Participation of Wild Species Genus Avena L. (Poaceae) of Different Ploidy in the Origin of Cultivated Species According to Data on Intragenomic Polymorphism of the ITS1-5.8S rRNA Region
Abstract
1. Introduction
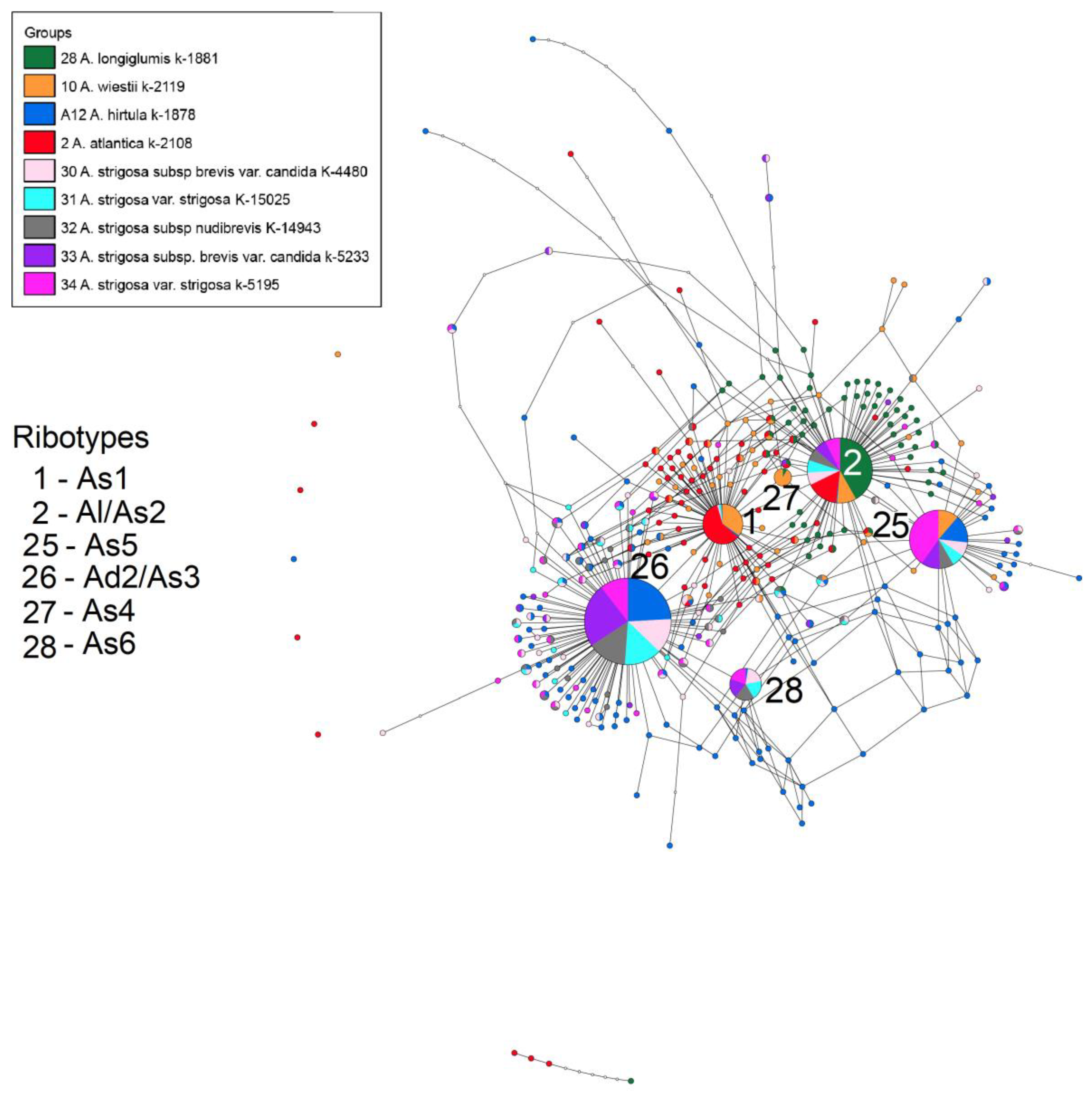
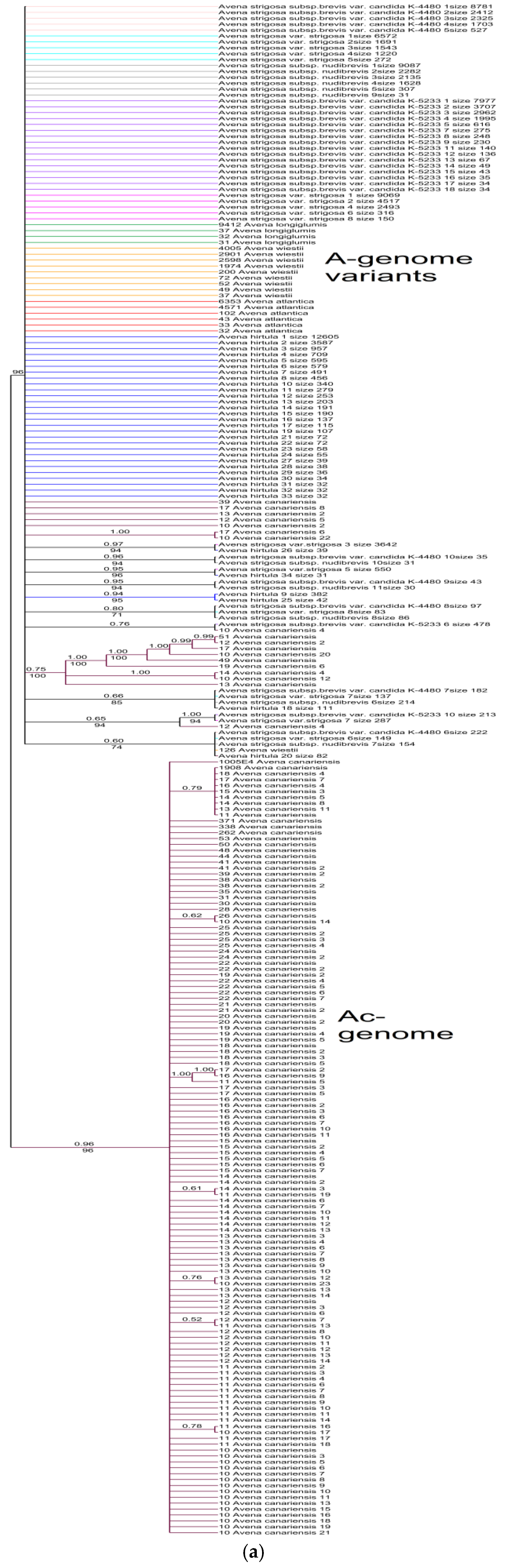
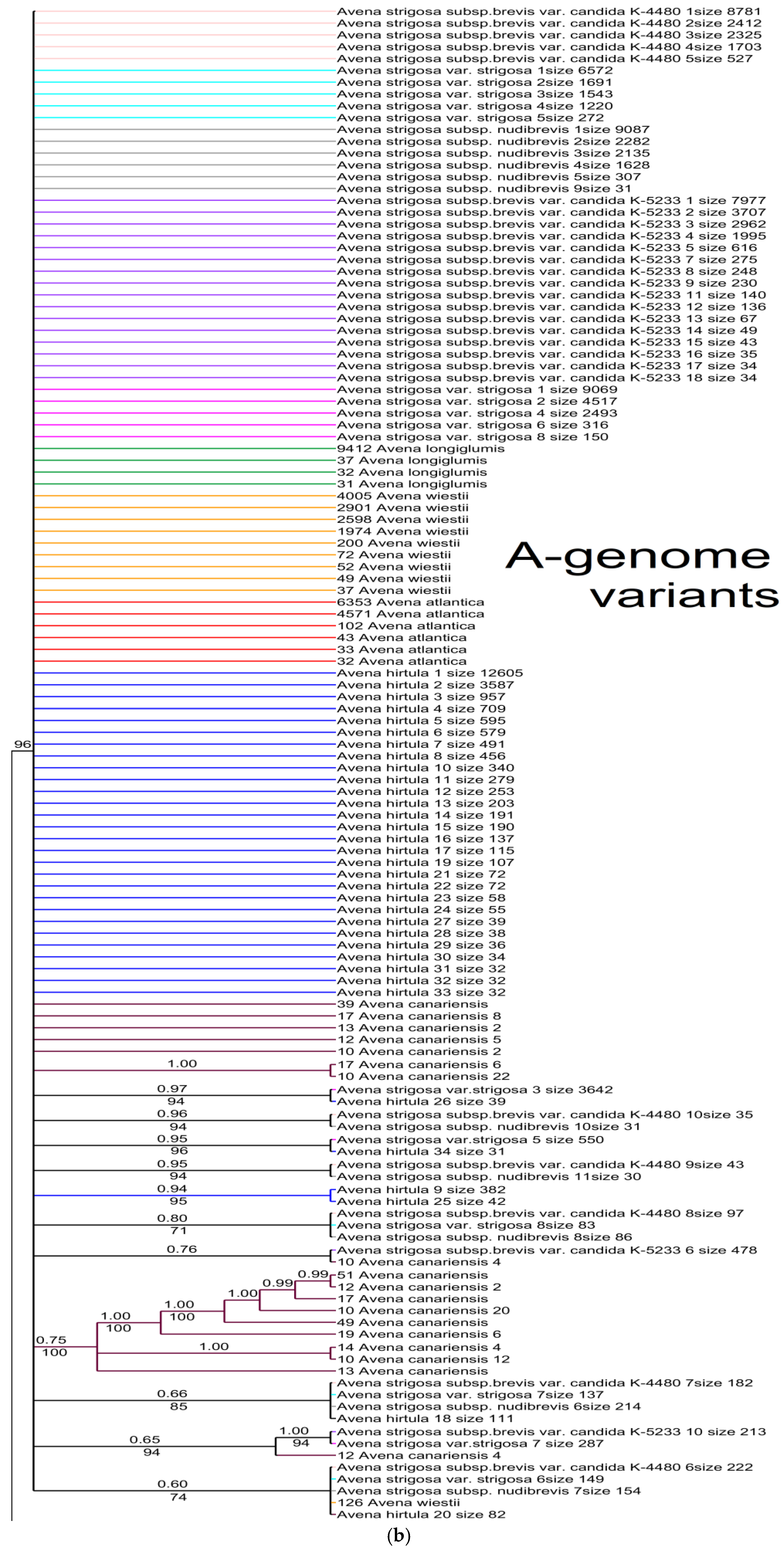
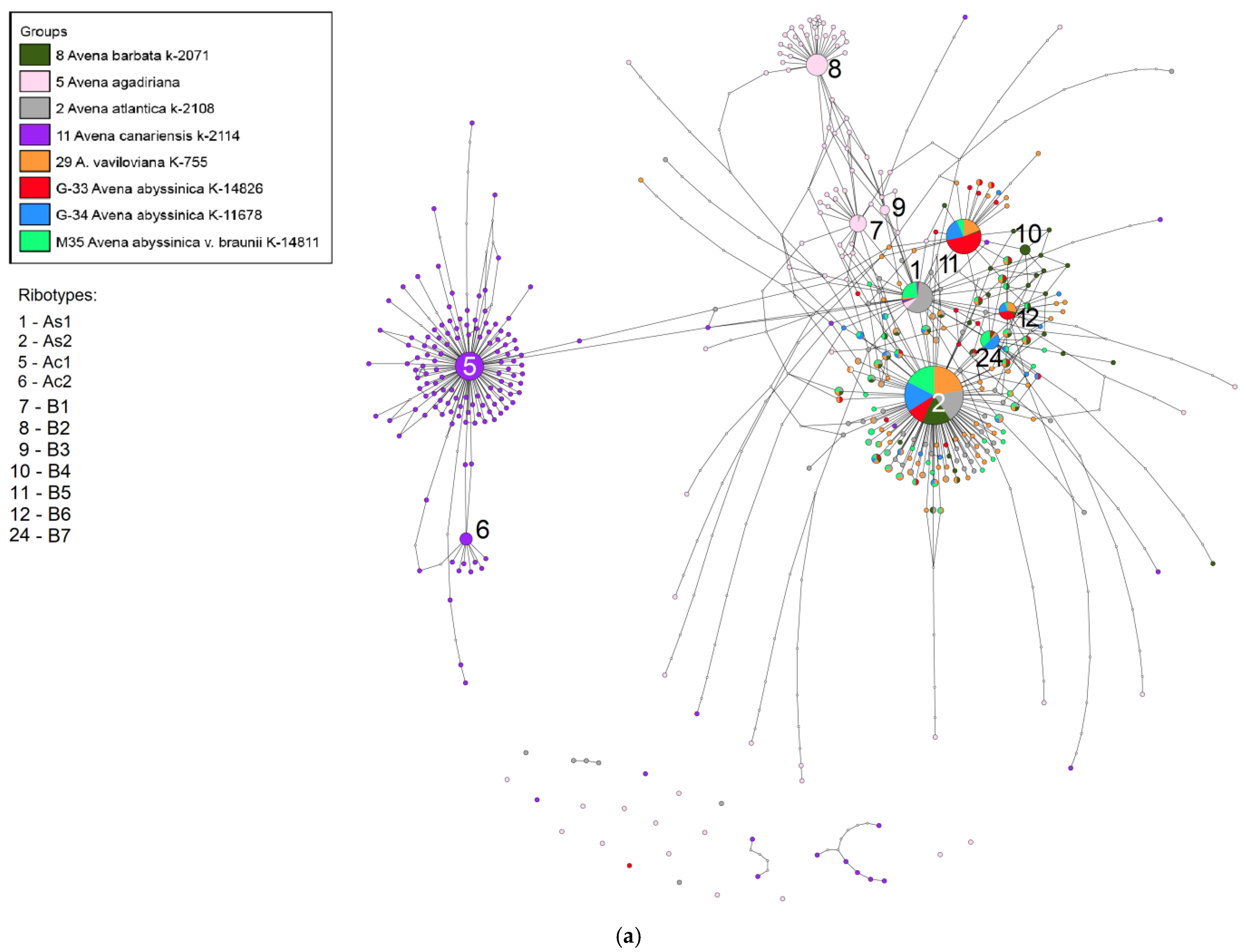
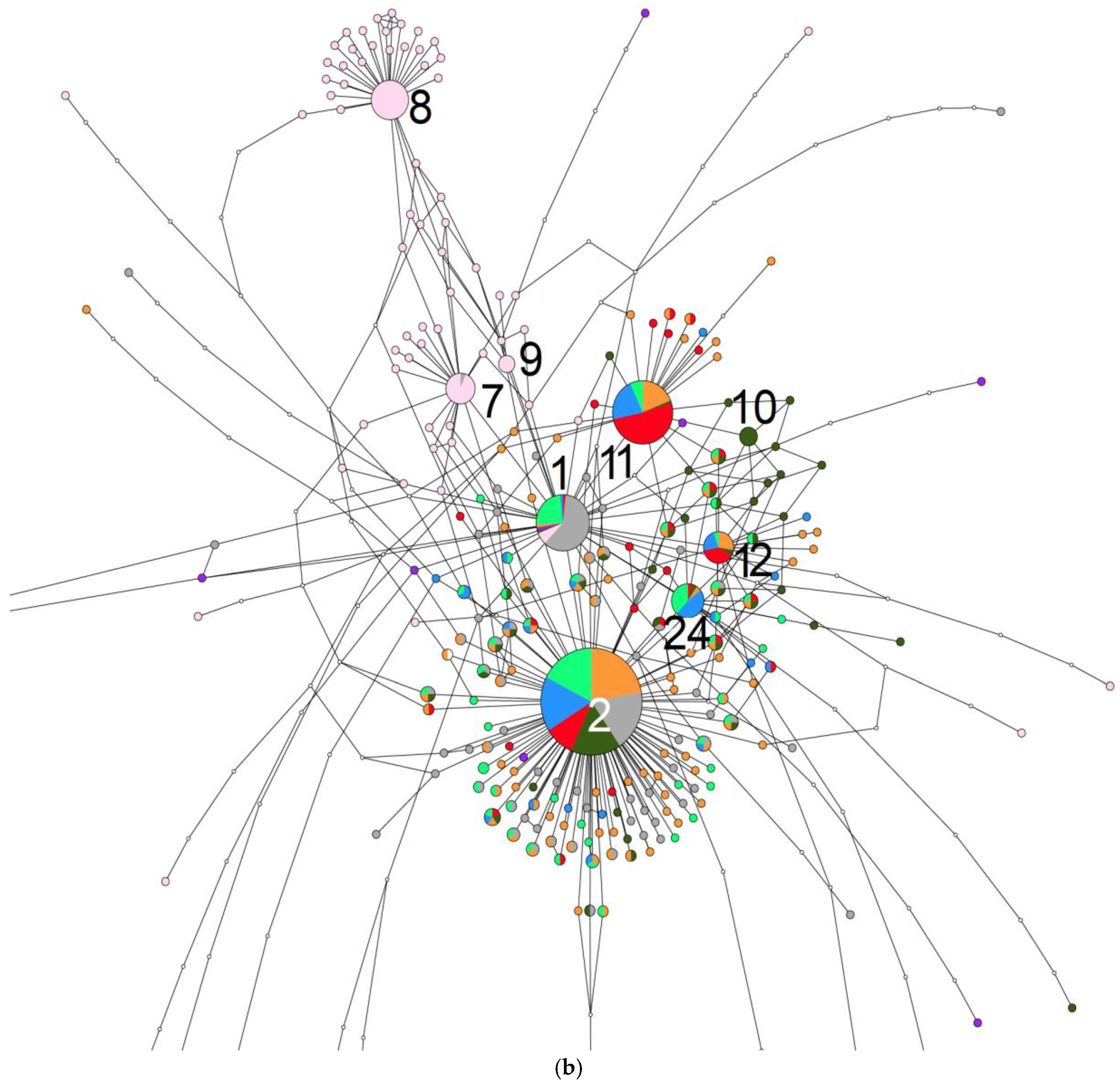
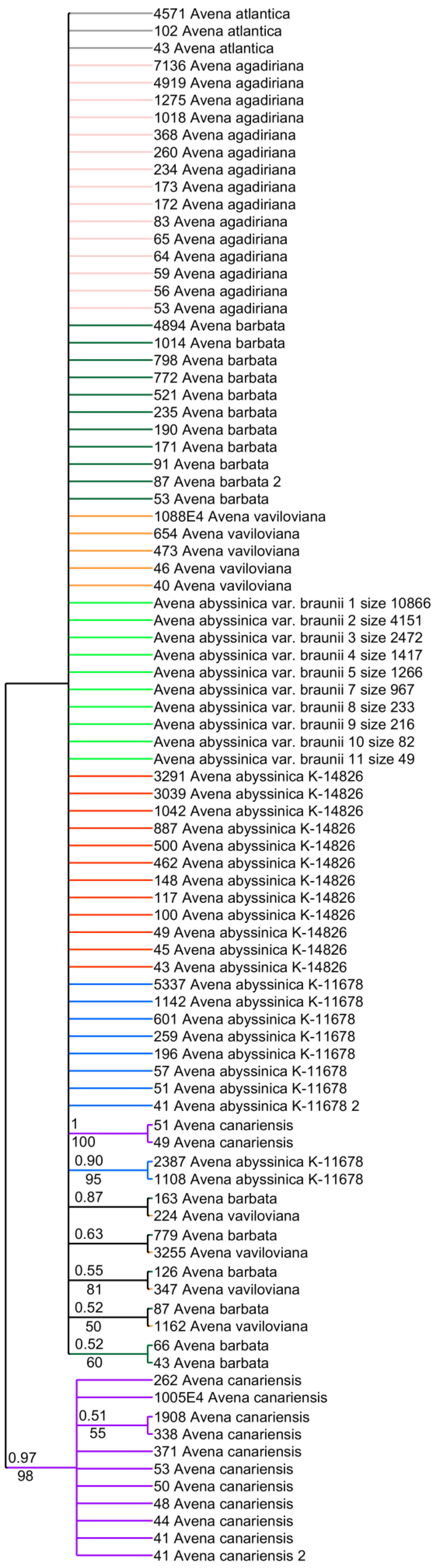
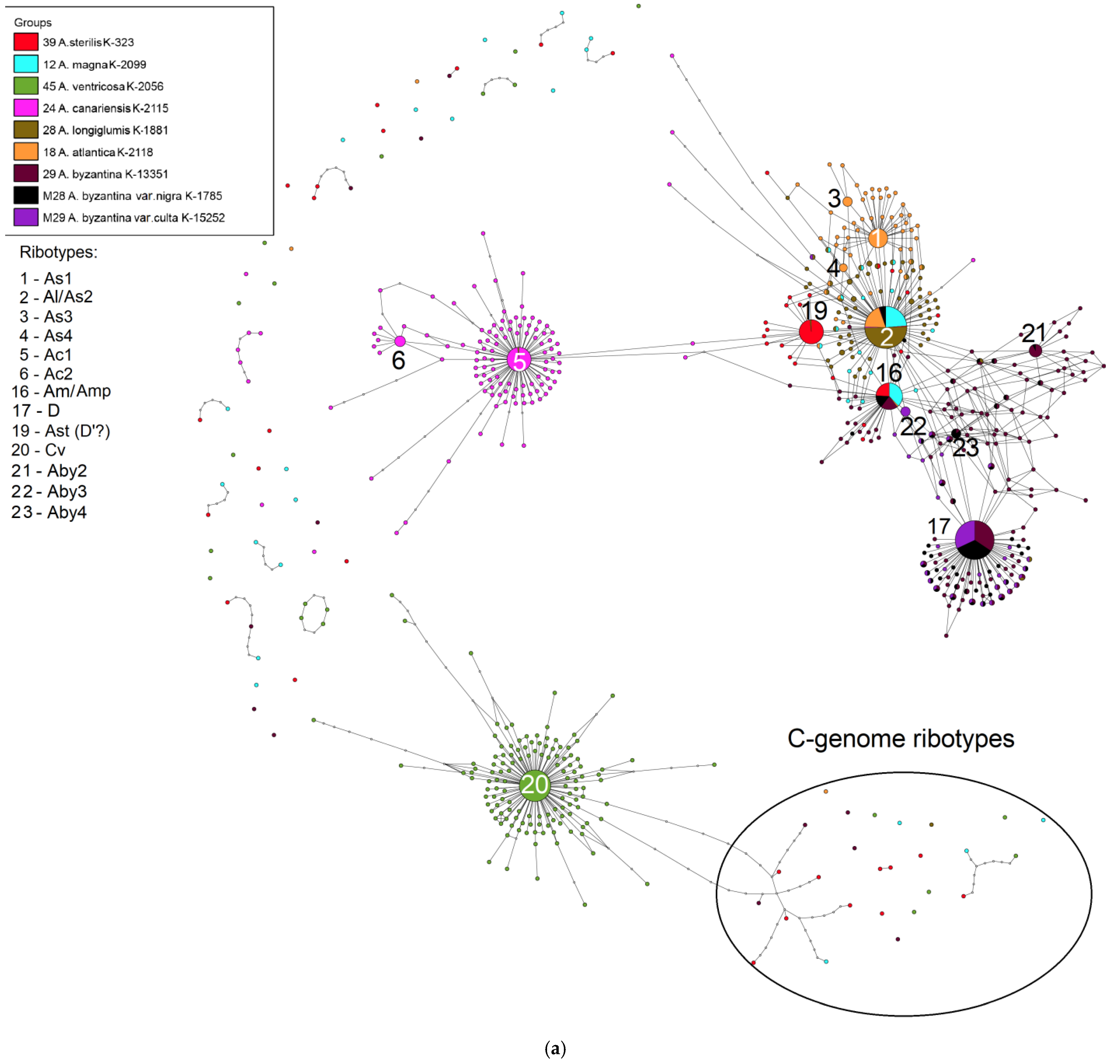
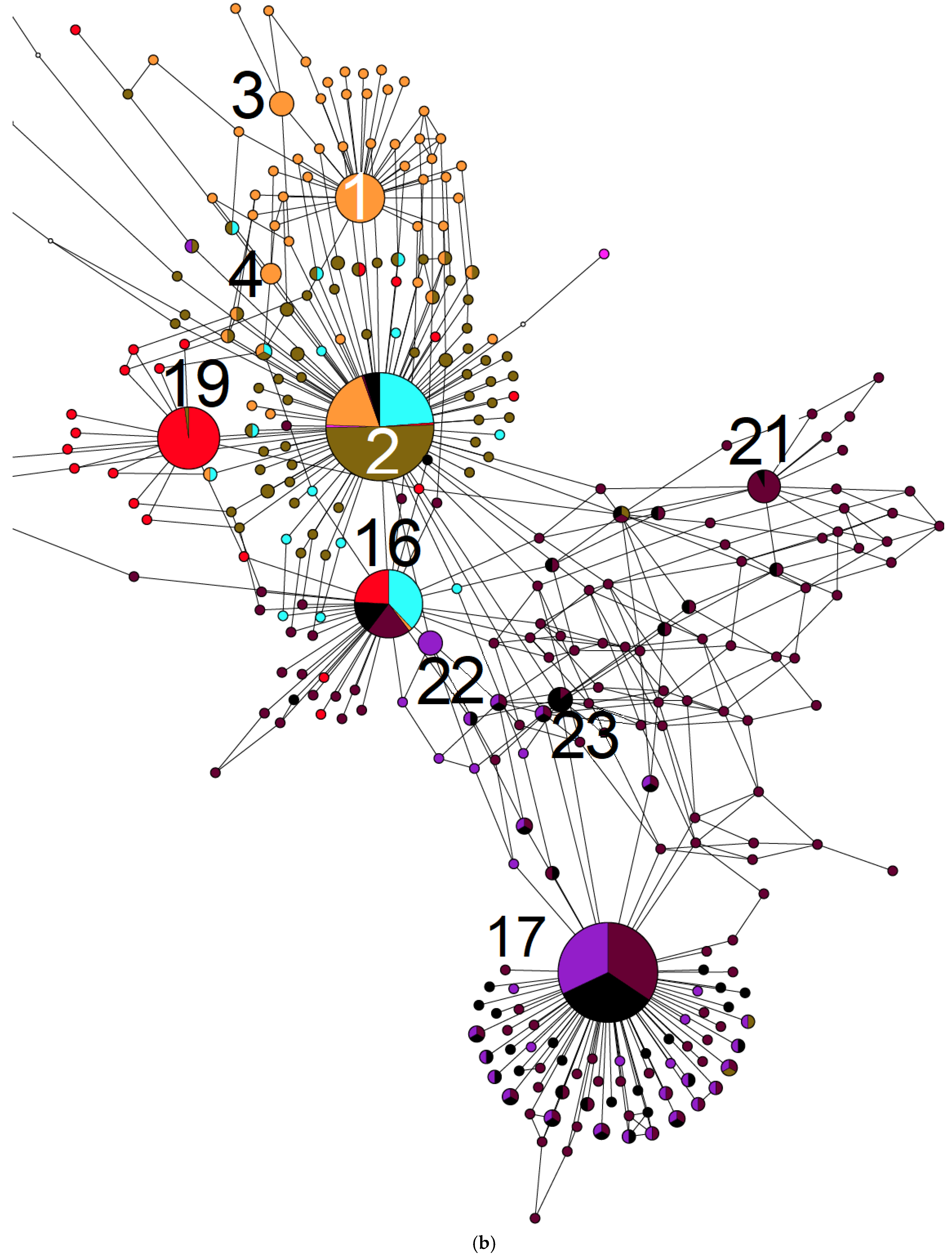
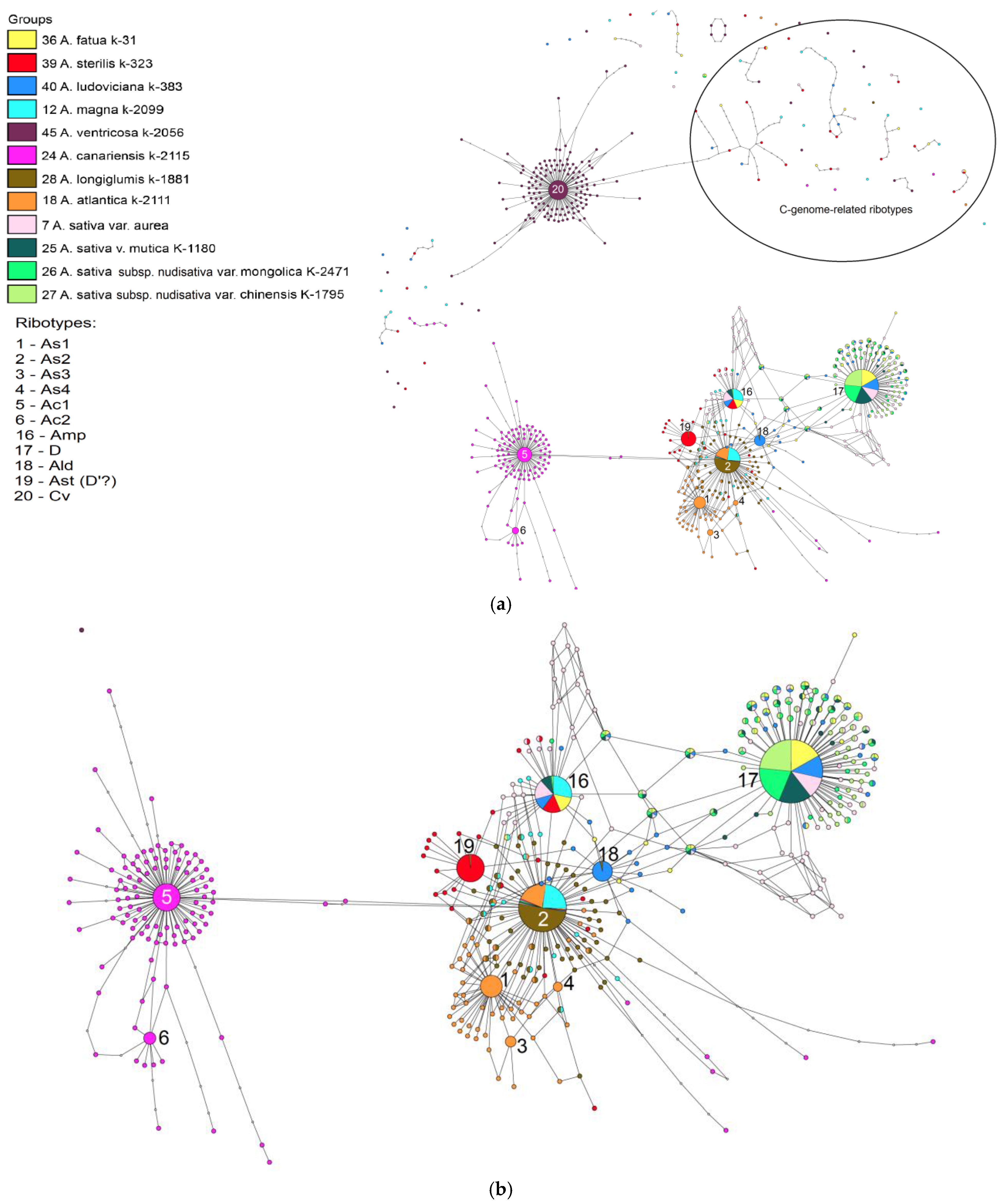
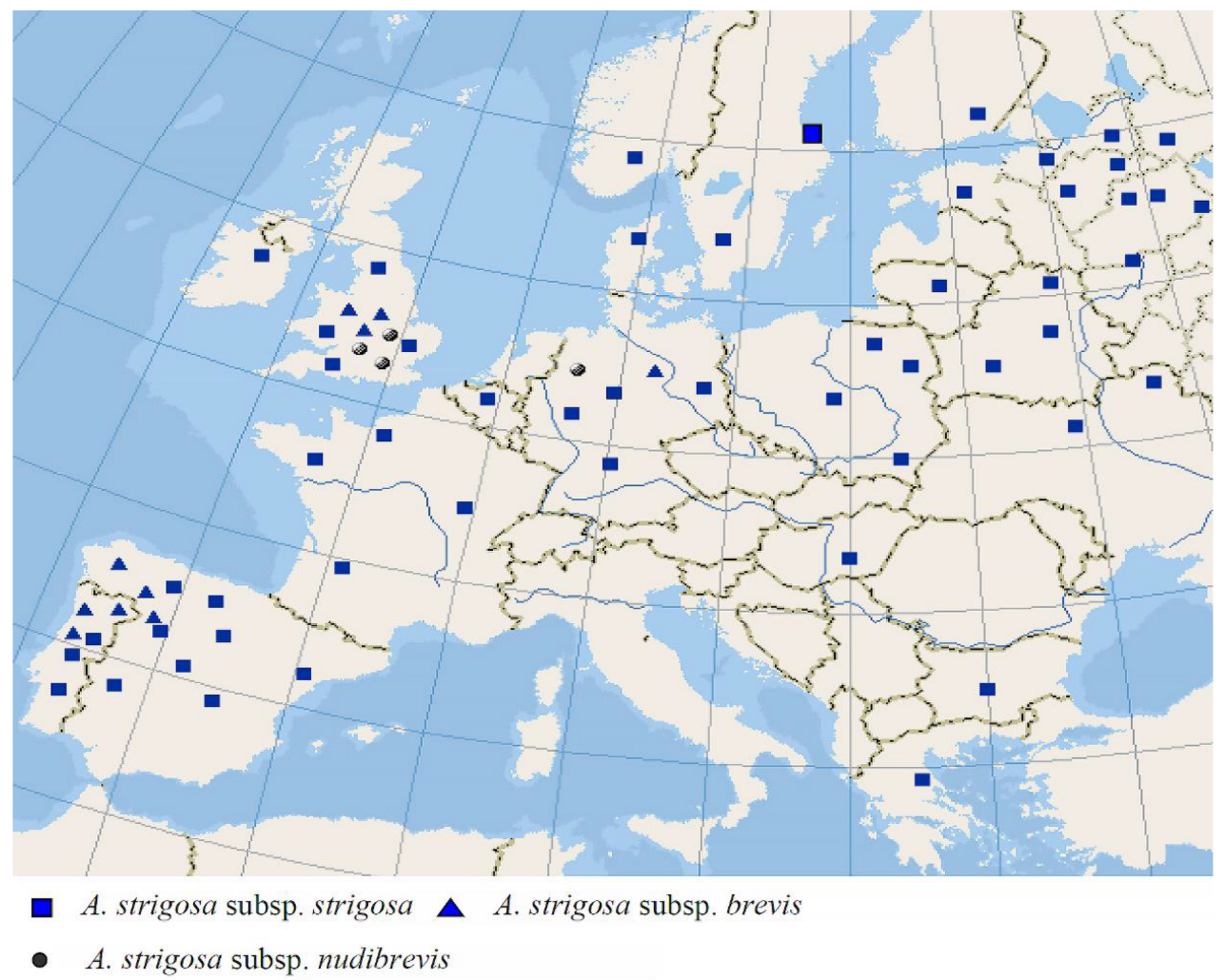
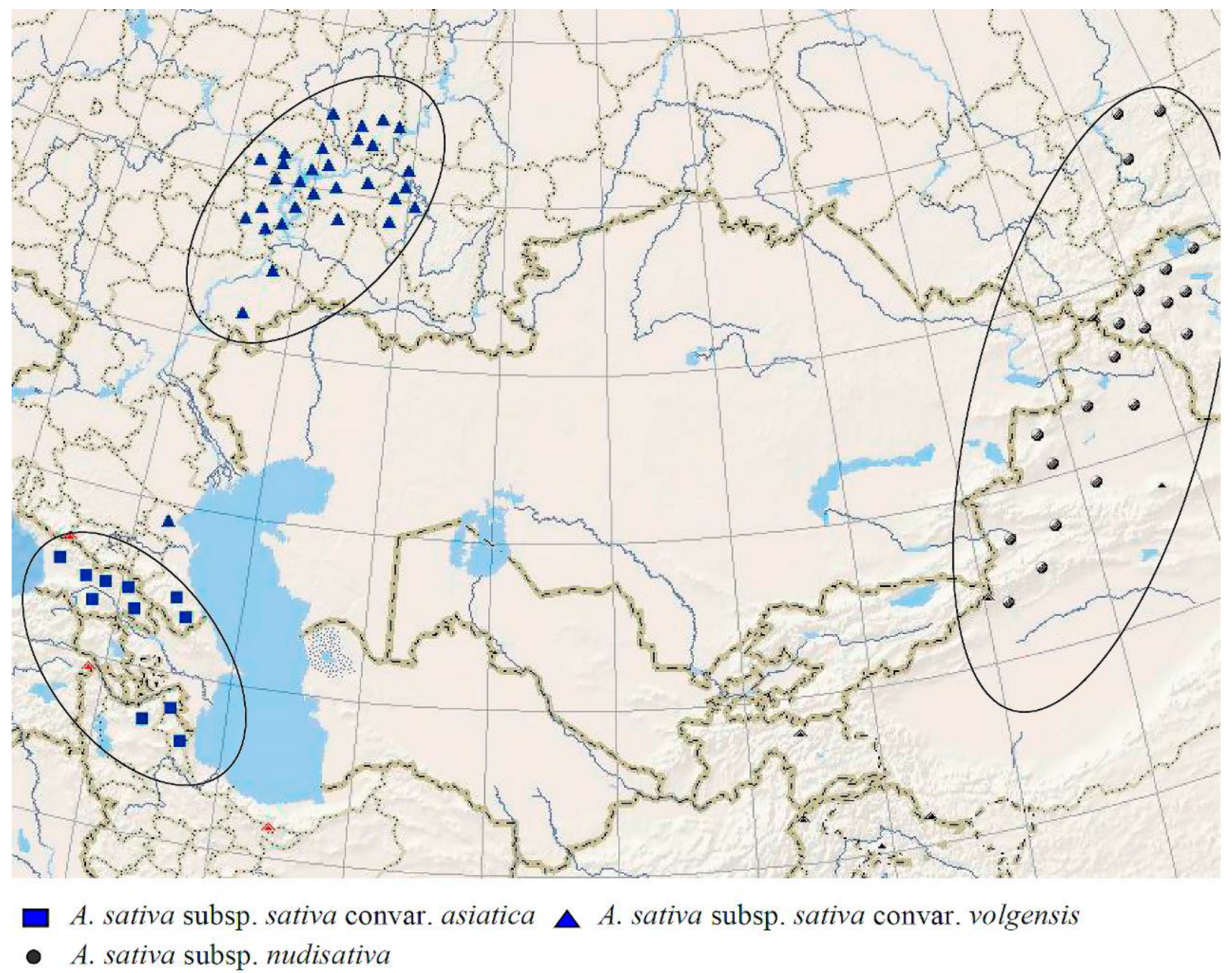
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Extraction, Amplification, and Sequencing
4.3. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodionova, N.A.; Soldatov, V.N.; Merezhko, V.E.; Yarosh, N.P.; Kobylyansky, V.D. Oat. In Cultivated Flora of the USSR; Soldatov, V.N., Kobylyansky, V.D., Eds.; Kolos: Moscow, Russia, 1994; 367p. (In Russian) [Google Scholar]
- Loskutov, I.G. Oat (Avena L.). Distribution, Taxonomy, Evolution and Breeding Value; VIR: St. Petersburg, Russia, 2007; 336p. (In Russian) [Google Scholar]
- Loskutov, I.G.; Rines, H.W. Avena. In Wild Crop Relatives: Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Chapter 3; pp. 109–183. [Google Scholar]
- Leggett, J.M.; Markland, G.S. The genomic structure of Avena revealed by GISH. In Kew Chromosome Conference IV; Brandham, P.E., Bennett, M.D., Eds.; Royal Botanic Gardens: Kew, UK, 1995; pp. 133–139. [Google Scholar]
- Mitchell, C.C.; Parkinson, S.E.; Baker, T.J.; Jellen, E.N. C-banding and localization of 18S–5.8S S-26S rDNA in tall oatgrass species. Crop. Sci. 2003, 43, 32–36. [Google Scholar] [CrossRef]
- Tomaszewska, P.; Schwarzacher, T.; Heslop-Harrison, J.S. Oat chromosome and genome evolution defined by widespread terminal intergenomic translocations in polyploids. Front. Plant Sci. 2022, 13, 1026364. [Google Scholar] [CrossRef]
