De Novo Assembly Discovered Novel Structures in Genome of Plastids and Revealed Divergent Inverted Repeats in Mammillaria (Cactaceae, Caryophyllales)
Abstract
1. Introduction
2. Results
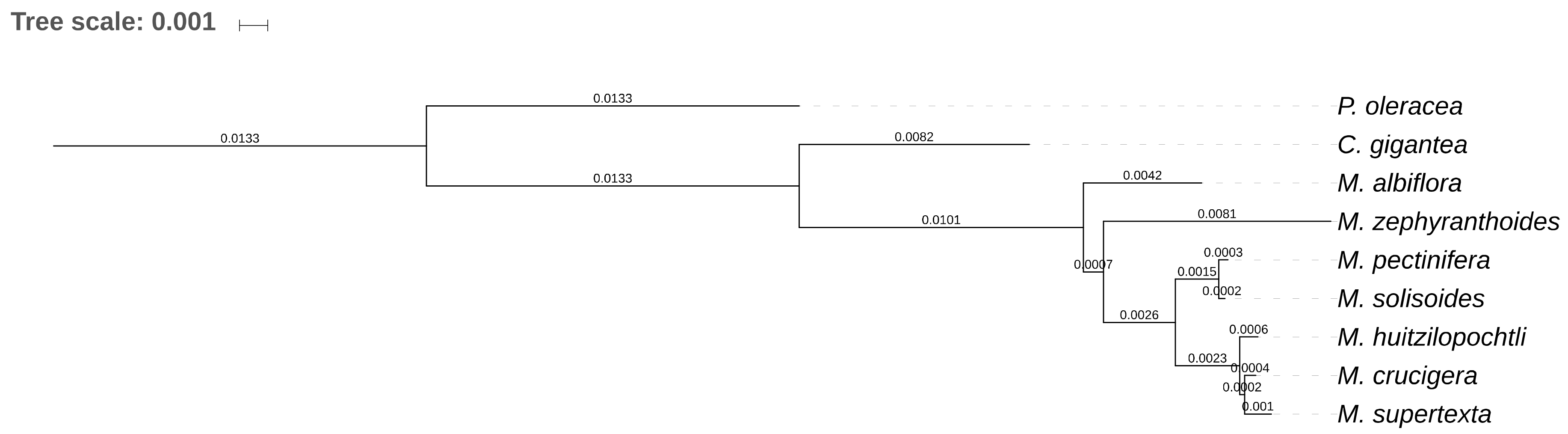
2.1. Gene Composition and Length Variation in Three Novel cpDNA Structures Identified in Mammillaria
2.2. Structure 1: Shortest IRs, Composed of Three Genes, rpl23-trnI-CAU-ycf2
2.3. Structure 2: IRs Composed by an Unusual Complete Battery of 11 Concatenated Genes and One Identical Intergenic Spacer.
2.4. Structure 3: Largest and Divergent IRs in Which Four Ribosomal Units are Included.
3. Discussion
4. Materials and Methods
4.1. Plant Sampling and DNA Extraction
4.2. High-Throughput Sequencing and Sanger Verification
4.3. Genome Assembly, Annotation, and Structural Alignment
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Morden, C.; Delwiche, C.; Kuhsel, M.; Palmer, D. Gene phylogenies and the endosymbiotic origin of plastids. Biosystems 1992, 28, 75–90. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. The origins of plastids. Biol. J. Linn. Soc. 1982, 17, 289–306. [Google Scholar] [CrossRef]
- Lemieux, C.; Otis, C.; Turnel, M. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature 2000, 403, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Xiao-Ming, Z.; Junrui, W.; Li, F.; Sha, L.; Hongbo, P.; Lan, Q.; Jing, L.; Yan, S.; Weihua, Q.; Lifang, Z.; et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci. Rep. 2017, 7, 1555. [Google Scholar] [CrossRef] [PubMed]
- Thorsness, P.E.; Weber, E.R. Escape and migration of nucleic acids between chloroplasts, mitochondria, and the nucleus. Int. Rev. Cytol. 1996, 165, 207–234. [Google Scholar] [CrossRef]
