Reactive Carbonyl Species: A Missing Link in ROS Signaling
Abstract
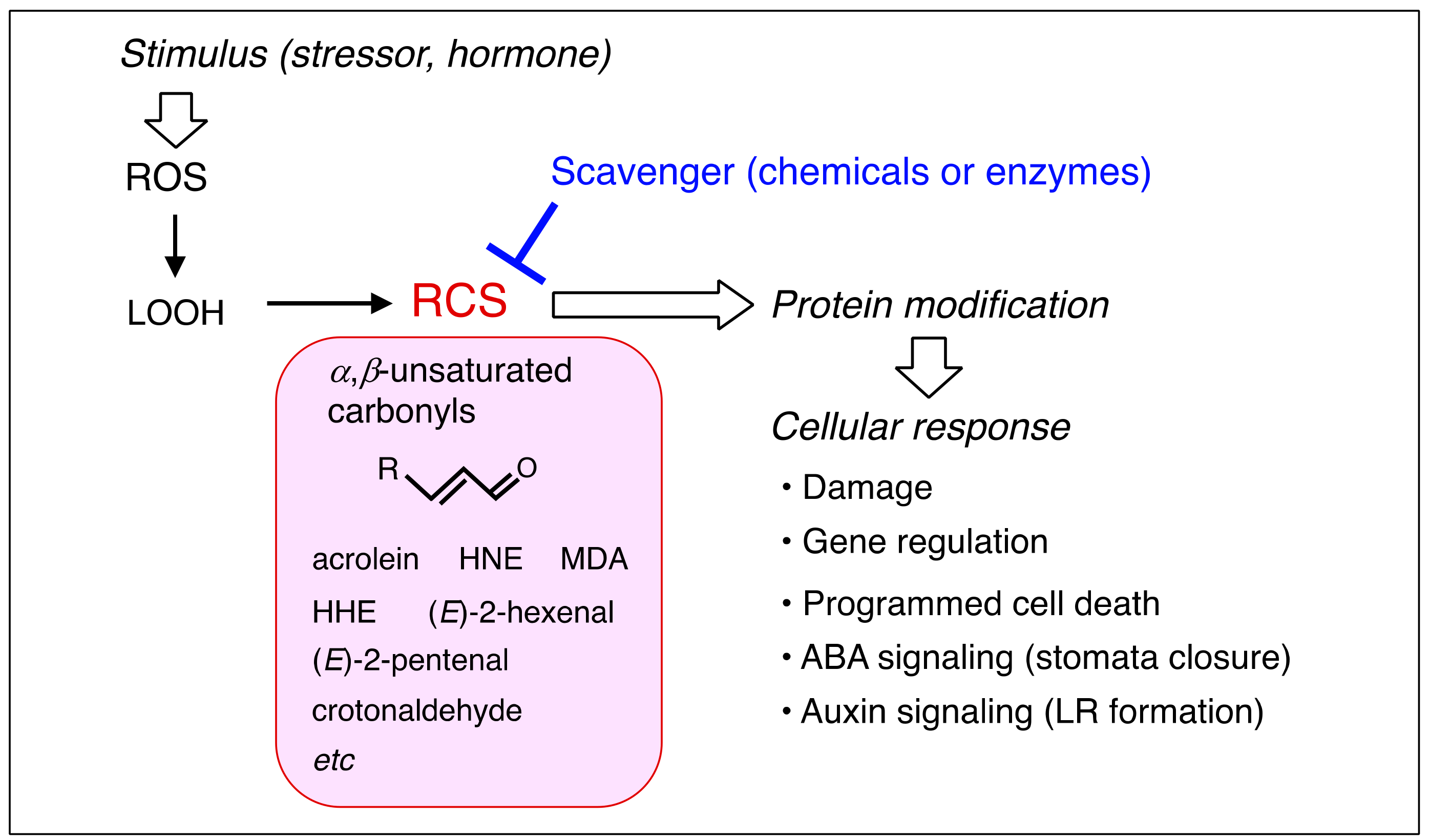
1. Introduction
2. Production of RCS in Plants
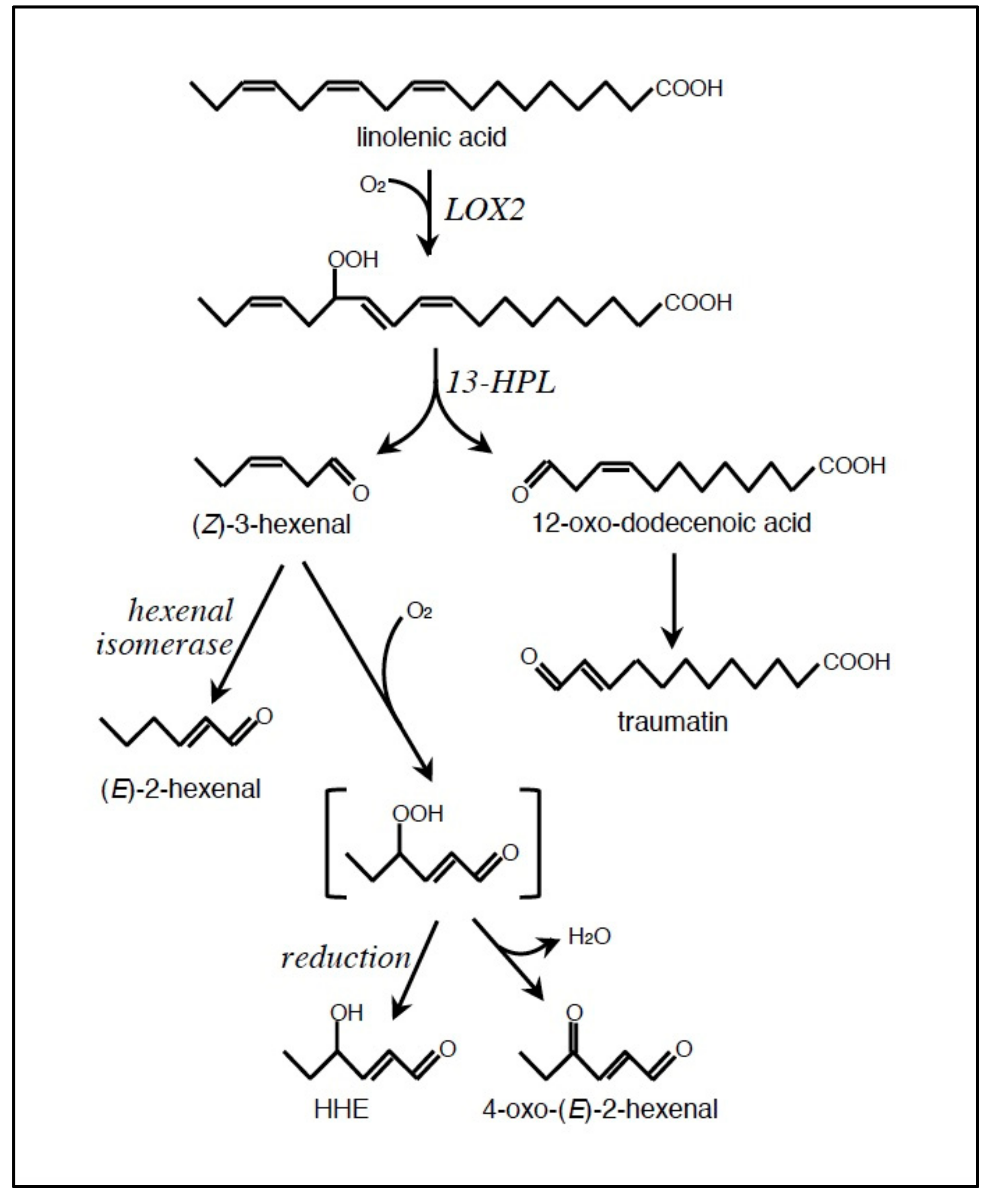
2.1. Formation of LOOH
2.2. Conversion of LOOH to RCS
2.3. Which Membrane(s) is the Source of RCS?
3. Response of Plants to Exogenously Added RCS
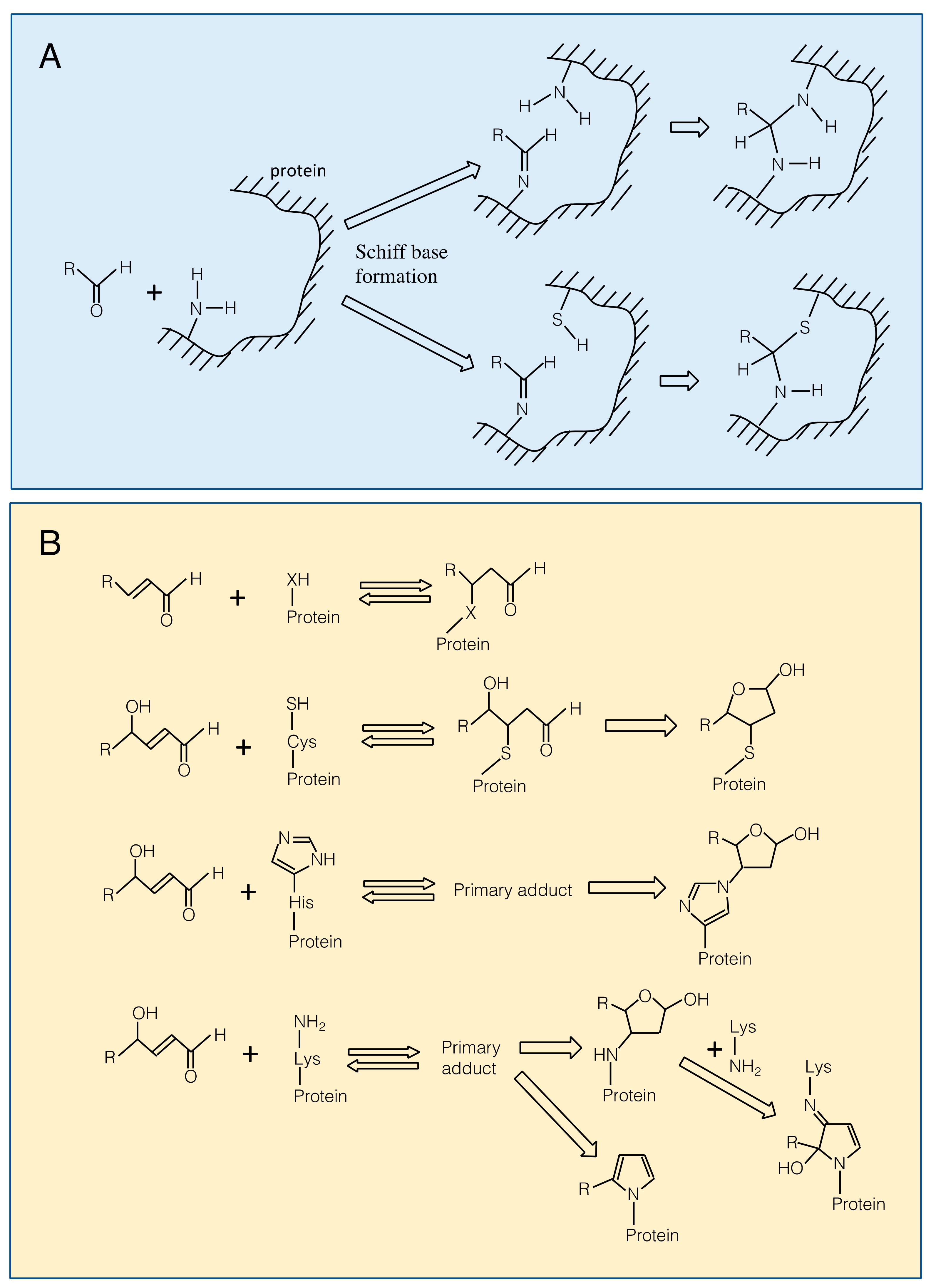
3.1. Reaction of RCS with Proteins
3.2. Cytotoxicity
3.3. Signaling Effects of Exogenously Added RCS
4. Evidence for Physiological Functions of Endogenously Produced RCS
4.1. Cytotoxicity of Endogenously Generated RCS
4.2. Signals Transmitted by Endogenously Generated RCS
4.2.1. Programmed Cell Death (PCD)
4.2.2. Senescence of Siliques
4.2.3. ABA Signaling for Stomata Closure
4.2.4. Auxin Signaling for Lateral Root Formation
4.2.5. Retrograde Signaling of β-Carotene Oxidation Products
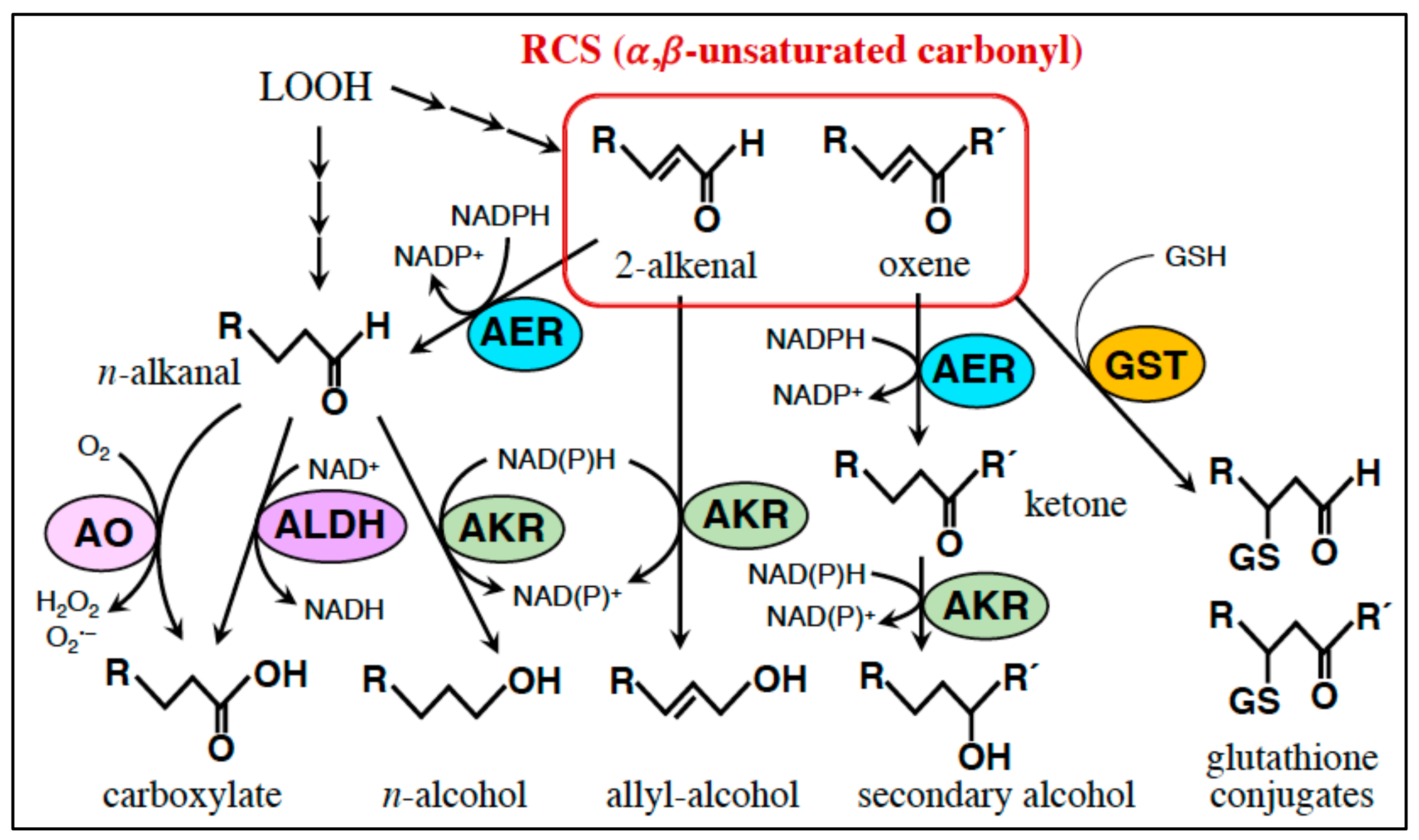
5. RCS-Scavenging System
5.1. Small Molecule Scavengers
5.2. Enzymes
5.2.1. AER and AOR: Reduction of the α,β-Unsaturated Bond.
5.2.2. Glutathione Transferase (GST): Conjugation of RCS with GSH
6. Perspectives
6.1. Contribution of Distinct Carbonyl Species
6.2. Regulation of Distinct Carbonyl Species
6.3. Action Mechanisms of RCS Signal
Funding
Conflicts of Interest
References
- Foyer, C.H.; Noctor, G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003, 119, 355–364. [Google Scholar] [CrossRef]
- Karpinski, S.; Reynolds, H.; Karpinska, B.; Wingsle, G.; Creissen, G.; Mullineaux, P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 1999, 284, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Orman-Ligeza, B.; Parizot, B.; de Rycke, R.; Fernandez, A.; Himschoot, E.; Breusegem, F.V.; Bennett, M.J.; Périlleux, C.; Beeckman, T.; Draye, X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 2016, 143, 3328–3339. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xue, L.; Xu, S.; Feng, H.; An, L. Hydrogen peroxide involvement in formation and development of adventitious roots in cucumber. Plant Growth Regul. 2007, 52, 173–180. [Google Scholar] [CrossRef]
- Bethke, P.C.; Jones, R.L. Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J. 2001, 25, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Pathogen-induced, NADPH oxidase–derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134. [Google Scholar] [CrossRef]
- Mano, J. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 2012, 59, 90–97. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Mène-Saffrané, L.; Davoine, C.; Stolz, S.; Majcherczyk, P.; Farmer, E.E. Genetic removal of tri-unsaturated fatty acids suppresses developmental and molecular phenotypes of an Arabidopsis tocopherol-deficient mutant. J. Biol. Chem. 2007, 282, 33749–33756. [Google Scholar] [CrossRef] [PubMed]
- Schauenstein, E.; Esterbauer, H.; Zollner, H. Aldehyde in Biological Systems: Their Natural Occurrence and Biological Activities; Pion: London, UK, 1977; pp. 589–590. [Google Scholar]
- Mano, J.; Miyatake, F.; Hiraoka, E.; Tamoi, M. Evaluation of the toxicity of stress-related aldehydes to photosynthesis in chloroplasts. Planta 2009, 230, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Mano, J.; Wang, S.; Tsuji, W.; Tanaka, K. The involvement of lipid peroxide-derived aldehydes in aluminum toxicity of tobacco roots. Plant Physiol. 2010, 152, 1406–1417. [Google Scholar] [CrossRef] [PubMed]
- Mano, J.; Tokushige, K.; Mizoguchi, K.; Fujii, H.; Khorobrykh, S. Accumulation of lipid peroxide-derived, toxic α,β,-unsaturated aldehyde (E)-2-pentenal, acrolein and (E)-2-hexenal in leaves under photoinhibitory illumination. Plant Biotechnol. 2010, 27, 193–197. [Google Scholar] [CrossRef]
