Investigation of the Jasmonate ZIM-Domain Family Reveals PavJAZ8 Regulates Fruit Aroma Traits in Sweet Cherry (Prunus avium L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Phytohormone Treatments
2.2. Identification and Classification of JAZ Gene Family in Sweet Cherry
2.3. Chromosomal Location and Phylogenetic Analysis of PavJAZs
2.4. Structural and Domain Analysis of PavJAZs
2.5. RNA Extraction and Quantitative Real-Time PCR (RT-qPCR) Analysis
2.6. Gas Chromatography-Mass Spectrometry Analysis of Volatile Metabolites
2.7. Transient Fruit Transformation
2.8. Subcellular Localization Analysis
2.9. Bimolecular Fluorescence Complementation
2.10. GST Pull-Down Assay
3. Results
3.1. Identification and Classification of the PavJAZ Genes
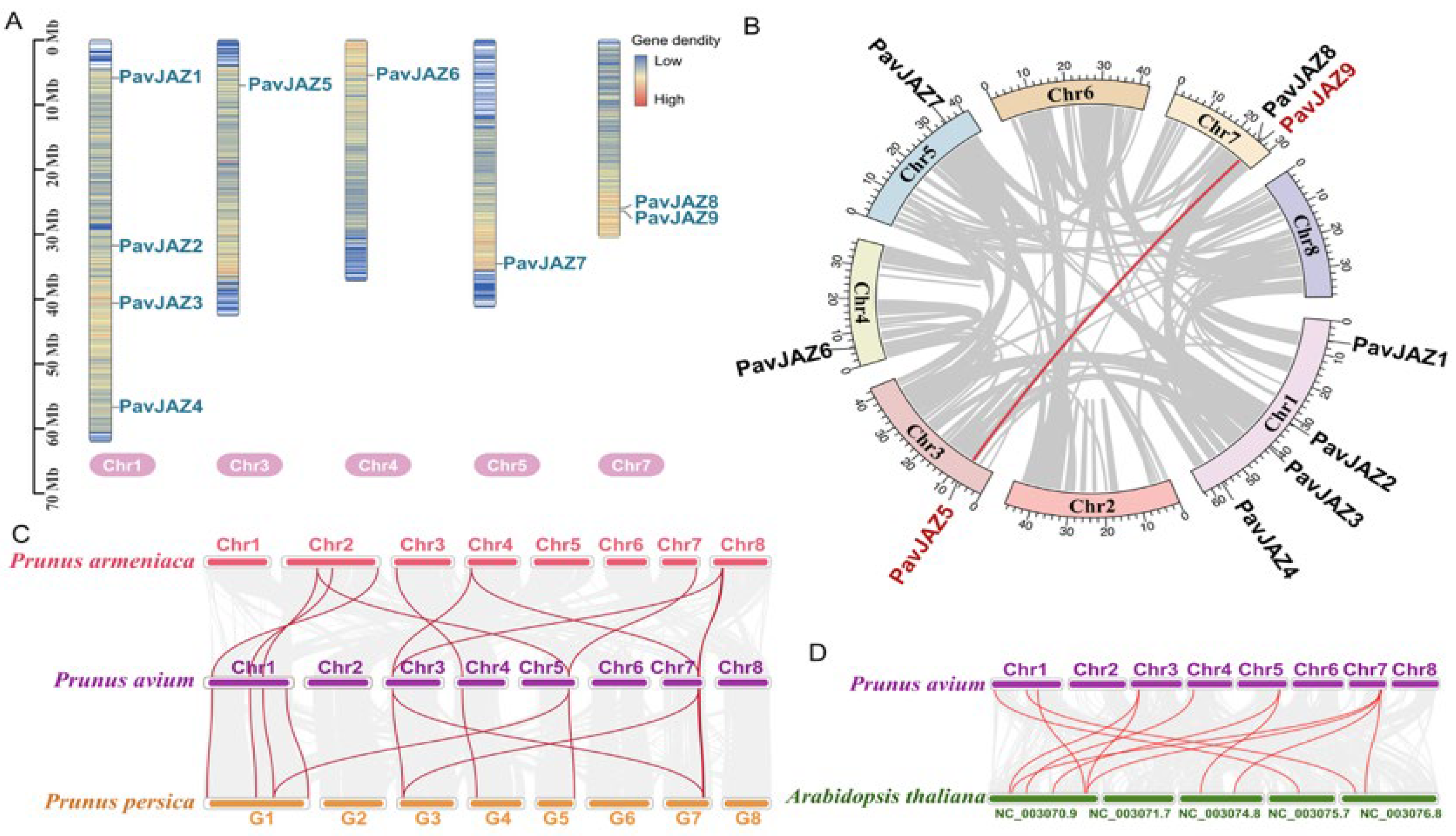
3.2. Chromosomal Locations and Collinearity Analysis of PavJAZs
3.3. Phylogenetic Analysis of JAZs
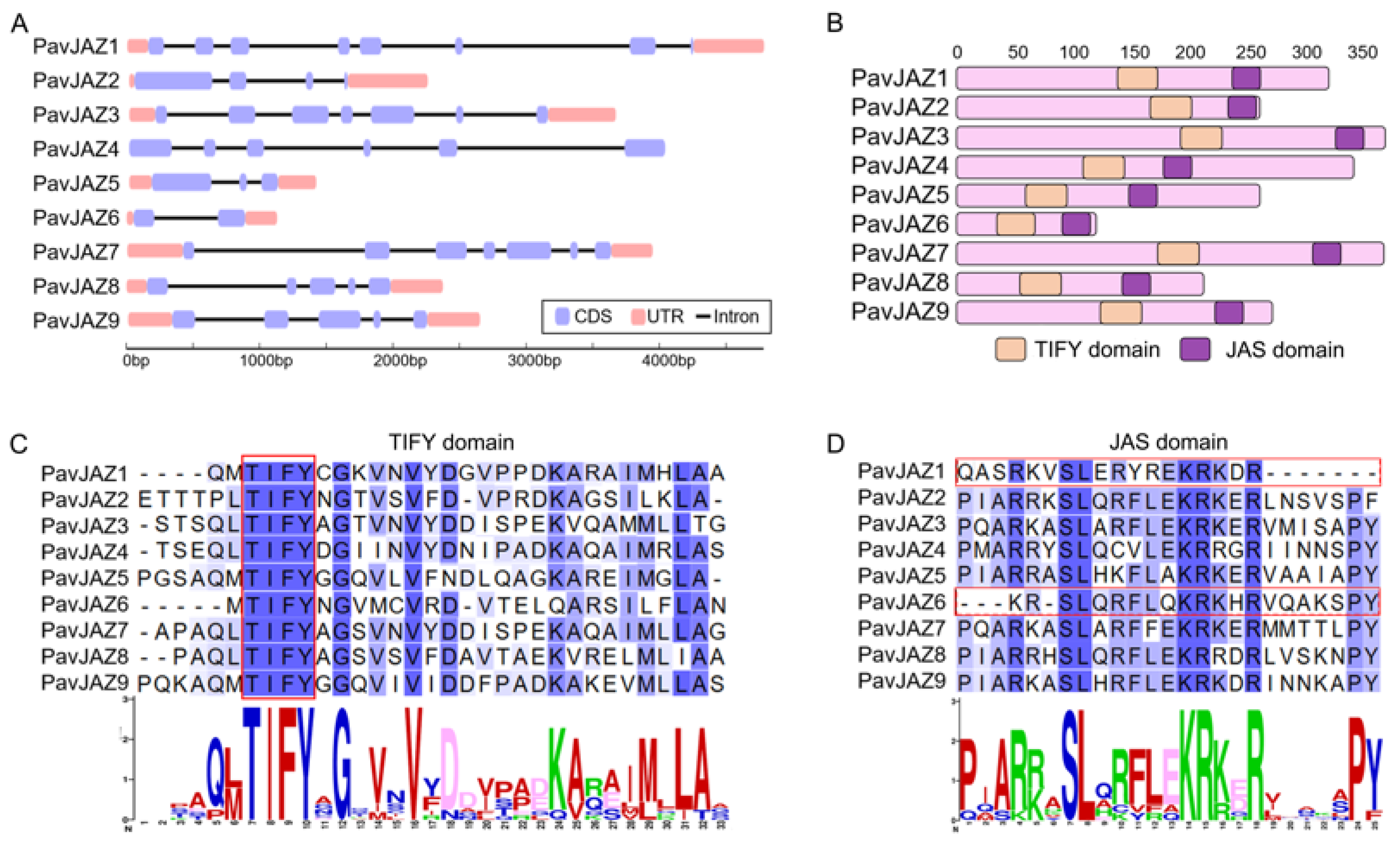
3.4. Protein Structure, Conserved Domains Analysis of PavJAZs
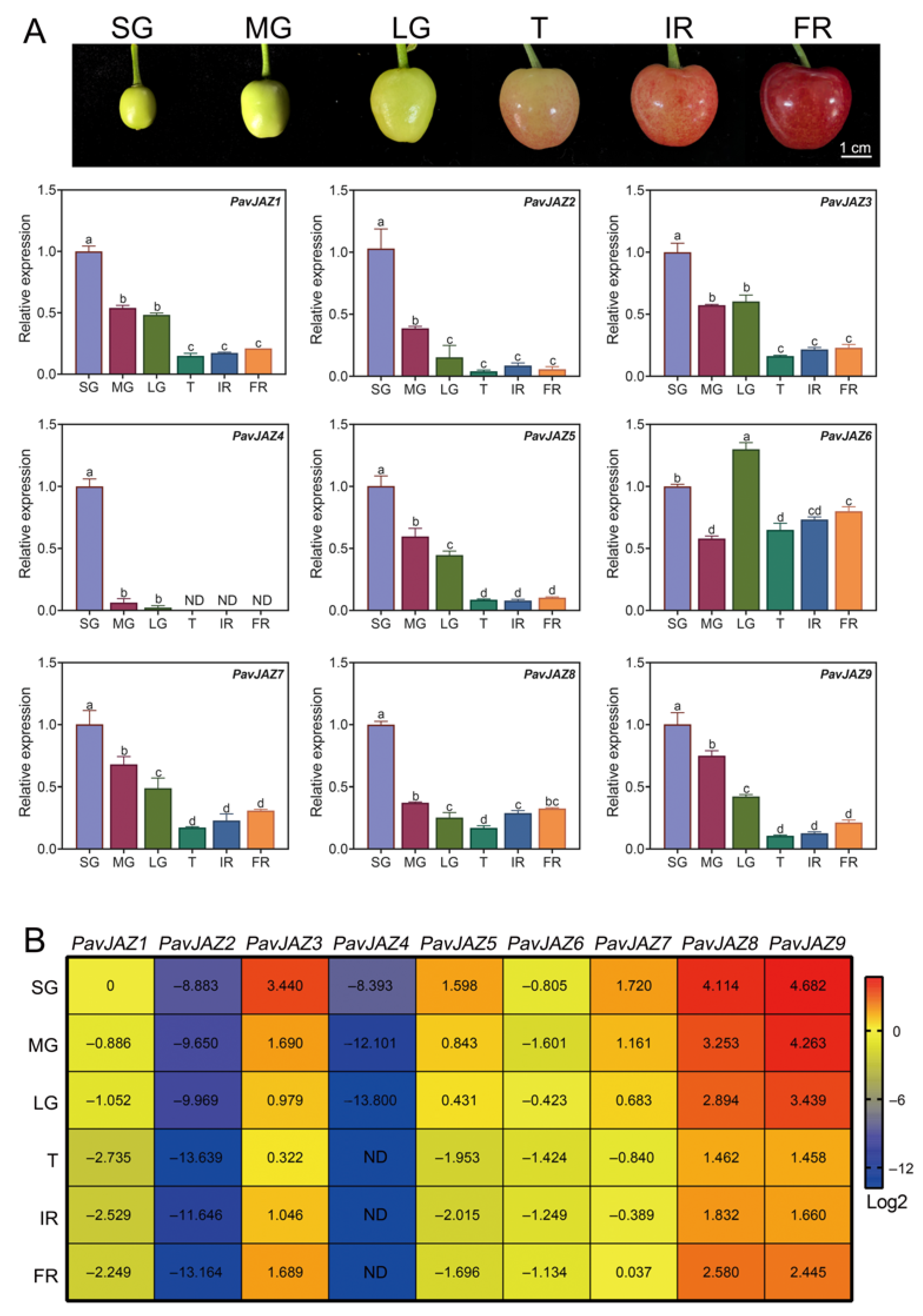
3.5. Expression Analysis of the PavJAZs in Different Fruit Stages
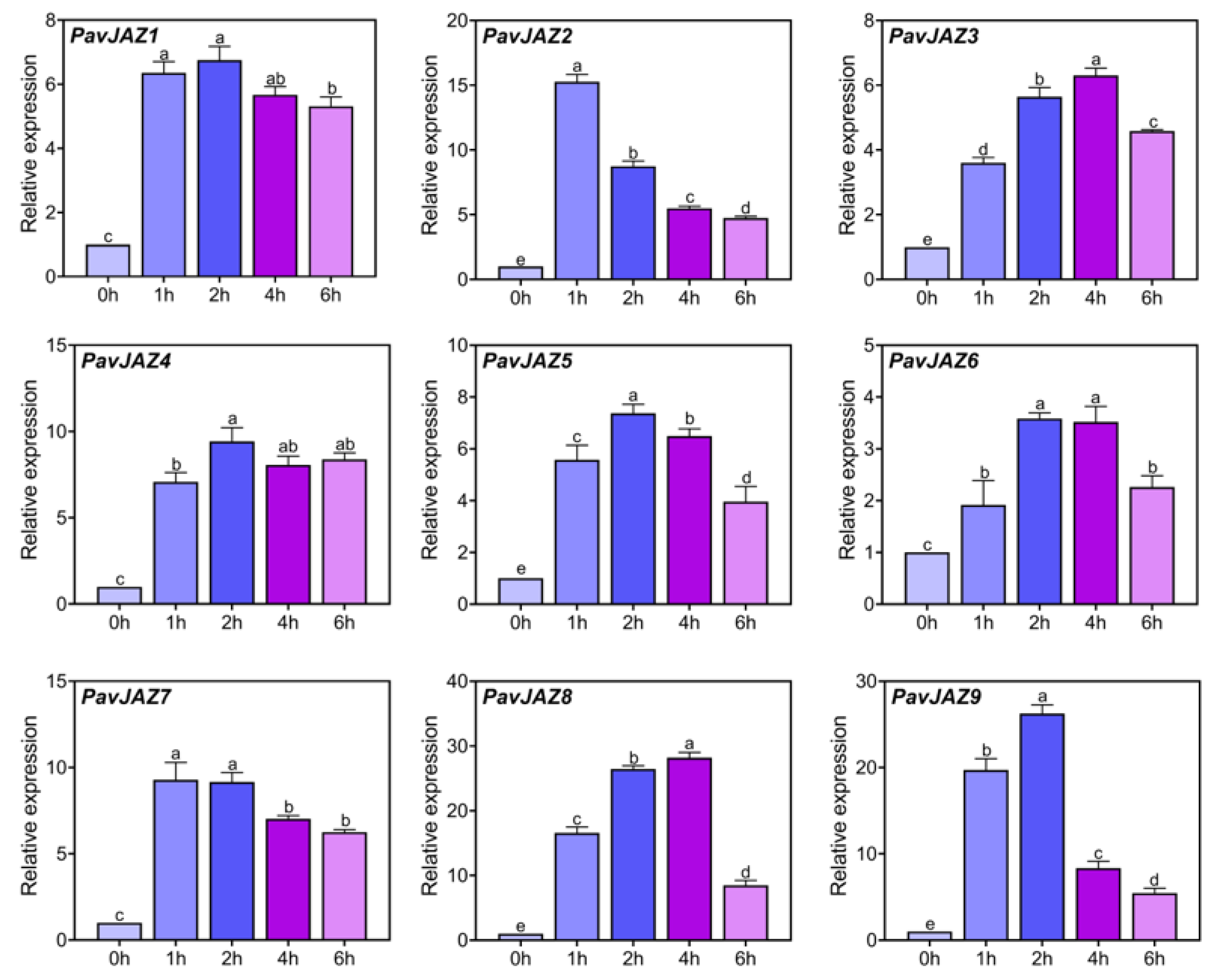
3.6. Relative Expression Levels of PavJAZs in Fruits Under MeJA Treatment
3.7. PavJAZ8 Regulates Sweet Cherry Fruit Aroma Traits
3.8. Expression Patterns of PavJAZ8 After Phytohormone Treatment
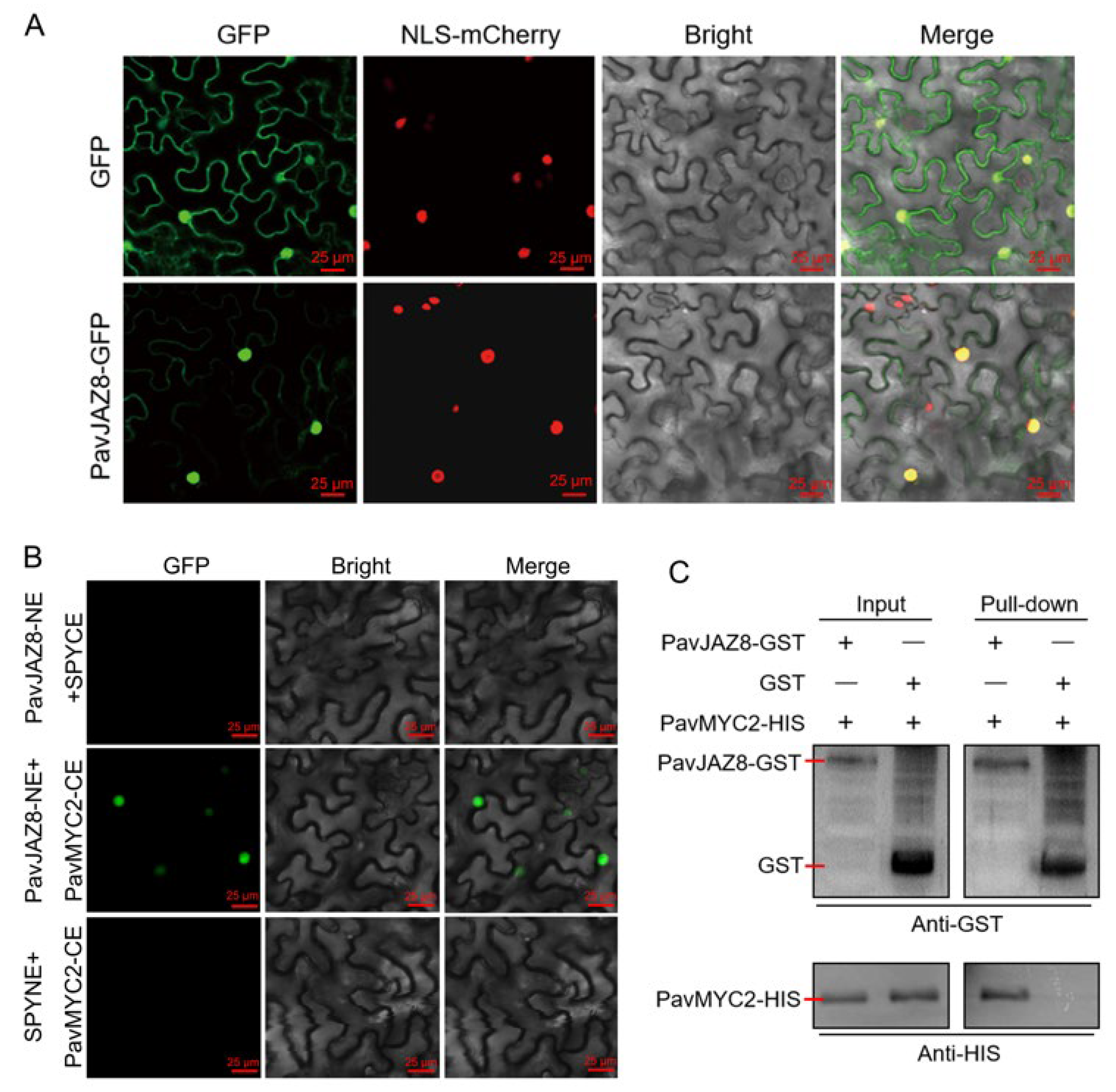
3.9. Subcellular Localization Analysis of PavJAZ8
3.10. PavJAZ8 Interacts with the PavMYC2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kelley, D.S.; Adkins, Y.; Laugero, K.D. A Review of the Health Benefits of Cherries. Nutrients 2018, 10, 368. [Google Scholar] [CrossRef]
- Feng, C.; Guo, Q.; Wu, C.; Zhang, X.; Wang, J.; Song, G.; Yan, G.; Zhou, Y.; Wang, W.; Zhang, K.; et al. Effect of bagging treatment on fruit anthocyanin biosynthesis in sweet cherry. Agric. Commun. 2025, 3, 100103. [Google Scholar] [CrossRef]