- Rajhathy, T.; Thomas, H. Cytogenetics of Oats (Avena L.) Miscellaneous Publications of the Genetics Society of Canada 2; Ontario (Canada) Genetics Society of Canada: Ottawa, ON, Canada, 1974; 90p. [Google Scholar]
- Fominaya, A.; Vega, C.; Ferrer, E. Giemsa C-banded karyotypes of Avena species. Genome 1988, 30, 627–632. [Google Scholar] [CrossRef]
- Drossou, A.; Katsiotis, A.; Leggett, J.M.; Loukas, M.; Tsakas, S. Genome and species relationships in genus Avena based on RAPD and AFLP molecular markers. Theor. Appl. Gen. 2004, 109, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Shelukhina, O.; Diederichsen, A.; Loskutov, I.G.; Pukhalskiy, V.A. Comparative cytogenetic analysis of Avena macrostachya and diploid C-genome Avena species. Genome 2010, 53, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Shelukhina, O.Y.; Goryunova, S.V.; Loskutov, I.G.; Pukhalskiy, V.A. Phylogenetic relationships of tetraploid AB genome Avena species evaluated by means of cytogenetic (C-banding and FISH) and RAPD analyses. J. Bot. 2010, 2010, 742307. [Google Scholar] [CrossRef]
- Yan, H.; Ren, Z.; Deng, D.; Yang, K. New evidence confirming the CD genomic constitutions of the tetraploid Avena species in the section Pachycarpa Baum. PLoS ONE 2021, 16, e0240703. [Google Scholar] [CrossRef]
- Murray, B.E.; Craig, J.L.; Rajhathy, T. A protein electrophoretic study of three amphiploids and eight species in Avena. Can. J. Genet. Cytol. 1970, 12, 651–655. [Google Scholar] [CrossRef]
- Ladizinsky, G. The cytogenetic position of Avena prostrata among the diploid oats. Can. J. Genet. Cytol. 1973, 15, 443–450. [Google Scholar] [CrossRef]
- Rajhathy, T. A standard kariotype for Avena sativa. Can. J. Genet. Cytol. 1963, 5, 127–132. [Google Scholar] [CrossRef]