- Yagi, Y.; Shiina, T. Recent advances in the study of chloroplast gene expression and its evolution. Front. Plant Sci. 2014, 5, 61. [Google Scholar] [CrossRef]
- Kung, S.D.; Lin, C.M. Chloroplast promoters from higher plants. Nucleic Acids Res. 1985, 13, 7543–7549. [Google Scholar] [CrossRef][Green Version]
- Mower, J.P.; Vickrey, T.L. Advances in Botanical Research Plastid Genome Evolution; Chaw, S.M., Jansen, R.K., Eds.; Academic Press of Elsevier: London, UK, 2018; Volume 85, Chapter 9; pp. 263–292. [Google Scholar]
- Zhu, A.; Guo, W.; Sakski, G.; Weishu, F.; Mover, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss of substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Lavin, M.; Doyle, J.J.; Palmer, J.D. Evolutionary significance of the loss of the chloroplast-DNA Inverted Repeat in the Leguminosae Subfamily Papilionoideae. Evolution 1990, 44, 390–402. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Copetti, D.; Búrquez, A.; Bustamante, E.; Charboneau, J.L.M.; Eguiarte, L.; Kumar, S.; Lee, H.O.; McMahon, M.; Steele, K.; et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea). Am. J. Bot. 2015, 102, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Jin, J.J.; Li, H.T.; Yang, J.B.; Mandala, V.S.; Croley, M.; Mostow, R.; Douglas, N.A.; Chase, M.W.; Christenhusz, M.J.M.; et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogenet. Evol. 2019, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.Y.; Gao, L.Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Tembrock, L.R. Two complete chloroplast genomes of white campion (Silene latifolia) from male and female individuals. Mitochondrial DNA Part A 2015, 28, 375–376. [Google Scholar] [CrossRef]