- Fan, D.; Wang, W.; Hao, Q.; Jia, W. Do Non-climacteric Fruits Share a Common Ripening Mechanism of Hormonal Regulation? Front. Plant Sci. 2022, 13, 923484. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, K.; Grierson, D. Molecular and Hormonal Mechanisms Regulating Fleshy Fruit Ripening. Cells 2021, 10, 1136. [Google Scholar] [CrossRef] [PubMed]
- Clayton-Cuch, D.; Yu, L.; Shirley, N.; Bradley, D.; Bulone, V.; Böttcher, C. Auxin Treatment Enhances Anthocyanin Production in the Non-Climacteric Sweet Cherry (Prunus avium L.). Int. J. Mol. Sci. 2021, 22, 10760. [Google Scholar] [CrossRef] [PubMed]
- Teribia, N.; Tijero, V.; Munné-Bosch, S. Linking hormonal profiles with variations in sugar and anthocyanin contents during the natural development and ripening of sweet cherries. New Biotechnol. 2016, 33, 824–833. [Google Scholar] [CrossRef]
- Tijero, V.; Teribia, N.; Muñoz, P.; Munné-Bosch, S. Implication of Abscisic Acid on Ripening and Quality in Sweet Cherries: Differential Effects during Pre- and Post-harvest. Front. Plant Sci. 2016, 7, 602. [Google Scholar] [CrossRef]
- Vignati, E.; Lipska, M.; Dunwell, J.M.; Caccamo, M.; Simkin, A.J. Fruit Development in Sweet Cherry. Plants 2022, 11, 1531. [Google Scholar] [CrossRef]
- Wang, W.; Fan, D.; Hao, Q.; Jia, W. Signal transduction in non-climacteric fruit ripening. Hortic. Res. 2022, 9, uhac190. [Google Scholar] [CrossRef]
- Concha, C.M.; Figueroa, N.E.; Poblete, L.A.; Oñate, F.A.; Schwab, W.; Figueroa, C.R. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol. Biochem. 2013, 70, 433–444. [Google Scholar] [CrossRef]
- Fenn, M.A.; Giovannoni, J.J. Phytohormones in fruit development and maturation. Plant J. 2021, 105, 446–458. [Google Scholar] [CrossRef]
- Garrido-Bigotes, A.; Figueroa, P.M.; Figueroa, C.R. Jasmonate metabolism and its relationship with abscisic acid during strawberry fruit development and ripening. J. Plant Growth Regul. 2018, 37, 101–113. [Google Scholar] [CrossRef]