- Farmer, E.E.; Mueller, M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.J.; Berger, S. Reactive electrophile oxylipins: Pattern recognition and signalling. Phytochemistry 2009, 70, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Liebler, D.C. Protein damage by reactive electrophiles: Targets and consequences. Chem. Res. Toxicol. 2008, 21, 117–128. [Google Scholar] [CrossRef]
- Hoque, T.S.; Hossain, M.A.; Mostofa, M.G.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Methylglyoxal: An emerging signaling molecule in plant abiotic stress responses and tolerance. Front. Plant Sci. 2016, 7, 1341. [Google Scholar] [CrossRef]
- Esterbauer, H.; Schauer, R.; Zollner, J.H. Chemistry and biochemistry of 4-hydroxynonenal, malondialdehyde and related aldehydes. Free Rad. Biol. Med. 1991, 11, 81–128. [Google Scholar] [CrossRef]
- West, J.D.; Marnett, L.J. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem. Res. Toxicol. 2006, 19, 173–194. [Google Scholar] [CrossRef]
- Wible, R.S.; Sutter, T.R. Soft cysteine signaling network: The functional significance of cysteine in protein function and the soft acids/bases thiol chemistry that facilitates cysteine modification. Chem. Res. Toxicol. 2017, 30, 729–762. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, F.J.; Cipollina, C.; Freeman, B.A. Formation and signaling actions of electrophilic lipids. Chem. Rev. 2011, 111, 5997–6021. [Google Scholar] [CrossRef] [PubMed]
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I.; Mano, J. Lipid peroxidation-derived reactive carbonyl species (RCS): Their interaction with ROS and cellular redox during environmental stresses. Environ. Exp. Bot. 2019, 165, 139–149. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Havaux, M. Sensing β-carotene oxidation in photosystem II to master plant stress tolerance. N. Phytol. 2019, in press. [Google Scholar] [CrossRef]
- Grosch, W. Reactions of hydroperoxides—Products of low molecular weight. In Autoxidation of Unsaturated Lipids; Chan, H.W.-S., Ed.; Academic Press: London, UK, 1987; pp. 95–139. [Google Scholar]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 5th ed.; Oxford Univ. Press: London, UK, 2015; pp. 257–265. [Google Scholar]
- Blée, E. Phytooxylipins and plant defense reactions. Prog. Lipid Res. 1998, 37, 33–72. [Google Scholar] [CrossRef]
- Schaller, F. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J. Exp. Bot. 2001, 52, 354. [Google Scholar] [CrossRef]
- Matsui, K. Green leaf volatiles: Hydroperoxide lyase pathway of oxylipin metabolism. Curr. Opin. Plant Biol. 2006, 9, 1–7. [Google Scholar] [CrossRef]
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; Van Breusegem, F.; Mueller, M.J. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008, 148, 960–968. [Google Scholar] [CrossRef]
- Boeglin, W.E.; Itoh, A.; Zheng, Y.; Coffa, G.; Howe, G.A.; Brash, A.R. Investigation of substrate binding and product stereochemistry issues in two linoleate 9-lipoxygenases. Lipids 2008, 43, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Bannenberg, G.; Martínez, M.; Hamberg, M.; Castresana, C. Diversity of the Enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 2009, 44, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, K.; Matsui, K.; Ozawa, R.; Takabayashi, J. Volatile C6-aldehydes and allo-ocimene activate defense genes and induce resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 2005, 47, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Shiojiri, K.; Kishimoto, K.; Ozawa, R.; Kugimiya, S.; Urashimo, S.; Arimura, G.; Horiuchi, J.; Nishioka, T.; Matsui, K.; Takabayashi, J. Changing green leaf volatile biosynthesis in plants: An approach for improving plant resistance against both herbivores and pathogens. Proc. Natl. Acad. Sci. USA 2006, 103, 16672–16676. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Matsui, K.; Iijima, K.; Akakabe, Y.; Muramoto, S.; Ozawa, R.; Uefune, M.; Sasaki, R.; Alamgir, K.Md.; Akitake, S.; et al. Intake and transformation to a glycoside of (Z)-3-hexenol from infested neighbors reveals a mode of plant odor reception and defense. Proc. Natl. Acad. Sci. USA 2014, 111, 7144–7149. [Google Scholar] [CrossRef] [PubMed]
