [PubMed]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. BARLEX—The barley draft genome explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef] [PubMed]
- Szurman-Zubrzycka, M.E.; Zbieszczyk, J.; Marzec, M.; Jelonek, J.; Chmielewska, B.; Kurowska, M.M.; Krok, M.; Daszkowska-Golec, A.; Guzy-Wrobelska, J.; Gruszka, D.; et al. HorTILLUS—A rich and renewable source of induced mutations for forward/reverse genetics and pre-breeding programs in barley (Hordeum vulgare L.). Front. Plant Sci. 2018, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.R.; Bowman, J.L.; Meyerowitz, E.M. Field guide to plant model systems. Cell 2016, 167, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Girin, T.; David, L.C.; Chardin, C.; Sibout, R.; Krapp, A.; Ferrario-Mery, S.; Daniel-Vedele, F. Brachypodium: A promising hub between model species and cereals. J. Exp. Bot. 2014, 65, 5683–5696. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.A. Brachypodium distachyon as a genetic model system. Annu. Rev. Genet. 2015, 49, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.G.; Irigoyen, S.; Catalan, P.; Mandadi, K.K. Brachypodium: A monocot grass model genus for plant biology. Plant Cell 2018, 30, 1673–1694. [Google Scholar] [CrossRef]
- Draper, J.; Mur, L.A.J.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P.M. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef]
- Mur, L.A.J.; Allainguillaume, J.; Catalan, P.; Hasterok, R.; Jenkins, G.; Lesniewska, K.; Thomas, I.; Vogel, J. Exploiting the brachypodium tool box in cereal and grass research. New Phytol. 2011, 191, 334–347. [Google Scholar] [CrossRef]
- Gill, U.S.; Lee, S.; Jia, Y.L.; Mysore, K.S. Exploring natural variation for rice sheath blight resistance in Brachypodium distachyon. Plant Signal. Behav. 2019, 14, 1–6. [Google Scholar] [CrossRef]
- Bragg, J.N.; Wu, J.J.; Gordon, S.P.; Guttman, M.E.; Thilmony, R.; Lazo, G.R.; Gu, Y.Q.; Vogel, J.P. Generation and characterization of the western regional research center brachypodium T-DNA insertional mutant collection. PLoS ONE 2012, 7, e41916. [Google Scholar] [CrossRef] [PubMed]
- Hsia, M.M.; O’Malley, R.; Cartwright, A.; Nieu, R.; Gordon, S.P.; Kelly, S.; Williams, T.G.; Wood, D.F.; Zhao, Y.J.; Bragg, J.; et al. Sequencing and functional validation of the JGI Brachypodium distachyon T-DNA collection. Plant J. 2017, 91, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Hill, T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 2008, 27, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Tuna, M.; Budak, H.; Huo, N.X.; Gu, Y.Q.; Steinwand, M.A. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biol. 2009, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Huo, N.X.; Garvin, D.F.; You, F.M.; McMahon, S.; Luo, M.C.; Gu, Y.Q.; Lazo, G.R.; Vogel, J.P. Comparison of a high-density genetic linkage map to genome features in the model grass Brachypodium distachyon. Theor. Appl. Genet. 