- Han, Y.; Chen, C.; Yan, Z.; Li, J.; Wang, Y. The methyl jasmonate accelerates the strawberry fruits ripening process. Sci. Hortic. 2019, 249, 250–256. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, C.; Pervaiz, T.; Zhao, P.; Liu, Z.; Wang, B.; Wang, C.; Zhang, L.; Fang, J.; Qian, J. Jasmonic acid involves in grape fruit ripening and resistant against Botrytis cinerea. Funct. Integr. Genom. 2016, 16, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Jiang, Z.; Sun, Q.; Wei, R.; Yin, Y.; Xie, Z.; Larkin, R.M.; Ye, J.; Chai, L.; Deng, X. Jasmonate activates a CsMPK6-CsMYC2 module that regulates the expression of β-citraurin biosynthetic genes and fruit coloration in orange (Citrus sinensis). Plant Cell 2023, 35, 1167–1185. [Google Scholar] [CrossRef]
- de la Pena Moreno, F.; Blanch, G.P.; Flores, G.; Ruiz del Castillo, M.L. Impact of postharvest methyl jasmonate treatment on the volatile composition and flavonol content of strawberries. J. Sci. Food Agric. 2010, 90, 989–994. [Google Scholar] [CrossRef]
- Delgado, L.D.; Zúñiga, P.E.; Figueroa, N.E.; Pastene, E.; Escobar-Sepúlveda, H.F.; Figueroa, P.M.; Garrido-Bigotes, A.; Figueroa, C.R. Application of a JA-Ile Biosynthesis Inhibitor to Methyl Jasmonate-Treated Strawberry Fruit Induces Upregulation of Specific MBW Complex-Related Genes and Accumulation of Proanthocyanidins. Molecules 2018, 23, 1433. [Google Scholar] [CrossRef]
- Paladines-Quezada, D.F.; Fernández-Fernández, J.I.; Moreno-Olivares, J.D.; Bleda-Sánchez, J.A.; Gómez-Martínez, J.C.; Martínez-Jiménez, J.A.; Gil-Muñoz, R. Application of elicitors in two ripening periods of Vitis vinifera L. cv Monastrell: Influence on anthocyanin concentration of grapes and wines. Molecules 2021, 26, 1689. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef]
- Fresno, D.H.; Munné-Bosch, S. Differential Tissue-Specific Jasmonic Acid, Salicylic Acid, and Abscisic Acid Dynamics in Sweet Cherry Development and Their Implications in Fruit-Microbe Interactions. Front. Plant Sci. 2021, 12, 640601. [Google Scholar] [CrossRef]