- Hammarberg, T.; Provost, P.; Persson, B.; Rådmark, O. The N-terminal domain of 5-lipoxygenase binds calcium and mediates calcium stimulation of enzyme activity. J. Biol. Chem. 2000, 275, 38787–38793. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Das, S.; Funk, C.D.; Murray, D.; Cho, W. Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J. Biol. Chem. 2002, 277, 13167–13174. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, S.; Matsui, K. Green leaf volatile-burst in Arabidopsis is governed by galactolipid oxygenation by a lipoxygenase that is under control of calcium ion. Biochem. Biophys. Res. Commun. 2018, 505, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Montillet, J.-L.; Chamnongpol, S.; Rustérucci, C.; Dat, J.; van de Cotte, J.; Agnel, J.-P.; Battesti, C.; Inzé, D.; Van Breusegem, F.; Triantaphylidès, C. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 2005, 138, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Montillet, J.-L.; Cacas, J.-L.; Lionel Garnier, L.; Montané, M.H.; Douki, T.; Bessoule, J.-J.; Polkowska-Kowalczyk, L.; Maciejewska, U.; Agnel, J.-P.; Vial, A.; et al. The upstream oxylipin profile of Arabidopsis thaliana: A toolto scan for oxidative stresses. Plant J. 2004, 40, 439–451. [Google Scholar] [CrossRef]
- Schneider, C.; Tallman, K.A.; Porter, N.A.; Brash, A.R. Two distinct pathways of formation of 4-hydroxynonenal. J. Biol. Chem. 2001, 276, 20381–20386. [Google Scholar] [CrossRef]
- Lee, S.H.; Oe, T.; Blair, I.A. Vitamin C-induced decomposition of lipid hydroperoxides to endogenous genotoxins. Science 2001, 292, 2083–2086. [Google Scholar] [CrossRef]
- Mochizuki, S.; Sugimoto, K.; Koeduka, T.; Matsui, K. Arabidopsis lipoxygenase 2 is essential for formation of green leaf volatiles and five-carbon volatiles. FEBS Lett. 2016, 590, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Kunishima, M.; Yamauchi, Y.; Mizutani, M.; Kuse, M.; Takikawa, H.; Sugimoto, Y. Identification of (Z)-3:(E)-2-hexenal isomerases essential to the production of the leaf aldehyde in plants. J. Biol. Chem. 2016, 291, 14023–14033. [Google Scholar] [CrossRef] [PubMed]
- Spyropoulou, E.A.; Dekker, H.L.; Steemers, L.; van Maarseveen, J.H.; de Koster, C.G.; Haring, M.A.; Schuurink, R.C.; Allmann, S. Identification and Characterization of (3Z):(2E)-Hexenal Isomerases from Cucumber. Front. Plant Sci. 2017, 8, 1342. [Google Scholar] [CrossRef] [PubMed]
- Gardner, H.W. Formation of (2E)-4-hydroxyl-2-nonenal and (2E)-4-hydroxy-2-hexenal by plant enzymes: A review suggests a role in the physiology in plants. Adv. Enzyme Res. 2016, 4, 56–61. [Google Scholar] [CrossRef][Green Version]
- Mano, J.; Khorobrykh, S.; Matsui, K.; Iijima, Y.; Sakurai, N.; Suzuki, H.; Shibata, D. Acrolein is formed from trienoic fatty acids in chloroplasts: A targeted metabolomics approach. Plant Biotechnol. 2014, 31, 535–544. [Google Scholar] [CrossRef]