- Kang, J.S.; Lee, B.Y.; Kwak, M. The complete chloroplast genome sequences of Lychnis wilfordii and Silene capitata and comparative analyses with other Caryophyllaceae genomes. PLoS ONE 2017, 27, e0172924. [Google Scholar] [CrossRef] [PubMed]
- Blazier, J.C.; Jansen, R.K.; Mower, J.P.; Govindu, M.; Zhang, J.; Weng, M.L.; Ruhlman, T.A. Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 2016, 117, 1209–1220. [Google Scholar] [CrossRef]
- Hunt, D.; Taylor, N.; Charles, G. The New Cactus Lexicon; DH Books: Milborne Port, UK, 2006. [Google Scholar]
- IUCN International Union for Conservation of Nature. Consulted for Mammillaria genus. Available online: https://www.iucnredlist.org/search/list?query=Mammillaria&searchType=species (accessed on 30 June 2019).
- Butterworth, C.A.; Wallace, R.S. Phylogenetic studies of Mammillaria (Cactaceae)—Insights from chloroplast sequence variation and hypothesis testing using the parametric bootstrap. Am. J. Bot. 2004, 91, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Crozier, B.S. Systematics of Cactaceae Juss: Phylogeny, cpDNA Evolution, and Classification, with Emphasis on the Genus Mammillaria Haw. Ph.D. Thesis, University of Texas, Austin, TX, USA, 2005. [Google Scholar]
- Hernández-Hernández, T.; Hernández, H.M.; De Nova, J.A.; Puente, R.; Eguiarte, L.; Magallón, S. Phylogenetic relationships and evolution of growth form in Cactaceae (Caryophyllales, Eudicotyledoneae). Am. J. Bot. 2011, 98, 44–61. [Google Scholar] [CrossRef]
- Arakaki, M.; Christin, P.A.; Nyffeler, R.; Lendel, A.; Eggli, U.; Ogburn, M.; Spriggs, E.; Moore, M.J.; Edwards, E.J. Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc. Natl. Acad. Sci. USA 2011, 108, 8379–8384. [Google Scholar] [CrossRef]