- Kondo, S.; Motoyama, M.; Michiyama, H.; Kim, M. Roles of jasmonic acid in the development of sweet cherries as measured from fruit or disc samples. Plant Growth Regul. 2002, 37, 37–44. [Google Scholar] [CrossRef]
- Balbontín, C.; Gutiérrez, C.; Schreiber, L.; Zeisler-Diehl, V.V.; Marín, J.C.; Urrutia, V.; Hirzel, J.; Figueroa, C.R. Alkane biosynthesis is promoted in methyl jasmonate-treated sweet cherry (Prunus avium) fruit cuticles. J. Sci. Food Agric. 2024, 104, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Katsir, L.; Chung, H.S.; Koo, A.J.; Howe, G.A. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 2008, 11, 428–435. [Google Scholar] [CrossRef]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021, 22, 256. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Bigotes, A.; Figueroa, N.E.; Figueroa, P.M.; Figueroa, C.R. Jasmonate signalling pathway in strawberry: Genome-wide identification, molecular characterization and expression of JAZs and MYCs during fruit development and ripening. PLoS ONE 2018, 13, e0197118. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ouyang, J.; Li, Y.; Zhai, C.; He, B.; Si, H.; Chen, K.; Rose, J.K.C.; Jia, W. A signaling cascade mediating fruit trait development via phosphorylation-modulated nuclear accumulation of JAZ repressor. J. Integr. Plant Biol. 2024, 66, 1106–1125. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Jia, H.; Wu, M.; Zhong, W.; Jia, D.; Wang, Z.; Huang, C. Genome-wide identification and characterization of the TIFY gene family in kiwifruit. BMC Genom. 2022, 23, 179. [Google Scholar] [CrossRef]
- Hu, Y.; Sun, H.; Han, Z.; Wang, S.; Wang, T.; Li, Q.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X. ERF4 affects fruit ripening by acting as a JAZ interactor between ethylene and jasmonic acid hormone signaling pathways. Hortic. Plant J. 2022, 8, 689–699. [Google Scholar] [CrossRef]