- Roach, T.; Baur, T.; Stöggl, W.; Krieger-Litszkay, A. Chlamydomonas reinhardtii responding to high light: A role for 2-propenal (acrolein). Physiol. Plant. 2017, 161, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Roach, T.; Stöggl, W.; Baur, T.; Kranner, I. Distress and eustress of reactive electrophiles and relevance to light stress acclimation via stimulation of thiol/disulfide-based defences. Free Rad. Biol. Med. 2018, 122, 65–73. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Furutera, A.; Seki, K.Y.; Toyoda, Y.; Tanaka, K.; Sugimoto, Y. Malondialdehyde generated from peroxidized linoleic acid causes protein modification in heat-stressed plants. Plant Physiol. Biochem. 2008, 46, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Winger, A.M.; Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Miller, A.H. The cytotoxic lipid peroxidation product 4-hydroxy-2-nonenal covalently modifies a selective range of proteins linked to respiratory function in plant mitochondria. J. Biol. Chem. 2007, 282, 37436–37447. [Google Scholar] [CrossRef]
- Mano, J.; Nagata, M.; Okamura, S.; Shiraya, T.; Mitsui, T. Identification of oxidative-modified proteins in salt-stressed Arabidopsis: A carbonyl-targeted proteomics approach. Plant Cell Physiol. 2014, 55, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Esterbauer, H.; Zollner, H.; Scholtz, N. Reaction of glutathione with conjugated carbonyls. Zeitschrift für Naturforschung C 1975, 30, 466–473. [Google Scholar] [CrossRef]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Bachi, A.; Dalle-Donne, I.; Scaloni, A. Redox proteomics: Chemical principles, methodological approaches and biological/biomedical promises. Chem. Rev. 2013, 113, 596–698. [Google Scholar] [CrossRef]
- LoPachin, R.M.; Gavin, T.; Peterson, D.R.; Barber, D.S. Molecular mechanisms of 4-hydroxyn-2-nonenal and acrolein toxidity: Nucleophilic targets and adduct formation. Chem. Research Toxicol. 2009, 22, 1499–1508. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, T.K.; Freeman, B.A. Transduction of redox signaling by electrophile-protein reactions. Sci. Signal. 2009, 2, re7. [Google Scholar] [CrossRef]
- Arnette, D.; Quillin, A.; Gledenhuys, W.J.; Menze, M.A.; Konkle, M. 4-Hydroxynonenal and 4-ocononenal differentially bind to the redox sensor mitoNEET. Chem. Res. Toxicol. 2019, 32, 977–981. [Google Scholar] [CrossRef]
- Mano, J.; Kanameda, S.; Kuramitsu, R.; Matsuura, N.; Yamauchi, Y. Detoxification of reactive carbonyl species by glutathione transferase Tau isozymes. Front. Plant Sci. 2019, 10, 487. [Google Scholar] [CrossRef]
- Mano, J.; Ishibashi, A.; Muneuchi, H.; Morita, C.; Sakai, H.; Biswas, S.; Kitajima, S. Acrolein-detoxifying isozymes of glutathione transferase in plants. Planta 2017, 245, 255–264. [Google Scholar] [CrossRef]
- Pampola, R. Advanced lipoxidation end-products. Chem. Biol. Interact. 2011, 192, 14–20. [Google Scholar] [CrossRef]
- Taylor, N.L.; Day, D.A.; Millar, A.H. Environmental stress causes oidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J. Biol. Chem. 2002, 277, 42662–42668. [Google Scholar] [CrossRef]
- Winger, A.M.; Millar, A.H.; Day, D.A. Sensitivity of plant mitochondrial terminal oxidases to the lipid peroxidation product 4-hydroxy-2-nonenal (HNE). Biochem. J. 2005, 387, 865–870. [Google Scholar] [CrossRef]
- Biswas, M.S.; Mano, J. Reactive carbonyl species activate caspase-3-like protease to initiate programmed cell death in plants. Plant Cell Physiol. 2016, 57, 1432–1442. [Google Scholar] [CrossRef]
- Park, S.-W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.R.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef]
- Viestra, R.D. The ubiquitin/26S proeasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003, 8, 135–142. [Google Scholar] [CrossRef]
- Zheng, J.; Bizzozero, O. Reduced proteasomal activity contributes to the accumulation of carbonylated proteins in chromic experimental autoimmune encephalomyelitis. J. Neurochem. 2010, 115, 1556–1567. [Google Scholar] [CrossRef]
- Kurepa, J.; Toh-e, A.; Smalle, J.A. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008, 53, 102–114. [Google Scholar] [CrossRef]