2011, 123, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Lee, M.Y.; Huo, N.X.; Bragg, J.; Yan, L.J.; Yuan, C.; Li, C.; Holditch, S.J.; Xie, J.Z.; Luo, M.C.; et al. Fine mapping of the Bsr1 barley stripe mosaic virus resistance gene in the model grass Brachypodium distachyon. PLoS ONE 2012, 7, e38333. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Ozdemir, B.S.; Budak, F.; Vogel, J.P.; Tuna, M.; Budak, H. Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 2009, 52, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, P.; Rodriguez-Quijano, M.; Vazquez, J.F.; Carrillo, J.M.; Benavente, E. Validation of microsatellite markers for cytotype discrimination in the model grass Brachypodium distachyon. Genome 2012, 55, 523–527. [Google Scholar] [CrossRef]
- Lopez-Alvarez, D.; Lopez-Herranz, M.L.; Betekhtin, A.; Catalan, P. A DNA barcoding method to discriminate between the model plant Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). PLoS ONE 2012, 7, e51058. [Google Scholar] [CrossRef]
- Contreras, R.; Figueiras, A.M.; Gallego, F.J.; Benavente, E.; Manzaneda, A.J.; Benito, C. Neutral molecular markers support common origin of aluminium tolerance in three congeneric grass species growing in acidic soils. AoB Plants 2017, 9, plx060. [Google Scholar] [CrossRef]
- Gordon, S.P.; Priest, H.; Marais, D.L.D.; Schackwitz, W.; Figueroa, M.; Martin, J.; Bragg, J.N.; Tyler, L.; Lee, C.R.; Bryant, D.; et al. Genome diversity in Brachypodium distachyon: Deep sequencing of highly diverse inbred lines. Plant J. 2014, 79, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Toal, T.W.; Burkart-Waco, D.; Howell, T.; Ron, M.; Kuppu, S.; Britt, A.; Chetelat, R.; Brady, S.M. Indel group in genomes (IGG) molecular genetic markers. Plant Physiol. 2016, 172, 38–61. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.P.; Ream, T.S.; Minevich, G.; Hobert, O.; Amasino, R.M. PHYTOCHROME C is an essential light receptor for photoperiodic flowering in the temperate grass, Brachypodium distachyon. Genetics 2014, 198, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Neji, M.; Guena, F.; Gordon, S.P.; Taamalli, W.; Vogel, J.P.; Ibrahim, Y.; Smaoui, A.E.; Abdelly, C.; Gandour, M. Insertion/deletion markers for assessing the genetic variation and the spatial genetic structure of Tunisian Brachypodium hybridum populations. Recent Res. Sci. Technol. 2016, 8, 14–23. [Google Scholar] [CrossRef]
- Cass, C.L.; Lavell, A.A.; Santoro, N.; Foster, C.E.; Karlen, S.D.; Smith, R.A.; Ralph, J.; Garvin, D.F.; Sedbrook, J.C. Cell wall composition and biomass recalcitrance differences within a genotypically diverse set of Brachypodium distachyon inbred lines. Front. Plant Sci. 2016, 7, 708. [Google Scholar] [CrossRef] [PubMed]
- Tyler, L.; Fangel, J.U.; Fagerstrom, A.D.; Steinwand, M.A.; Raab, T.K.; Willats, W.G.T.; Vogel, J.P. Selection and phenotypic characterization of a core collection of Brachypodium distachyon inbred lines. BMC Plant Biol. 2014, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Tran, Q.-G.; Han, Y.-J.; Hwang, O.-J.; Hoang, Q.T.N.; Kim, J.-I. Exploring responses to light in the monocot model plant, Brachypodium distachyon. Korean J. Plant Res. 2018, 31, 522–530. [Google Scholar] [CrossRef]