- Li, L.; Ji, G.; Guan, W.; Qian, F.; Li, H.; Cai, G.; Wu, X. Genome-Wide Identification and Variation Analysis of JAZ Family Reveals BnaJAZ8.C03 Involved in the Resistance to Plasmodiophora brassicae in Brassica napus. Int. J. Mol. Sci. 2022, 23, 12862. [Google Scholar] [CrossRef]
- Li, Y.G.; Zhang, J.; Cai, X.X.; Fan, L.P.; Zhu, Z.H.; Zhu, X.J.; Guo, D.L. Genome-wide survey and expression analysis of JAZ genes in watermelon (Citrullus lanatus). Mol. Biol. Rep. 2024, 52, 24. [Google Scholar] [CrossRef]
- Song, M.; Wang, H.; Ma, H.; Zheng, C. Genome-wide analysis of JAZ family genes expression patterns during fig (Ficus carica L.) fruit development and in response to hormone treatment. BMC Genom. 2022, 23, 170. [Google Scholar] [CrossRef]
- Ye, L.; Cao, L.; Zhao, X.; Guo, X.; Ye, K.; Jiao, S.; Wang, Y.; He, X.; Dong, C.; Hu, B. Investigation of the JASMONATE ZIM-DOMAIN gene family reveals the canonical JA-signaling pathway in Pineapple. Biology 2022, 11, 445. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X.; et al. Chromosome-scale genome assembly of sweet cherry (Prunus avium L.) cv. Tieton obtained using long-read and Hi-C sequencing. Hortic. Res. 2020, 7, 122. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, D.; Yu, K.; Ji, J.; Liu, N.; Zhang, Y.; Xu, M.; Zhang, Y.J.; Ma, X.; Liu, S.; et al. Frequent germplasm exchanges drive the high genetic diversity of Chinese-cultivated common apricot germplasm. Hortic. Res. 2021, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Zhang, H.; Jiang, C.; Gao, F.; Yan, L.; Zheng, X.; Cheng, J.; Wang, W.; Wang, X.; Ye, X.; et al. De novo chromosome-level genome of a semi-dwarf cultivar of Prunus persica identifies the aquaporin PpTIP2 as responsible for temperature-sensitive semi-dwarf trait and PpB3-1 for flower type and size. Plant Biotechnol. J. 2022, 20, 886–902. [Google Scholar] [CrossRef]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. Biorxiv 2019. Biorxiv:767764. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Wang, W.; Dai, Z.; Li, J.; Ouyang, J.; Li, T.; Zeng, B.; Kang, L.; Jia, K.; Xi, Z.; Jia, W. A Method for Assaying of Protein Kinase Activity In Vivo and Its Use in Studies of Signal Transduction in Strawberry Fruit Ripening. Int. J. Mol. Sci. 2021, 22, 10495. [Google Scholar] [CrossRef]