- Fabijanski, S.; Fedak, G.; Armstrong, K.; Altosaar, I. A repeated sequence probe for the C genome in Avena (oats). Theor. Appl. Genet. 1990, 79, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Katsiotis, A.; Hagidimitriou, M.; Heslop-Harrison, J.S. The close relationship between the A and B genomes in Avena L. (Poaceae) determined by molecular cytogenetic analysis of total genomic, tandemly and dispersed repetitive DNA sequences. Ann. Bot. 1997, 79, 103–109. [Google Scholar] [CrossRef]
- Yan, H.-H.; Baum, B.R.; Zhou, P.-P.; Zhao, J.; Wei, Y.-M.; Ren, C.-Z.; Xiong, F.-Q.; Liu, G.; Zhong, L.; Zhao, G.; et al. Phylogenetic analysis of the genus Avena based on chloroplast intergenic spacer psbA-trnH and single-copy nuclear gene Acc1. Genome 2014, 57, 267–277. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, P.; Zhao, J.; Li, J.; Lai, S.; Tinker, N.A.; Liao, S.; Yan, H. Phylogenetic relationships in the genus Avena based on the nuclear Pgk1 gene. PLoS ONE 2018, 13, e0200047. [Google Scholar] [CrossRef]
- Ladizinsky, G. A new species of Avena from Sicily, possible the tetraploid progenitor of hexaploid oats. Genet. Resour. Crop Evol. 1998, 45, 263–269. [Google Scholar] [CrossRef]
- Li, C.D.; Rossnagel, B.G.; Scoles, G.J. The development of oar microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor. Appl. Genet. 2000, 101, 1259–1268. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Blinova, E.V.; Shneyer, V.S.; Rodionov, A.V. Origin of Wild Polyploid Avena Species Inferred from Polymorphism of the ITS1 rDNA in Their Genomes. Diversity 2023, 15, 717. [Google Scholar] [CrossRef]
- Brassac, J.; Blattner, F.R. Species-Level Phylogeny and Polyploid Relationships in Hordeum (Poaceae) Inferred by Next-Generation Sequencing and In Silico Cloning of Multiple Nuclear Loci. Syst. Biol. 2015, 64, 792–808. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, A.V.; Gnutikov, A.A.; Nosov, N.N.; Machs, E.M.; Mikhaylova, Y.V.; Shneyer, V.S.; Punina, E.O. Intragenomic Polymorphism of the ITS 1 Region of 35S rRNA Gene in the Group of Grasses with Two-Chromosome Species: Different Genome Composition in Closely Related Zingeria Species. Plants 2020, 9, 1647, Erratum in Plants 2021, 10, 463. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Koroleva, T.M.; Punina, E.O.; Probatova, N.S.; Shneyer, V.S.; Rodionov, A.V. Origin of the rare hybrid genus ×Trisetokoeleria Tzvelev (Poaceae) according to molecular phylogenetic data. Plants 2022, 11, 3533. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Punina, E.O.; Loskutov, I.G.; Shneyer, V.S.; Chekrygin, S.A.; Rodionov, A.V. Hybridization in the subtribe Alopecurinae Dumort. (Poaceae) according to molecular phylogenetic analysis: Different ploidy level tells different origin of the groups. Plants 2024, 13, 919. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, B.G.; Sanderson, M.J.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S.; Donoghue, M.J. The ITS Region of Nuclear Ribosomal DNA: A Valuable Source of Evidence on Angiosperm Phylogeny. Ann. Mis. Bot. Gard. 1995, 82, 247–277. [Google Scholar] [CrossRef]
- Mahelka, V.; Krak, K.; Kopecký, D.; Fehrer, J.; Šafář, J.; Bartoš, J.; Hobza, R.; Blavet, N.; Blattner, F.R. Multiple horizontal transfers of nuclear ribosomal genes between phylogenetically distinct grass lineages. Biol. Sci. 2017, 114, 1726–1731. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.D.P.N.; Riina, R.; Valduga, E.; Caruzo, M.B.R. A new species of Croton (Euphorbiaceae) endemic to the Brazilian Pampa and its phylogenetic affinities. Pl. Syst. Evol. 2022, 308, 14. [Google Scholar] [CrossRef]
- Suyama, Y.; Hirota, S.K.; Matsuo, A.; Tsunamoto, Y.; Mitsuyuki, C.; Shimura, A.; Okano, K. Complementary combination of multiplex high-throughput DNA sequencing for molecular phylogeny. Ecol. Res. 2022, 37, 171–181. [Google Scholar] [CrossRef]