- Smakowska, E.; Czarna, M.; Janska, H. Mitochondrial ATP-dependent proteases in protection against accumulation of carbonylated proteins. Mitochondrion 2014, 19B, 245–251. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Okada, G.; Kobayashi, S.; Chikuma, T.; Hojo, H. 4-Hydroxy-2-nonenal-modified glyceraldehyde-3-phosphate dehydrogenase is degraded by cathepsin G in rat neutrophils. Oxid. Med. Cell. Longev. 2011, 2011, 213686. [Google Scholar] [CrossRef]
- Weber, H.; Chételat, A.; Reymond, P.; Farmer, E.E. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 2004, 37, 877–888. [Google Scholar] [CrossRef]
- Reynolds, T. Comparative effects of aliphatic compounds on inhibition of lettuce fruit germination. Ann. Bot. 1977, 41, 637–648. [Google Scholar] [CrossRef]
- Alméras, E.; Stolz, S.; Vollenweider, S.; Reymond, P.; Mène-Saffrané, L.; Farmer, E.E. Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 2003, 34, 205–216. [Google Scholar] [CrossRef]
- Matsui, K.; Sugimoto, K.; Mano, J.; Ozawa, R.; Takabayashi, J. Differential metabolism of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements. PLoS ONE 2012, 7, e36433. [Google Scholar] [CrossRef]
- Mano, J.; Belles-Boix, E.; Babiychuk, E.; Inzé, D.; Torii, Y.; Hiraoka, E.; Takimoto, K.; Slooten, L.; Asada, K.; Kushnir, S. Protection against photooxidative injury of tobacco leaves by 2-alkenal reductase. Detoxication of lipid peroxide-derived reactive carbonyls. Plant Physiol. 2005, 139, 1773–1783. [Google Scholar] [CrossRef]
- Millar, A.H.; Leaver, C.L. The cytotoxic lipid peroxidation product, 4-hydroxy-2-nonenal, specifically inhibits decarboxylating dehydrogenases in the matrix of plant mitochondria. FEBS Lett. 2000, 481, 117–121. [Google Scholar] [CrossRef]
- Shimakawa, G.; Iwamoto, T.; Mabuchi, T.; Saito, R.; Yamamoto, H.; Amako, K.; Sugimoto, T.; Makino, A.; Miyake, C. Acrolein, an α,β-unsaturated carbonyl, inhibits both growth and PSII activity in the cyanobacterium Synechocystis sp. PCC 6803. Biosci. Biotechnol. Biochem. 2013, 77, 1655–1660. [Google Scholar] [CrossRef]
- Bate, N.J.; Rothstein, S.J. C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 1998, 16, 561–569. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Kunishima, M.; Mizutani, M.; Sugimoto, Y. Reactive short-chain leaf volatiles act as powerful inducers of abiotic stress-related gene expression. Sci. Rep. 2015, 5, 8030. [Google Scholar] [CrossRef]
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I. The roles of reactive carbonyl species in induction of antioxidant defence and ROS signalling in extreme halophytic model Eutrema parvulum and glycophytic model Arabidopsis thaliana. Exp. Environ. Bot. 2019, 160, 81–91. [Google Scholar] [CrossRef]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef]
- Eckardt, N.A. Oxylipin signaling in plant stress responses. Plant Cell 2008, 20, 495–497. [Google Scholar] [CrossRef]
- Stotz, H.U.; Mueller, S.; Zoeller, M.; Mueller, M.J.; Berger, S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 2013, 64, 963–975. [Google Scholar] [CrossRef]
- Matsui, K.; Sugimoto, K.; Kakumyan, P.; Khorobrykh, S.A.; Mano, J. Volatile oxylipins formed under stress in plants. In Lipidomics, Methods in Molecular Biology; Armstrong, D., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 580, pp. 17–28. [Google Scholar] [CrossRef]
- Mano, J.; Biswas, M.S. Analysis of reactive carbonyl species generated under oxidative stress. In Plant Programmed Cell Death: Methods and Protocols; De Gara, L., Locato, V., Eds.; Springer: New York, NY, USA, 2018; pp. 117–124. [Google Scholar]