- Qi, X.; Liu, C.; Song, L.; Dong, Y.; Chen, L.; Li, M. A Sweet Cherry Glutathione S-Transferase Gene, PavGST1, Plays a Central Role in Fruit Skin Coloration. Cells 2022, 11, 1170. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Schütze, K.; Harter, K.; Chaban, C. Bimolecular Fluorescence Complementation (BiFC) to Study Protein-Protein Interactions in Living Plant Cells. In Plant Signal Transduction; Springer: Berlin/Heidelberg, Germany, 2009; pp. 189–202. [Google Scholar] [CrossRef]
- Feng, C.; Zhang, X.; Wang, J.; Wu, C.; Wang, W.; Yan, G.; Zhou, Y.; Zhang, K.; Duan, X. Characterization of volatile profiles in cherry fruits: Integration of E-nose and HS-SPME-GC-MS. Food Chem. X 2025, 28, 102632. [Google Scholar] [CrossRef]
- Villavicencio, J.D.; Zoffoli, J.P.; O’Brien, J.A.; Contreras, C. Methyl jasmonate mediated modulation of lipoxygenase pathway modifies aroma biosynthesis during ripening of ’Regina’ sweet cherry (Prunus avium L.). Postharvest Biol. Technol. 2025, 225, 113511. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Yang, W.; Li, J.; Tang, W.; Gong, R. Transcriptomic and Metabolomic Analysis of Quality Changes during Sweet Cherry Fruit Development and Mining of Related Genes. Int. J. Mol. Sci. 2022, 23, 7402. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef]
- Sherif, S.; El-Sharkawy, I.; Mathur, J.; Ravindran, P.; Kumar, P.; Paliyath, G.; Jayasankar, S. A stable JAZ protein from peach mediates the transition from outcrossing to self-pollination. BMC Biol. 2015, 13, 11. [Google Scholar] [CrossRef]