- Turmel, M.; Otis, C.; Lemieux, C. Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci. Rep. 2017, 7, 994. [Google Scholar] [CrossRef]
- Ocampo, G.; Columbus, J.T. Molecular phylogenetics of suborder Cactineae (Caryophyllales), including insight into photosynthetic diversification and historical biogeography. Am. J. Bot. 2010, 97, 1827–1847. [Google Scholar] [CrossRef] [PubMed]
- Alkatib, S.; Fleischmann, T.T.; Scharff, L.B.; Bock, R. Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acid Res. 2012, 40, 6713–6724. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Ye, Y.; Bal, T.; Xu, M.; Xu, L.A. Complete chloroplast genome of Pinus massoniana (Pinaceae): Gene rearrangements, loss of ndh genes, and short inverted repeats contraction, expansion. Molecules 2017, 22, 1528. [Google Scholar] [CrossRef]
- Hao, D.C.; Chen, S.L.; Huang, B.L. Evolution of the chloroplast trnL-trnF region in the Gymnosperm lineages Taxaceae and Cephalotaxaceae. Biochem. Genet. 2009, 47, 351–369. [Google Scholar] [CrossRef] [PubMed]
- Drescher, A.; Ruf, S.; Calsa, T., Jr.; Carrer, H.; Bock, R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000, 22, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; An, B.; Park, S. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci. Rep. 2018, 8, 13568. [Google Scholar] [CrossRef] [PubMed]
- Barthet, M.M.; Hilu, K.W. Expression of matK: Functional and evolutionary implications. Am. J. Bot. 2007, 94, 1402–1412. [Google Scholar] [CrossRef]