- Kropać, Z. Avena strigosa – a disappearing synanthropic species in Czechoslovakia. Preslia 1981, 53, 305–321. [Google Scholar]
- Podyma, W.; Bolc, P.; Nocen, J.; Puchta, M.; Wlodarczyk, S.; Lapinski, B.; Boczkowska, M. A multilevel exploration of Avena strigosa diversity as a prelude to promote alternative crop. BMC Plant Biol. 2019, 19, 291. [Google Scholar] [CrossRef]
- Schreber, I. Spicilegium Florae Lipsicae; Prostat in Bibliopolio Dykiano: Leipzig, Germany, 1771; p. 52. [Google Scholar]
- Badaeva, E.D.; Loskutov, I.G.; Shelukhina, O.Y.; Pukhalskiy, V.A. Cytogenetic analysis of diploid Avena L. species containing the as genome. Rus. J. Genet. 2005, 41, 1428–1433. [Google Scholar] [CrossRef]
- Emme, E.K. Evolution of the cultivated and wild oats. Bot. Zhurn. 1938, 7, 91–122. (In Russian) [Google Scholar]
- Rajhathy, T. Evidence and an hypothesis for the origin of the C genome of hexaploid Avena. Can. J. Gen. Cytol. 1966, 8, 774–779. [Google Scholar] [CrossRef]
- Steer, M.W.; Holden, J.H.W.; Gunning, B.E.S. Avena chloroplasts: Species relationships and the occurrence of stromacentres. Can. J. Gen. Cytol. 1970, 12, 21–28. [Google Scholar] [CrossRef]
- Tomás, D.; Rodrigues, J.; Varela, A.; Veloso, M.M.; Viegas, W.; Silva, M. Use of repetitive sequences for molecular and cytogenetic characterization of Avena species from Portugal. Int. J. Mol. Sci. 2016, 17, 203. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Machs, E.M.; Blinova, E.V.; Probatova, N.S.; Langdon, T.; Rodionov, A.V. New insights into the genomic structure of the oats (Avena L., Poaceae): Intragenomic polymorphism of ITS1 sequences of rare endemic species Avena bruhnsiana Gruner and its relationship to other species with C-genomes. Euphytica 2022, 218, 3. [Google Scholar] [CrossRef]