- Schwartz, C.J.; Doyle, M.R.; Manzaneda, A.J.; Rey, P.J.; Mitchell-Olds, T.; Amasino, R.M. Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon. Bioenergy Res. 2010, 3, 38–46. [Google Scholar] [CrossRef]
- Garvin, D.F.; Gu, Y.Q.; Hasterok, R.; Hazen, S.P.; Jenkins, G.; Mockler, T.C.; Mur, L.A.J.; Vogel, J.P. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci. 2008, 48, S69–S84. [Google Scholar] [CrossRef]
- Ream, T.S.; Woods, D.P.; Schwartz, C.J.; Sanabria, C.P.; Mahoy, J.A.; Walters, E.M.; Kaeppler, H.F.; Amasino, R.M. Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 2014, 164, 694–709. [Google Scholar] [CrossRef]
- Dell’Acqua, M.; Zuccolo, A.; Tuna, M.; Gianfranceschi, L.; Pe, M.E. Targeting environmental adaptation in the monocot model Brachypodium distachyon: A multi-faceted approach. BMC Genom. 2014, 15, 801. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, Y.; Stritt, C.; Walser, J.C.; Gordon, S.P.; Vogel, J.P.; Roulin, A.C. Genome-wide scans of selection highlight the impact of biotic and abiotic constraints in natural populations of the model grass Brachypodium distachyon. Plant J. 2018, 96, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef] [PubMed]

| Marker Name | Chromosome Location | Expected Size | Amplicon Size Difference | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | |
|---|---|---|---|---|---|---|
| Bd21 | Bd21-3 | |||||
| BdindelWSU_1 | 1 | 419 | 1065 | 646 | AAGCGCATCCTTCACGCTTTC | GTGCAGAGCTGCAGAGTAGAGATTAG |
| BdindelWSU_2 | 1 | 1522 | 1096 | 426 | GGAACCTGTGTTCGGACTCG | TGCAGGAACGGAGAAATGGA |
| BdindelWSU_3 | 1 | 992 | 287 | 705 | CCGTATCAGAGCACGCCAGAAT | CTTGGGAGGAACTAATACACCAGATCG |
| BdindelWSU_4 | 1 | 402 | 159 | 243 | CGCCCTTCTTTTGAAACCCAAA | TGAGCACTTGACAGTTTTTGGTGA |
| BdindelWSU_5 | 2 | 288 | 826 | 538 | TCTCCTTGAAATTGCATGTTGGA | CCTGACAGTGGGCCATAGCA |
| BdindelWSU_6 | 2 | 490 | 285 | 205 | CCCCCATGGTTGAATGGTTC | CACATCACGTCGCACCAGAA |
| BdindelWSU_7 | 2 | 1103 | 866 | 237 | CTGCAGAGCTTCAGCCTTGG | TTGCCAGCGGAACGATGTTA |
| BdindelWSU_8 | 3 | 213 | 940 | 727 | AAGGCCTCCGCCAAGCTATC | GACCTCGGGACGCGACAG |
| BdindelWSU_9 | 3 | 509 | 683 | 174 | CAAGCAAAAAGCCTTGGAGCA | CACACCATACAAAAACGTCCAGGAT |
| BdindelWSU_10 | 3 | 1160 | 206 | 954 | TTCAGCGCTTTCGGTACTTTG | GAGATGCGATGGGGAAGCAT |
| BdindelWSU_11 | 3 | 246 | 410 | 164 | TGAGAGCACCGGACAGGACA | CTGGCTCGGATTGGATTGATTTTC |
| BdindelWSU_12 | 3 | 600 | 782 | 182 | GGTTAGCGAGGCGTCAATGG | CCCATGGACCCATGCATATTG |
| BdindelWSU_13 | 3 | 547 | 1388 | 841 | CAGATCTGGGGGCTCGTTGA | CCAATCAAATTGCGGAGCTG |
| BdindelWSU_14 | 3 | 317 | 1908 | 1591 | GGGACCCCATGTTACCCTGA | CGATGCCAATCCAAGAGACG |
| BdindelWSU_15 | 4 | 500 | 906 | 406 | AAACAGCAAGGTTGGCCTTATCC | GGGGTCAGCAATTGAATGTGT |
| BdindelWSU_16 | 5 | 1413 | 827 | 586 | ATGCCGGCTTAGCGTTAGTTG | CTGACCCTTGTACACACTCGTAGCA |