- Wang, W.; Dai, Z.; Wang, P.; Zhang, X.; Wang, J.; Wu, C.; Feng, C.; Yan, G.; Zhang, K.; Zhou, Y.; et al. Jasmonate ZIM-domain (JAZ) proteins regulate fruit ripening and quality traits: Mechanisms and advances. Food Qual. Saf. 2025, 9, fyaf022. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernández-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Bigotes, A.; Torrejón, M.; Solano, R.; Figueroa, C.R. Interactions of JAZ repressors with anthocyanin biosynthesis-related transcription factors of Fragaria × ananassa. Agronomy 2020, 10, 1586. [Google Scholar] [CrossRef]
- Grunewald, W.; Vanholme, B.; Pauwels, L.; Plovie, E.; Inzé, D.; Gheysen, G.; Goossens, A. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009, 10, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Lee, L.Y.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; A, M.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Luo, H.; Dai, S.; Ren, J.; Zhang, C.; Ding, Y.; Li, Z.; Sun, Y.; Ji, K.; Wang, Y.; Li, Q. The role of ABA in the maturation and postharvest life of a nonclimacteric sweet cherry fruit. J. Plant Growth Regul. 2014, 33, 373–383. [Google Scholar] [CrossRef]
- Zhai, Z.; Xiao, Y.; Wang, Y.; Sun, Y.; Peng, X.; Feng, C.; Zhang, X.; Du, B.; Zhou, X.; Wang, C.; et al. Abscisic acid-responsive transcription factors PavDof2/6/15 mediate fruit softening in sweet cherry. Plant Physiol. 2022, 190, 2501–2518. [Google Scholar] [CrossRef]
- Ozkan, Y.; Ucar, M.; Yildiz, K.; Ozturk, B. Pre-harvest gibberellic acid (GA3) treatments play an important role on bioactive compounds and fruit quality of sweet cherry cultivars. Sci. Hortic. 2016, 211, 358–362. [Google Scholar] [CrossRef]
- Zhang, C.; Whiting, M.D. Improving ‘Bing’ sweet cherry fruit quality with plant growth regulators. Sci. Hortic. 2011, 127, 341–346. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | CDS (bp) | Protein (aa) | Molecular Weight (Kd) | Isoelectric Point | GRAVY | Instability Index |
|---|---|---|---|---|---|---|---|
| PavJAZ1 | FUN_000255-T1 | 1002 | 333 | 36.78 | 9.19 | −0.853 | 46.60 |
| PavJAZ2 | FUN_003441-T1 | 801 | 266 | 29.23 | 9.79 | −0.357 | 57.83 |