- Baum, B.R. Avena occidentalis, a hitherto overlooked species of oats. Can. J. Bot. 1971, 49, 1055–1057. [Google Scholar] [CrossRef]
- Rajhathy, T.; Morrison, J.W. Chromosome morphology in the genus Avena. Can. J. Bot. 1959, 37, 331–337. [Google Scholar] [CrossRef]
- Linares, C.; Vega, C.; Ferrer, E.; Fominaya, A. Identification of C-banded chromosomes in meiosis and the analysis of nucleolar activity in Avena byzantina C. Koch cv ‘Kanota’. Theor. Appl. Genet. 1992, 83, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Shelukhina, O.Y.; Dedkova, O.S.; Loskutov, I.G.; Pukhalskyi, V.A. Comparative cytogenetic analysis of hexaploid Avena L. species. Rus. J. Gen. 2011, 47, 691–702. [Google Scholar] [CrossRef]
- Chaffin, A.S.; Huang, Y.F.; Smith, S.; Bekele, W.A.; Babiker, E.; Gnanesh, B.N.; Foresman, B.J.; Blanchard, S.G.; Jay, J.J.; Reid, R.W.; et al. A Consensus Map in Cultivated Hexaploid Oat Reveals Conserved Grass Synteny with Substantial Subgenome Rearrangement. Pl. Genome 2016, 9, 2. [Google Scholar] [CrossRef]
- Trabut, L. Contribution a l’etude de l’origine des Avoines cultivees. Compt.-Rend. Acad. Sc. Paris 1909, CXLIX, 227. [Google Scholar]
- Trabut, L. Origin of cultivated oats. J. Hered. 1914, 5, 74–85. [Google Scholar] [CrossRef]
- Vavilov, N.I. Immunity of Plants to Infectious Diseases; Printing House of the Ryabushinsky Partnership: Moscow, Russia, 1918; 239p. (In Russian) [Google Scholar]
- Vavilov, N.I. World centers of varietal wealth (genes) of cultivated plants. News State Inst. Exp. Agron. 1927, 5, 339–351. (In Russian) [Google Scholar]
- Malzew, A.I. Wild and Cultivated Oat. Sectio Euavena Griseb; Publishers of the All-Union Institute of Applied Botany and New Cultures under the Council of People’s Commissars of the USSR: Leningrad, Russia, 1930; 522p. (In Russian) [Google Scholar]
- Coffman, F.A. Oat History, Identification and Classification; USDA: Washington, DC, USA, 1977; 357p.