- Mano, J.; Torii, Y.; Hayashi, S.; Takimoto, K.; Matsui, K.; Nakamura, K.; Inzé, D.; Babyichuk, E.; Kushnir, S.; Asada, K. The NADPH:quinone oxidoreductase P1-ζ-cryatallin in Arabidopsis catalyzes the α,β-hydrogenation of 2-alkenals: Detoxication of the lipid peroxide-derived reactive aldehydes. Plant Cell Physiol. 2002, 23, 1445–1455. [Google Scholar] [CrossRef]
- Biswas, M.S.; Mano, J. Lipid peroxide-derived short-chain carbonyls mediate hydrogen peroxide-induced and salt-induced programmed cell death in plants. Plant Physiol. 2015, 168, 885–898. [Google Scholar] [CrossRef]
- Kai, H.; Hirashima, K.; Matsuda, O.; Ikegami, H.; Winkelmann, T.; Nakahara, T.; Iba, K. Thermotolerant cyclamen with reduced acrolein and methyl vinyl ketone. J. Expt. Bot. 2012, 63, 4143–4150. [Google Scholar] [CrossRef]
- Srivastava, S.; Brychkova, C.; Yarmolinsky, D.; Soltabayeva, A.; Samani, T.; Sagi, M. Aldehyde oxidase 4 plays a critical role in delaying silique senescence by catalyzing aldehyde detoxification. Plant Physiol. 2017, 173, 1977–1997. [Google Scholar] [CrossRef]
- Islam, M.M.; Ye, W.; Matsushima, D.; Munemasa, S.; Okuma, E.; Nakamura, Y.; Biswas, M.S.; Mano, J.; Murata, Y. Reactive carbonyl species mediate abscisic acid signaling in guard cells. Plant Cell Physiol. 2016, 57, 2552–2563. [Google Scholar] [CrossRef]
- Islam, M.M.; Ye, W.; Matsushima, D.; Rhaman, M.S.; Munemasa, S.; Okuma, E.; Nakamura, Y.; Biswas, M.S.; Mano, J.; Murata, Y. Reactive carbonyl species function as signal mediators downstream of H2O2 production and regulate [Ca2+]cyt elevation in ABA signal pathway in Arabidopsis guard cells. Plant Cell Physiol. 2019, 60, 1146–1159. [Google Scholar] [CrossRef]
- Biswas, M.S.; Fukaki, H.; Mori, I.C.; Nakahara, K.; Mano, J. Reactive oxygen species and reactive carbonyl species constitute a feed-forward loop in the auxin signaling for lateral root formation. Plant J. 2019, in press. [Google Scholar] [CrossRef]
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540. [Google Scholar] [CrossRef]
- Yin, L.; Mano, J.; Tanaka, K.; Wang, S.; Zhang, M.; Deng, X.; Zhang, S. High level of reduced glutathione contributes to detoxification of lipid peroxide-derived reactive carbonyl species in transgenic Arabidopsis overexpressing glutathione reductase under aluminum stress. Physiol. Plant. 2017, 161, 211–223. [Google Scholar] [CrossRef]
- Ohkama-Ohtsu, N.; Zhao, P.; Xiang, C.; Oliver, D.J. Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 2007, 49, 878–888. [Google Scholar] [CrossRef]
- Grzam, A.; Martin, M.N.; Hell, R.; Meyer, A.J. γ-Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S-conjugates in Arabidopsis. FEBS Lett. 2007, 581, 3131–3138. [Google Scholar] [CrossRef]
- Aldini, G.; Carini, M.; Beretta, C.; Bradamante, S.; Facino, R.M. Carnosine is a quencher of 4-hydroxy-nonenal: Through what mechanism of reaction? Biochem. Biophys. Res. Commun. 2002, 298, 699–706. [Google Scholar] [CrossRef]
- Carini, M.; Aldini, G.; Beretta, G.; Arlandini, E.; Facino, R.M. Acrolein-sequestering ability of endogenous dipeptides: Characterization of carnosine and homocarnosine/acrolein adducts by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2003, 38, 996–1006. [Google Scholar] [CrossRef]
- Zhu, Q.; Zheng, Z.-P.; Cheng, K.-W.; Wu, J.-J.; Zhang, S.; Tang, Y.S.; Sze, K.-H.; Chen, J.; Chen, F.; Wang, M. Natural polyphenols as direct trapping agents of lipid peroxidation-derived acrolein and 4-hydroxy-trans-2-nonenal. Chem. Res. Toxicol. 2009, 22, 1721–1727. [Google Scholar] [CrossRef]
- Colzani, M.; Regazzoni, L.; Criscuolo, A.; Baron, G.; Carini, M.; Vistoli, G.; Lee, Y.-M.; Han, S.-I.; Aldini, G.; Yeum, K.-J. Isotopic labelling for the characterisation of HNE-sequestering agents in plant-based extracts and its application for the identification of anthocyanidins in black rice with giant embryo. Free Rad. Res. 2018, 52, 896–906. [Google Scholar] [CrossRef]