| PavJAZ3 | FUN_004843-T1 | 1134 | 377 | 40.50 | 8.61 | −0.433 | 43.38 |
| PavJAZ4 | FUN_007360-T1 | 1047 | 348 | 40.24 | 9.80 | −0.636 | 55.00 |
| PavJAZ5 | FUN_013768-T1 | 801 | 266 | 28.83 | 9.15 | −0.391 | 50.91 |
| PavJAZ6 | FUN_032462-T1 | 363 | 124 | 14.66 | 9.60 | −0.959 | 103.28 |
| PavJAZ7 | FUN_026609-T1 | 1125 | 374 | 39.32 | 9.40 | −0.222 | 53.13 |
| PavJAZ8 | FUN_039026-T1 | 651 | 216 | 23.21 | 6.18 | −0.667 | 66.52 |
| PavJAZ9 | FUN_039096-T1 | 837 | 278 | 30.09 | 9.21 | −0.564 | 50.10 |
| Gene Pair | Ka | Ks | Ka/Ks | Effective Len | Average S Sites | Average N Sites |
|---|---|---|---|---|---|---|
| PavJAZ5/PavJAZ9 | 0.5468 | 2.0797 | 0.2629 | 654 | 143.1667 | 510.8333 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Shi, T.; Dai, Z.; Zhang, X.; Wang, J.; Wu, C.; Feng, C.; Yan, G.; Zhang, K.; Yang, Y.; et al. Investigation of the Jasmonate ZIM-Domain Family Reveals PavJAZ8 Regulates Fruit Aroma Traits in Sweet Cherry (Prunus avium L.). Biomolecules 2025, 15, 1721. https://doi.org/10.3390/biom15121721
Wang W, Shi T, Dai Z, Zhang X, Wang J, Wu C, Feng C, Yan G, Zhang K, Yang Y, et al. Investigation of the Jasmonate ZIM-Domain Family Reveals PavJAZ8 Regulates Fruit Aroma Traits in Sweet Cherry (Prunus avium L.). Biomolecules. 2025; 15(12):1721. https://doi.org/10.3390/biom15121721
Chicago/Turabian StyleWang, Wei, Tianle Shi, Zhengrong Dai, Xiaoming Zhang, Jing Wang, Chuanbao Wu, Chen Feng, Guohua Yan, Kaichun Zhang, Yuan Yang, and et al. 2025. "Investigation of the Jasmonate ZIM-Domain Family Reveals PavJAZ8 Regulates Fruit Aroma Traits in Sweet Cherry (Prunus avium L.)" Biomolecules 15, no. 12: 1721. https://doi.org/10.3390/biom15121721
APA StyleWang, W., Shi, T., Dai, Z., Zhang, X., Wang, J., Wu, C., Feng, C., Yan, G., Zhang, K., Yang, Y., & Duan, X. (2025). Investigation of the Jasmonate ZIM-Domain Family Reveals PavJAZ8 Regulates Fruit Aroma Traits in Sweet Cherry (Prunus avium L.). Biomolecules, 15(12), 1721. https://doi.org/10.3390/biom15121721