- Koernicke, F.; Werner, H. Die Arten und Varietaten des Getreides; Handbuch des Getrei-debaues I.: Berlin, Germany, 1885; 738p. [Google Scholar]
- Romero Zarco, C. Sinopsis del genero Avena L. (Poaceae, Avenae) en España peninsular y Baleares. Lagascalia 1996, 18, 171–198. [Google Scholar]
- Loskutov, I.G.; Gnutikov, A.A.; Blinova, E.V.; Rodionov, A.V. The application of Vavilov’s approaches to the phylogeny and evolution of cultivated species of the genus Avena L. Vavilov J. Gen. Sel. 2023, 27, 921–932. [Google Scholar] [CrossRef]
- Linnaeus, C. Species Plantarum. Vol. I; L. Salvius: Stockholm, Sweden, 1753; 560p. [Google Scholar]
- Haussknecht, C. Über die Abstammung des Saathabers. Mitteil. d. geogr. Gesellsch. Jena 1885, 3, 231–242. [Google Scholar]
- Shepeleva, E.M. Karyological research of cultivated and wild oat species. Rep. AS USSR 1939, 25, 215–218. [Google Scholar]
- Nikoloudakis, N.; Skaracis, G.A.; Katsiotis, A. Evolutionary insights inferred by molecular analysis of the ITS1-5.8S-ITS2 and IGS Avena sp. sequences. Mol. Phyl. Evol. 2008, 46, 102–115. [Google Scholar] [CrossRef]
- Fu, Y.B. Oat evolution revealed in the maternal lineages of 25 Avena species. Sci. Rep. 2018, 8, 4252. [Google Scholar] [CrossRef]
- Rajhathy, T. Chromosome polymorphism in Avena ventricosa. Chromosoma 1971, 35, 206–216. [Google Scholar] [CrossRef]
- Musaev, S.G.; Isaev, Y.M. Bruns’ Oat – an endemic species of the flora of Azerbaijan. Rep. Azerb. SSR 1971, 27, 64–65. (In Russian) [Google Scholar]
- Zhang, M.; Tang, Y.W.; Xu, Y.; Yonezawa, T.; Shao, Y.; Wang, Y.G.; Song, Z.-P.; Yang, J.; Zhang, W.J. Concerted and birth-and-death evolution of 26S ribosomal DNA in Camellia L. Ann. Bot. 2021, 127, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Rajhathy, T.; Zillinsky, F.J.; Hayes, J.D. A Collection of Wild Oat Mediterranean Region; Can Depart. Agric. Res.: Ottawa, ON, Canada, 1966; 25p.