| BdindelWSU_17 | 5 | 900 | 323 | 577 | CGGCCCTCCAGACATTGTTC | TGCATCCCCTATCTTGGACGA |
| BdindelWSU_18 | 5 | 987 | 619 | 368 | CGTATCAATCACGATCGGTCCA | ATAGGCAGCAGCGTGGTGGT |
| BdindelWSU_19 | 5 | 1170 | 522 | 648 | CGATACAAATTCATTTGACGGGTGT | CGCGGGAAACGTCTTTACAA |
| BdindelWSU_20 | 5 | 775 | 1165 | 390 | CAGAACTACGGCAGTTAATTTGGATTG | CGGGATTATTTGCGCGAGA |
| BdindelWSU_21 | 5 | 862 | 632 | 230 | CAACCCATCCGCAGACACAC | TGGATGAAGCCCATTGCAGA |
| BdindelWSU_22 | 5 | 300 | 569 | 269 | GGTCAGTGCGATGTGGTGTTTC | GTGGGAATCCATCTGCTTTGTTCTT |
| Marker | Bd21 | Bd21-3 | Bd3-1 | Bd2-3 | Bd30-1 | Bd1-1 |
|---|---|---|---|---|---|---|
| BdindelWSU_1 | 419 | 1065 | 419 | 419 | 419 | 1065 |
| BdindelWSU_2 | 1522 | 1096 | 1096 | 1522 | 1096 | 1522 |
| BdindelWSU_3 | 1000 * | 300 * | 1000 | 300 | 300 | 1000 |
| BdindelWSU_4 | 402 | 159 | 159 | 159 | 159 | 159 |
| BdindelWSU_5 | 288 | 826 | 826 | 826 | 826 | 288 |
| BdindelWSU_6 | 490 | 285 | 285 | 285 | 285 | 490 |
| BdindelWSU_7 | 1103 | 866 | 1103 | 1103 | 1103 | 866 |
| BdindelWSU_8 | 213 | 940 | 213 | 940 | 940 | 213 |
| BdindelWSU_9 | 509 | 683 | 509 | 509 | 683 | 683 |
| BdindelWSU_10 | 1160 | 206 | 1160 | 1160 | 1160 | 1160 |
| BdindelWSU_11 | 246 | 410 | 246 | 246 | 246 | 410 |
| BdindelWSU_12 | 600 | 782 | 600 | 900 | 600 | 750 |
| BdindelWSU_13 | 547 | 1388 | 547 | 1388 | 547 | 1388 |
| BdindelWSU_14 | 317 | 1908 | 1908 | 1908 | 317 | 1908 |
| BdindelWSU_15 | 500 | 906 | 906 | 500 | 500 | 906 |
| BdindelWSU_16 | 1413 | 827 | 1413 | 827 | 827 | 827 |
| BdindelWSU_17 | 900 | 323 | 900 | 323 | 323 | 323 |
| BdindelWSU_18 | 987 | 619 | 619 | 619 | 619 | 987 |
| BdindelWSU_19 | 1170 | 522 | 522 | 522 | 522 | 1170 |
| BdindelWSU_20 | 775 | 1165 | 775 | 775 | 1165 | 775 |
| BdindelWSU_21 | 862 | 632 | 862 | 862 | 632 | 862 |
| BdindelWSU_22 | 300 | 569 | 569 | 569 | 300 | 569 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brew-Appiah, R.A.T.; Peracchi, L.M.; Sanguinet, K.A. Never the Two Shall Mix: Robust Indel Markers to Ensure the Fidelity of Two Pivotal and Closely-Related Accessions of Brachypodium distachyon. Plants 2019, 8, 153. https://doi.org/10.3390/plants8060153
Brew-Appiah RAT, Peracchi LM, Sanguinet KA. Never the Two Shall Mix: Robust Indel Markers to Ensure the Fidelity of Two Pivotal and Closely-Related Accessions of Brachypodium distachyon. Plants. 2019; 8(6):153. https://doi.org/10.3390/plants8060153
Chicago/Turabian StyleBrew-Appiah, Rhoda A. T., Luigi M. Peracchi, and Karen A. Sanguinet. 2019. "Never the Two Shall Mix: Robust Indel Markers to Ensure the Fidelity of Two Pivotal and Closely-Related Accessions of Brachypodium distachyon" Plants 8, no. 6: 153. https://doi.org/10.3390/plants8060153
APA StyleBrew-Appiah, R. A. T., Peracchi, L. M., & Sanguinet, K. A. (2019). Never the Two Shall Mix: Robust Indel Markers to Ensure the Fidelity of Two Pivotal and Closely-Related Accessions of Brachypodium distachyon. Plants, 8(6), 153. https://doi.org/10.3390/plants8060153