- Sengupta, D.; Naik, D.; Reddy, A.R. Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: A structure-function update. J. Plant Physiol. 2015, 179, 40–55. [Google Scholar] [CrossRef]
- Kirch, H.-H.; Bartels, D.; Wei, Y.; Schnable, P.S.; Wood, A.J. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004, 9, 1360–1385. [Google Scholar] [CrossRef]
- Brocker, C.; Vasiliou, M.; Carpenter, S.; Carpenter, C.; Zhang, Y.; Wang, X.; Kotchoni, S.O.; Wood, A.W.; Kirch, H.-H.; Kopecˇny´, D.; et al. Aldehyde dehydrogenase (ALDH) superfamily in plants: Gene nomenclature and comparative genomics. Planta 2012, 237, 189–210. [Google Scholar] [CrossRef]
- Stiti, N.; Missihoun, T.D.; Kotchoni, S.O.; Kirch, H.-H.; Bartels, D. Aldehyde dehydrogenases in Arabidopsis thaliana: Biochemical requirements, metabolic pathways, and functional analysis. Front. Plant Sci. 2011, 2, 65. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Hasegawa, A.; Taninaka, A.; Mizutani, M.; Sugimoto, Y. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 2011, 286, 6999–7009. [Google Scholar] [CrossRef]
- Nordling, E.; Jörnvall, H.; Persson, B. Medium-chain dehydrogenases/reductases (MDR) Family characterizations including genome comparisons and active site modelling. FEBS J. 2002, 269, 4267–4276. [Google Scholar] [CrossRef]
- Youn, B.; Kim, S.-J.; Moinuddin, S.G.A.; Lee, C.; Bedgar, D.L.; Harper, A.R.; Davin, L.B.; Lewis, N.G.; Kang, C.H. Mechanistic and structural studies of apoform, binary, and ternary complexes of the arabidopsis alkenal double bond reductase At5g16970. J. Biol. Chem. 2006, 281, 40076–40088. [Google Scholar] [CrossRef]
- Koeduka, T.; Watanabe, B.; Suzuki, S.; Hiratake, J.; Mano, J.; Yazaki, K. Characterization of raspberry ketone/zongerone synthase, catalyzing the alpha,beta-hydrogenation of phenylbutanones in raspberry fruits. Biochem. Biophys. Res. Commun. 2011, 412, 104–108. [Google Scholar] [CrossRef]
- Papdi, C.; Ábrahám, E.; Joseph, M.P.; Popescu, C.; Koncz, C.; Szabados, L. Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 2008, 147, 528–542. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Hasegawa, A.; Mizutani, M.; Sugimoto, Y. Chloroplastic NADPH-dependent alkenal/one oxidoreductase contributes to the detoxification of reactive carbonyls produced under oxidative stress. FEBS Lett. 2012, 586, 1208–1213. [Google Scholar] [CrossRef]
- Takagi, D.; Ifuku, K.; Ikeda, K.; Inoue, K.I.; Park, P.; Tamoi, M.; Inoue, H.; Sakamoto, K.; Saito, R.; Miyake, C. Suppression of chloroplastic alkenal/one oxidoreductase represses the carbon catabolic pathway in Arabidopsis leaves during night. Plant Physiol. 2016, 170, 2024–2039. [Google Scholar] [CrossRef]
- Monticolo, F.; Colantuono, C.; Chusano, M.L. Shaping the evolutionary tree of green plants: Evidence from the GST family. Sci. Rep. 2017, 7, 14363. [Google Scholar] [CrossRef]
- Dixon, D.P.; Hawkins, T.; Hussey, P.J.; Edwards, R. Enzyme activities an subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 2009, 60, 1207–1218. [Google Scholar] [CrossRef]
- Chen, J.H.; Jiang, H.-W.; Hsieh, E.-J.; Chen, H.-Y.; Chien, C.-T.; Hsieh, H.-L.; Lin, T.-P. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2012, 158, 340–351. [Google Scholar] [CrossRef]
- Xu, J.; Tian, Y.-S.; Xing, X.-J.; Peng, R.-H.; Zhu, B.; Gao, J.-J.; Yao, Q.-H. Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol. Plant. 2016, 156, 164–175. [Google Scholar] [CrossRef]
- Sharma, R.; Sahoo, A.; Devendran, R.; Jain, M. Over-expression of a rice Tau class glutathione-S-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS ONE 2014, 9, e92900. [Google Scholar] [CrossRef]