- Arias, S.; Gama-López, S.; Guzmán-Cruz, L.U.; Vázquez-Benítez, B. Cactaceae. In Flora del Valle de Tehuacán-Cuicatlán; Medina, L.R., Ed.; Instituto de Biología, Universidad Nacional Autónoma de México: Mexico City, Mexico, 2012; Volume 95. [Google Scholar]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Love, R.R.; Weisenfeld, N.I.; Jaffe, D.B.; Besansky, N.J.; Neafsey, D.E. Evaluation of DISCOVAR de novo using a mosquito simple for cost-effective short-read genome assembly. BMC Genom. 2016, 17, 18. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Raney, B.; Paten, B.; Pham, S. Ragout-a reference-assisted assembly tool for bacterial genomes. Bioinformatics 2014, 30, i302–i309. [Google Scholar] [CrossRef] [PubMed]
- Soto-Jiménez, L.M.; Estrada, K.; Sanchez-Flores, A. GARM: Genome assembly, reconciliation and merging pipeline. Curr. Top. Med. Chem. 2014, 14, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; De Sila, N.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Kikuchi, T.; Sanders, M.; Newbold, C.; Berriman, M.; Otto, T.D. REAPR: A universal tool for genome assembly evaluation. Genome Biol. 2013, 14, R47. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Liu, X.; Yang, H.; Zhao, J.; Zhou, B.; Li, T.; Xiang, B. The complete chloroplast genome sequence of the folk medicinal and vegetable plant purslane (Portulaca oleracea L.). J. Hortic. Sci. Biotech. 2018, 93, 356–365. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Katoh, K.; Daron, M.S. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Posada, D. JModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
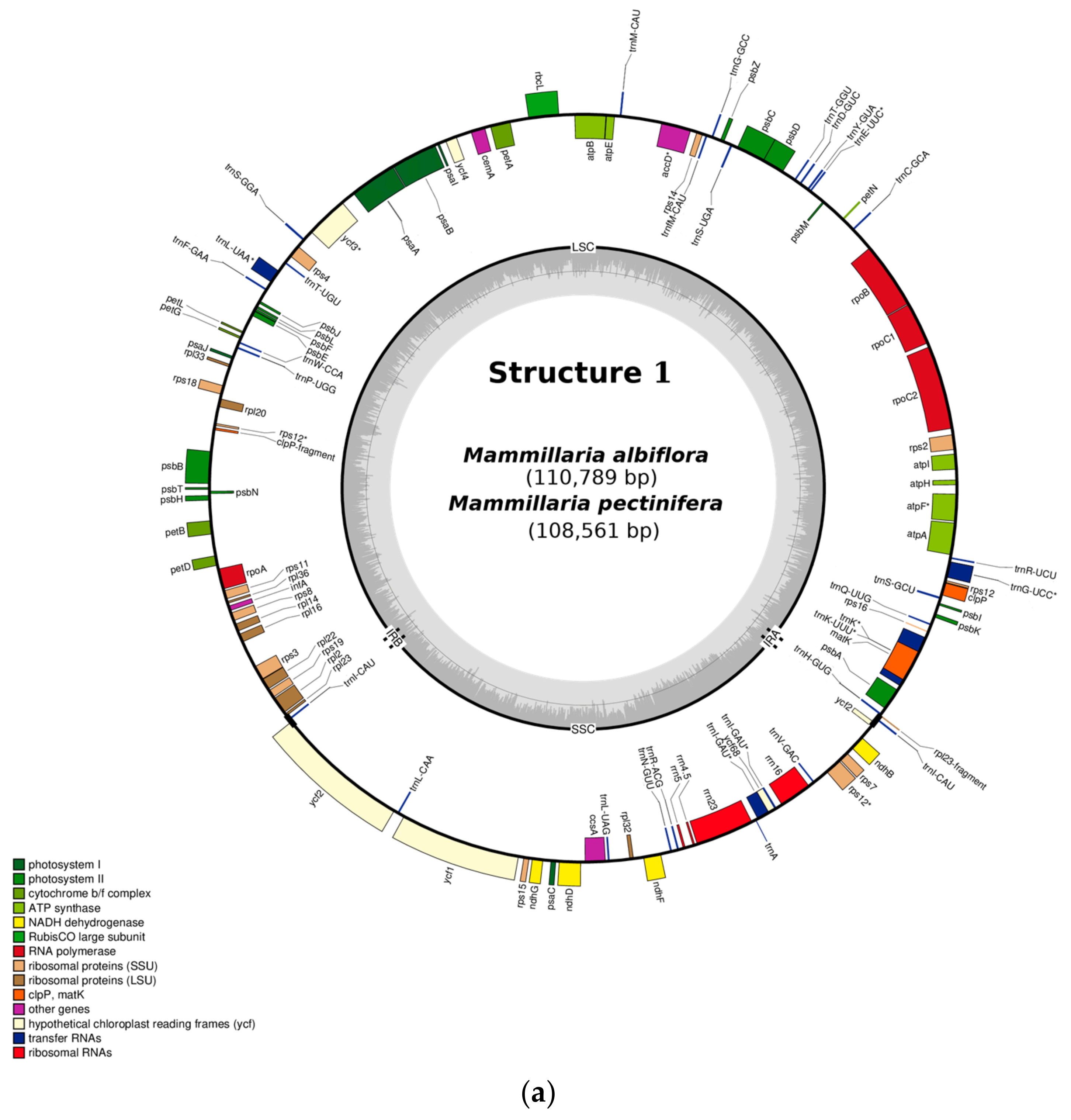
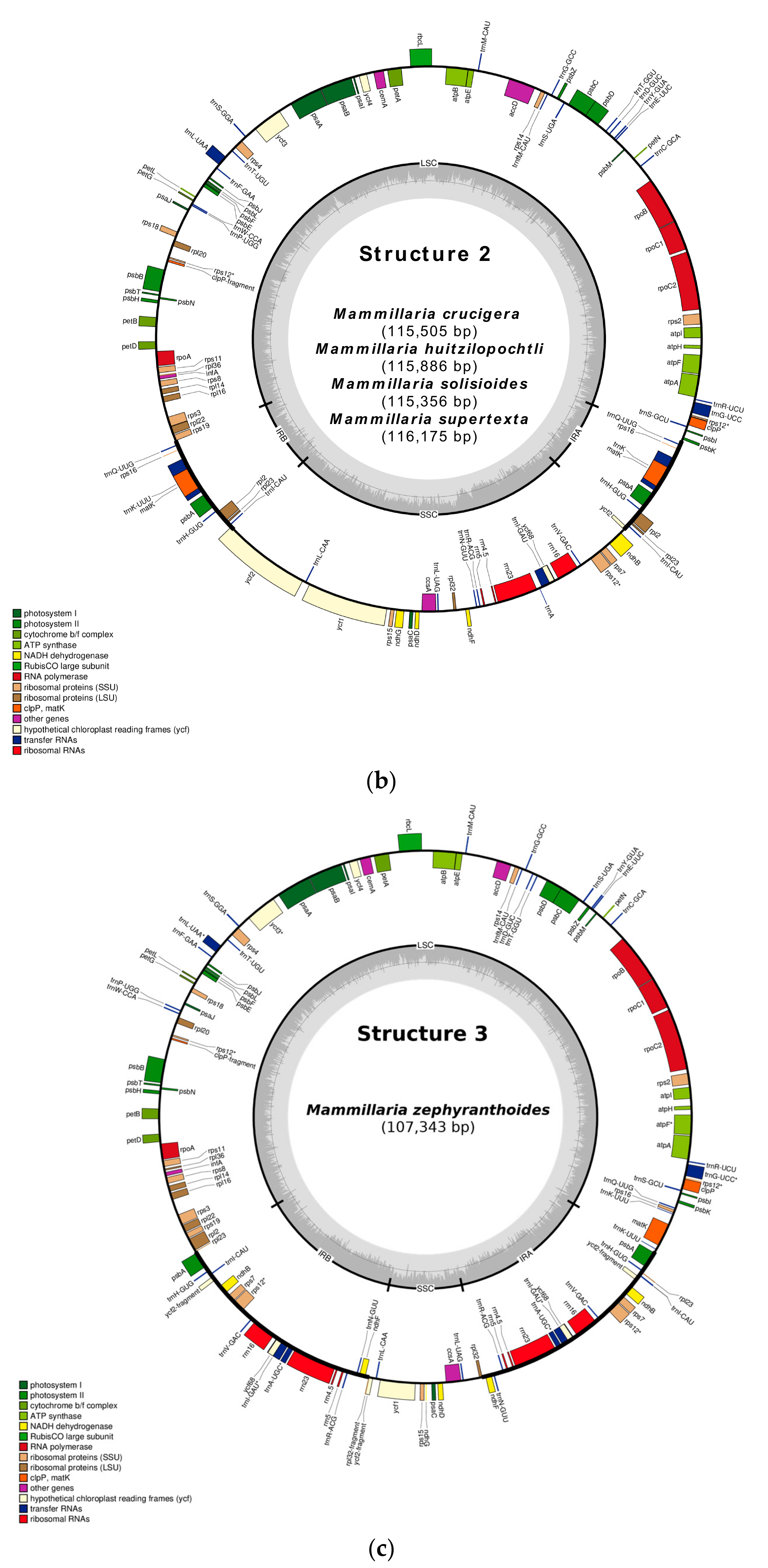
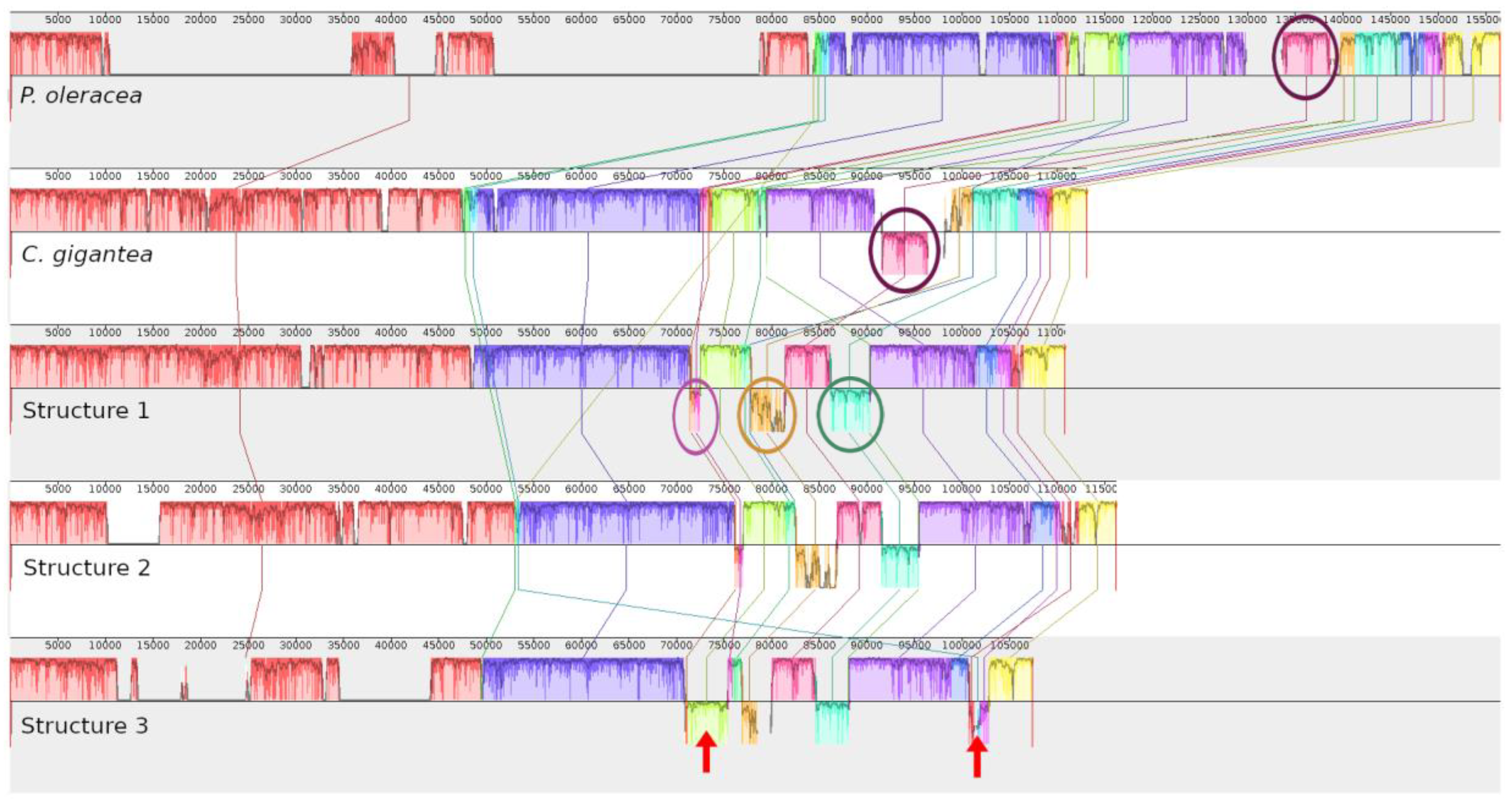
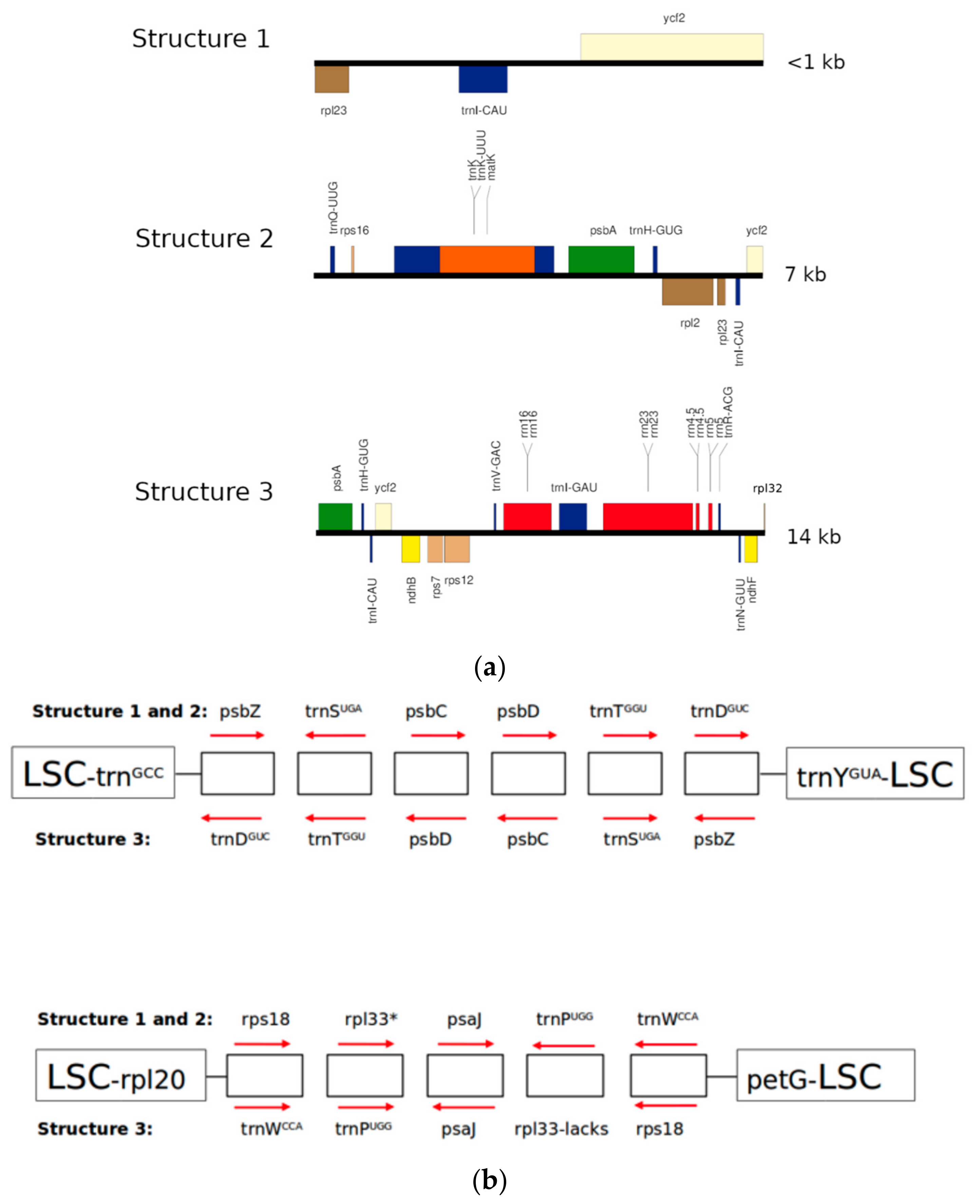
| Type of Structure | Total Length | IRs | LSC | SCC | Total Number of Genes | Access Number 1 |
|---|---|---|---|---|---|---|
| I. Structure 1 | ||||||
| 1.1 M. albiflora | 110789 | 1348 | 78380 | 31061 | 113 | MN517610 |