- Perez de la Vega, M. Plant genetic adaptedness to climatic and edaphic environment. In Adaptation in Plant Breeding; Tigerstedt, P.M.A., Ed.; Kluwer Academic Publishers: Amsterdam, The Netherlands, 1997; pp. 27–38. [Google Scholar]
- Thomas, H. 29. Oats. Avena spp. (Gramineae–Aveneae). In Evolution of Crop Plants; Smartt, J., Simmonds, N.W., Eds.; Longman Group: New York, NY, USA, 1995; pp. 132–137. [Google Scholar]
- Holden, J.H.W. 28. Oats. Avena spp. (Gramineae–Aveneae). In Evolution of Crop Plants; Simmonds, N.W., Ed.; Longman Group: New York, NY, USA, 1979; pp. 86–90. [Google Scholar]
- Ridgway, K.P.; Duck, J.M.; Young, J.P.W. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol. 2003, 3, 8. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]






| Species | Sample ID | Country of Origin | Accession Number |
|---|---|---|---|
| Avena strigosa subsp. brevis var. candida | K-4480 | United Kingdom: Wales | PV391964–PV392013 |
| Avena strigosa var. strigosa | K-15025 | Canada | PV392014–PV392035 |
| Avena strigosa subsp. nudibrevis | K-14943 | United Kingdom | PV392036–PV392071 |
| Avena strigosa subsp. brevis var. candida | K-5233 | Portugal: Aveiro, north from Coimbra | PV392072–PV392136 |
| Avena strigosa var. strigosa | K-5195 | Spain: Los Nogales, Lugo, Galicia | PV392137–PV392197 |
| Avena abyssinica var. schimperi | K-14826 | Ethiopia | PV392232–PV392262 |
| Avena abyssinica var. braunii | K-11678 | Ethiopia | PV392263–PV392291 |
| Avena abyssinica var. braunii | K-14811 | Ethiopia | PV392292–PV392371 |
| Avena byzantina | K-13351 | Spain | PV397121–PV397264 |
| Avena byzantina var. nigra | K-1785 | USA | PV397265–PV397313 |
| Avena byzantina var. culta | K-15252 | Tunisia | PV397314–PV397353 |
| Avena sativa var. aurea | K-14787 | Russia: Moscow Oblast | PV429768–PV429858 |
| Avena sativa var. mutica | K-1180 | Russia: Tambov Oblast | PV429859–PV429881 |
| Avena sativa subsp. nudisativa var. mongolica | K-2471 | Mongolia | PV429882–PV429916 |
| Avena sativa subsp. nudisativa var. chinensis | K-1795 | USA | PV429917–PV429978 |
| Species | Genome | Total Number of Reads | Ribotype Symbol (of the Major Ribotypes) | Number of Reads | % from the Total Number of Reads |
|---|---|---|---|---|---|
| Avena strigosa subsp. brevis var. candida | As | 21,927 | Ad2/As3 | 8781 | 40 |
| Al/As2 | 2412 | 11 | |||
| As5 | 2325 | 11 | |||
| As6 | 1703 | 8 | |||
| Avena strigosa var. strigosa | As | 15,373 | Ad2/As3 | 6572 | 43 |
| Al/As2 | 1691 | 11 | |||
| As5 | 1543 | 10 | |||
| As6 | 1220 | 8 | |||
| Avena strigosa subsp. nudibrevis | As | 20,746 | Ad2/As3 | 9087 | 44 |
| Al/As2 | 2282 | 11 | |||
| As5 | 2135 | 10 | |||
| As6 | 1628 | 8 | |||
| Avena strigosa subsp. brevis var. candida | As | 24,714 | Ad2/As3 | 7977 | 32 |
| As5 | 3707 | 15 | |||
| Al/As2 | 2962 | 12 | |||
| As6 | 1995 | 8 | |||
| Avena strigosa var. strigosa | As | 15,576 | Ad2/As3 | 9069 | 58 |
| As5 | 4517 | 29 | |||
| Al/As2 | 3642 | 23 | |||
| As6 | 2493 | 16 | |||
| Avena abyssinica var. schimperi | AB | 14,399 | B5 | 3291 | 23 |
| As1 | 3039 | 21 | |||
| B6 | 1042 | 7 | |||
| Avena abyssinica var. braunii K-11678 | AB | 15,270 | As1 | 5337 | 35 |
| B5 | 2387 | 16 | |||
| B7 | 1142 | 5 | |||
| B6 | 1108 | 4 | |||
| Avena abyssinica var. braunii K-14811 | AB | 21,862 | As1 | 10,877 | 49 |
| As2 | 4151 | 19 | |||
| B7 | 2472 | 11 | |||
| B5 | 1266 | 6 | |||
| B6 | 1012 | 5 | |||
| Avena byzantina | ACD | 23,292 | D | 9935 | 43 |
| Aby2 | 2716 | 12 | |||
| Am/Amp | 2619 | 12 | |||
| Avena byzantina var. nigra | ACD | 18,956 | D | 7961 | 42 |
| Am/Amp | 1458 | 8 | |||
| Al/As2 | 1054 | 6 | |||
| Aby4 | 1010 | 5 | |||
| Avena byzantina var. culta | ACD | 18,761 | D | 7425 | 40 |
| Aby3 | 1269 | 7 | |||
| Avena sativa var. aurea | ACD | 23,663 | D | 7623 | 32 |
| Am/Amp | 3786 | 16 | |||
| Avena sativa var. mutica | ACD | 13,886 | D | 5971 | 43 |
| Am/Amp | 1138 | 8 | |||
| Avena sativa subsp. nudisativa var. mongolica | ACD | 16,539 | D | 8600 | 52 |
| Avena sativa subsp. nudisativa var. chinensis | ACD | 19,272 | D | 11,563 | 60 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | |||||||||
| 2 | 3 | 4 | 5 | 6 | 8 | 9 | 9 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 3 | 3 | 3 | 3 | 4 | 6 | 7 | 7 | 7 | 7 | 8 | 9 | 9 | 0 | 0 | 0 | 1 | 2 | 2 | 3 | 4 | 4 | 4 | 4 | 5 | 5 | 5 | 6 | 6 | 6 | 7 | 7 | 0 | 0 | 1 | |
| 9 | 6 | 3 | 6 | 2 | 1 | 4 | 6 | 2 | 1 | 5 | 6 | 8 | 2 | 6 | 0 | 5 | 7 | 8 | 3 | 1 | 1 | 4 | 5 | 7 | 4 | 4 | 8 | 3 | 4 | 8 | 4 | 3 | 5 | 7 | 0 | 3 | 4 | 7 | 2 | 7 | 9 | 2 | 5 | 9 | 7 | 9 | 4 | 7 | 6 | |
| As1 | C | A | C | G | G | G | C | A | C | T | T | G | T | T | G | A | D | T | C | A | C | T | A | G | G | T | G | C | G | C | G | G | C | C | T | G | C | A | A | C | T | G | G | C | T | A | G | C | D | T |
| Al/As2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | D | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| Ad2/As3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| As5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | D | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| As6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| B5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | D | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| B6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| B7 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | D | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| D | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | C | D | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| Aby2 | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | C | D | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| Aby3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | D | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| Aby4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | D | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| Am/Amp | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | D | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . |
| A. byzantina C-genome-related | T | G | T | A | T | A | T | T | T | D | D | . | C | C | A | C | C | . | T | G | A | C | G | A | . | C | T | A | A | T | A | A | T | T | C | T | T | G | G | A | C | T | A | T | G | C | A | D | C | C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Rodionov, A.V.; Shneyer, V.S. Participation of Wild Species Genus Avena L. (Poaceae) of Different Ploidy in the Origin of Cultivated Species According to Data on Intragenomic Polymorphism of the ITS1-5.8S rRNA Region. Plants 2025, 14, 1550. https://doi.org/10.3390/plants14101550
Gnutikov AA, Nosov NN, Loskutov IG, Rodionov AV, Shneyer VS. Participation of Wild Species Genus Avena L. (Poaceae) of Different Ploidy in the Origin of Cultivated Species According to Data on Intragenomic Polymorphism of the ITS1-5.8S rRNA Region. Plants. 2025; 14(10):1550. https://doi.org/10.3390/plants14101550
Chicago/Turabian StyleGnutikov, Alexander A., Nikolai N. Nosov, Igor G. Loskutov, Alexander V. Rodionov, and Victoria S. Shneyer. 2025. "Participation of Wild Species Genus Avena L. (Poaceae) of Different Ploidy in the Origin of Cultivated Species According to Data on Intragenomic Polymorphism of the ITS1-5.8S rRNA Region" Plants 14, no. 10: 1550. https://doi.org/10.3390/plants14101550
APA StyleGnutikov, A. A., Nosov, N. N., Loskutov, I. G., Rodionov, A. V., & Shneyer, V. S. (2025). Participation of Wild Species Genus Avena L. (Poaceae) of Different Ploidy in the Origin of Cultivated Species According to Data on Intragenomic Polymorphism of the ITS1-5.8S rRNA Region. Plants, 14(10), 1550. https://doi.org/10.3390/plants14101550