- Gronwald, J.W.; Plaisance, K.L. Isolation and characterization of glutathione S-transferase isozymes from sorghum. Plant Physiol. 1998, 117, 877–892. [Google Scholar] [CrossRef]
- Kobayashi, H.; Takase, H.; Suzuki, Y.; Tanzawa, F.; Tanaka, R.; Fujita, K.; Kohno, M.; Mochizuki, M.; Suzuki, S.; Konno, T. Environment stress enhances biosynthesis of flavor precursors, S-3-(hexan-1-ol)-glutathione and S-3-(hexan-1-ol) L-cysteine, in grapevine through glutathione S-transferase activation. J. Exp. Bot. 2011, 62, 1325–1336. [Google Scholar] [CrossRef]
- Simpson, P.J.; Tantitadapitak, C.; Reed, A.M.; Mather, O.C.; Bunce, C.M.; White, S.A.; Ride, J.P. Characterization of two novel aldo-keto reductases from Arabidopsis: Expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. J. Mol. Biol. 2009, 392, 465–480. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- LoPachin, R.M.; Gavin, T.; DeCaprio, A.; Barber, D.S. Application of the hard and soft, acids and bases (HSAB) theory to toxicant-target interactions. Chem. Res. Toxicol. 2012, 25, 239–251. [Google Scholar] [CrossRef]
- Ge, Y.; Cai, Y.-M.; Bonneau, L.; Rotari, V.; Danon, A.; MacKenzie, E.A.; McLellan, H.; Mach, L.; Gallois, P. Inhibition of cathepsin B by caspase-3 inhibitors blocks programmed cell death in Arabidopsis. Cell Death Differ. 2016, 23, 1493–1501. [Google Scholar] [CrossRef]
- Hatsugai, N.; Iwasaki, S.; Tamura, K.; Kondo, M.; Fuji, K.; Ogasawara, K.; Nishimura, M.; Hara-Nishimura, I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009, 22, 2496–2506. [Google Scholar] [CrossRef]
- Møller, I.M.; Sweetlove, L.J. ROS signaling—Specificity is required. Trends Plant Sci. 2010, 15, 370–374. [Google Scholar] [CrossRef]
- Higdon, A.; Diers, A.R.; Oh, J.Y.; Lander, A.; Darley-Usmar, M. Cell signalling by reactive lipid species: A new concepts and molecular mechanisms. Biochem. J. 2012, 442, 453–464. [Google Scholar] [CrossRef]
| Compartment | Protein | RCS | Effect | Ref |
|---|---|---|---|---|
| Mitochondrion | GDC H-SU and other lipoate enzymes | HNE | Modified, Inactivated | [63] |
| Alternative oxidase | HNE | Inactivated | [64] | |
| Succinate dehydrogenase α-SU ATP synthase β-SU Pyruvate dehydrogenase E1 β-SU Elongation factor Tu Voltage-dependent anion channel Adenine nucleotide translocator | HNE | Modified | [52] | |
| Chloroplast | OEC33 LHCII | MDA | Modified | [51] |
| Phosphoribulokinase Glyceraldehyde-3-phosphate dehydrogenase Fructose-1,6-bisphosphatase | acrolein | Inactivated | [13] | |
| Cyclophilin 20-3 | HNE | Modified | [53] | |
| OPDA | Modified, Activated | [66] | ||
| Cytosol | Triosephosphate isomerase Cysteine synthase Ascorbate peroxidase Heat shock cognate 70 kDa protein 3 | HNE | Modified | [53] |
| C1LP, C3LP | acrolein, HNE | Activated | [65] | |
| Peroxisome | Nitrile-specifier protein 5 Gly-rich RNA binding protein 7 Nucleotide diphosphate kinase | HNE | Modified | [53] |
| Apoplast | Germin-like protein subfamily 3 number 1 Peroxidase 34 | HNE | Modified | [53] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mano, J.; Biswas, M.S.; Sugimoto, K. Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants 2019, 8, 391. https://doi.org/10.3390/plants8100391
Mano J, Biswas MS, Sugimoto K. Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants. 2019; 8(10):391. https://doi.org/10.3390/plants8100391
Chicago/Turabian StyleMano, Jun’ichi, Md. Sanaullah Biswas, and Koichi Sugimoto. 2019. "Reactive Carbonyl Species: A Missing Link in ROS Signaling" Plants 8, no. 10: 391. https://doi.org/10.3390/plants8100391
APA StyleMano, J., Biswas, M. S., & Sugimoto, K. (2019). Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants, 8(10), 391. https://doi.org/10.3390/plants8100391