| 1.2. M. pectinifera | 108561 | 1544 | 72273 | 29744 | 113 | MN519716 |
| II. Structure 2 | ||||||
| 1. M. crucigera | 115505 | 14522 | 71565 | 29418 | 120 | MN517613 |
| 2. M. huitzilopochtli | 115886 | 14488 | 71997 | 29401 | 120 | MN517612 |
| 3. M. solisioides | 115356 | 14428 | 71690 | 29238 | 120 | MN518341 |
| 4. M. supertexta | 116175 | 14490 | 72240 | 29445 | 119 | MN508963 |
| III. Structure 3 | ||||||
| 1. M. zephyranthoides | 107343 | 28252 | 71811 | 7281 | 131 | MN517611 |
| Gene Type/Structure | Region | Structure 1 | Structure 2 | Structure 3 |
|---|---|---|---|---|
| 1. Ribosomal RNA (rrn) | SSC | rrn4.5, 5,16, 23 | rrn4.5, 5,16, 23 | |
| IRs | rrn4.5, 5,16, 23 (2X) | |||
| 2. Transfer RNA (trn) | LSC | trnCGCA, trnDGUC, trnEUUC, trnFGAA, trnGGCC, trnGUCC, trnHGUG, trnKUUU, trnLUAA, trnMCAU, trnMCAU, trnPUGG, trnQUUG, trnRUCU, trnSGGA, trnSGGU, trnSUGA, trnTGGU, trnTUGU, trnYGUA | trnCGCA, trnDGUC, trnEUUC, trnFGAA, trnGGCC, trnGUCC, trnLUAA, trnMCAU, trnfMCAU, trnPUGG, trnRUCU, trnSGGA, trnSGCU, trnSUGA, trnTGGU, trnTUGU, trnWCCA, trnYGUA | trnCGCA, trnDGUC, trnEUUC, trnFGAA, trnGGCC, trnGUCC, trnKUUU, trnLUAA, trnMCAU, trnfMCAU, trnPUGG, trnQUUG, trnRUCU, trnSGGA, trnSGCU, trnSUGA, trnTGGU, trnTUGU, trnWCCA, trnYGUA |
| SSC | trnA-f, trnIGAU, trnIGAU, trnLCAA, trnLUAG, trnNGUU, trnRACG, trnVGAG | trnA-f, trnIGAU, trnIUAG, trnNGUU, trnLCAA, trnRACG, trnVGAG | trnLUAG, trnLCAA | |
| IRs | trnICAU (2X) | trnHGUG,tmICAU, trnKUUU, trnQUUG (2X) | trnAUGC, trnHGUG, trnICAU, trnIGAU, trnNGUU, trnRACG, trnVGAC (2X) | |
| 3. Proteins of small subunits of the ribosome (rps) | LSC | rps2, 3, 4, 8, 11, 12 (2), 14, 16Ψ, 18Ψ, 19 | rps2, 3, 4, 8, 11, 12 (2), 14, 18Ψ, 19 | rps2, 3, 4, 8 11, 12, 12Ψ, 14, 16Ψ, 18Ψ, 19 |
| SSC | rps7, 12, 15 | rps7, 12, 15 | rps15 | |
| IRs | rps16Ψ (2X) | rps7, 12, (2X) | ||
| 4. Proteins of large subunits of the ribosome (rpl) | LSC | rpl2, 14, 16, 20, 22, 33Ψ, 36Ψ | rpl14, 16Ψ, 20, 22,33Ψ*, 36Ψ | rpl2, 14, 16Ψ, 20, 22, 23Ψ, 36 |
| SSC | rpl32 | rpl32 | ||
| IRs | rpl23Ψ (2X) | rpl2, 23Ψ (2X) | rpl32 (2X), 23Ψ (IRA) | |
| 5. DNA dependent RNA polymerase (rpo) | LSC | rpoA, B, C1, C2 | rpoA, B, C1, C2 | rpoA, B, C1, C2, |
| 6. NADH dehydrogenase (ndh) | SSC | ndhBΨ, DΨ, FΨ, GΨ*** | ndhBΨ, DΨ, FΨ, G*** | |
| IRs | ndhBΨ, DΨ, FΨ, GΨ (2X) | |||
| 7. Photosystem I (psa) | LSC | psaA, B, I, J | psaA, B, I, J | psaA, B, I, J |
| SSC | psaC | psaC | psaC | |
| 8. Photosystem II (psb) | LSC | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z | psbB, C, D, E, F, H, I, J, K, L, M, N, T, Z | psbB, C, D, E, F, H, I, J, L, K, M, N T, Z |
| IRs | psbA (2X) | psbA (2X) | ||
| 9. Cytochrome b/f complex (pet) | LSC | petA, B, D, G, L, N | petA, B, D, G, L, N | petA, B, D, G, L, N |
| 10. ATP synthase (atp) | LSC | atpA, B, E, F, H, I | atpA, B, E, F, H, I | atpA, B, E, F, H, I |
| 11. Rubisco (rbc) | LSC | rbcL | rbcL | rbcL |
| 12. Maturase K | LSC | matK | matK | |
| IRs | matK (2X) | |||
| 13. Protease (clp) | LSC | clpPΨ, clpP | clpPΨ, clpP | clpPΨ, clpP |
| 14. Envelope membrane protein (cem) | LSC | cemA | cemA | cemA |
| 15. Subunit of acetil-CoA-carboxylase (acc) | LSC | accDΨ | accDΨ | accDΨ |
| 16. c-type cytochrome synthesis (ccs) | SSC | ccsA | SSC: ccsA | SSC: ccsA |
| 17. Translational initiation factor (inf) | LSC | infA | infA | infA |
| 18. Hypothetical chloroplast reading frames (ycf) | LSC | ycf3, ycf4Ψ | ycf3, ycf4Ψ** | ycf3, ycf4 |
| SSC | ycf1, ycf2, ycf68Ψ | ycf1, ycf2, ycf68Ψ | ycf1Ψ, ycf2Ψ | |
| IRs | ycf2-p (2X) | ycf2-p (2X) | ycf2Ψ, ycf68Ψ (2X) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solórzano, S.; Chincoya, D.A.; Sanchez-Flores, A.; Estrada, K.; Díaz-Velásquez, C.E.; González-Rodríguez, A.; Vaca-Paniagua, F.; Dávila, P.; Arias, S. De Novo Assembly Discovered Novel Structures in Genome of Plastids and Revealed Divergent Inverted Repeats in Mammillaria (Cactaceae, Caryophyllales). Plants 2019, 8, 392. https://doi.org/10.3390/plants8100392
Solórzano S, Chincoya DA, Sanchez-Flores A, Estrada K, Díaz-Velásquez CE, González-Rodríguez A, Vaca-Paniagua F, Dávila P, Arias S. De Novo Assembly Discovered Novel Structures in Genome of Plastids and Revealed Divergent Inverted Repeats in Mammillaria (Cactaceae, Caryophyllales). Plants. 2019; 8(10):392. https://doi.org/10.3390/plants8100392
Chicago/Turabian StyleSolórzano, Sofía, Delil A. Chincoya, Alejandro Sanchez-Flores, Karel Estrada, Clara E. Díaz-Velásquez, Antonio González-Rodríguez, Felipe Vaca-Paniagua, Patricia Dávila, and Salvador Arias. 2019. "De Novo Assembly Discovered Novel Structures in Genome of Plastids and Revealed Divergent Inverted Repeats in Mammillaria (Cactaceae, Caryophyllales)" Plants 8, no. 10: 392. https://doi.org/10.3390/plants8100392
APA StyleSolórzano, S., Chincoya, D. A., Sanchez-Flores, A., Estrada, K., Díaz-Velásquez, C. E., González-Rodríguez, A., Vaca-Paniagua, F., Dávila, P., & Arias, S. (2019). De Novo Assembly Discovered Novel Structures in Genome of Plastids and Revealed Divergent Inverted Repeats in Mammillaria (Cactaceae, Caryophyllales). Plants, 8(10), 392. https://doi.org/10.3390/plants8100392





