Abstract
In this research, we proposed a Deep Convolutional Neural Network (DCNN) model for image-based plant leaf disease identification using data augmentation and hyperparameter optimization techniques. The DCNN model was trained on an augmented dataset of over 240,000 images of different healthy and diseased plant leaves and backgrounds. Five image augmentation techniques were used: Generative Adversarial Network, Neural Style Transfer, Principal Component Analysis, Color Augmentation, and Position Augmentation. The random search technique was used to optimize the hyperparameters of the proposed DCNN model. This research shows the significance of choosing a suitable number of layers and filters in DCNN development. Moreover, the experimental outcomes illustrate the importance of data augmentation techniques and hyperparameter optimization techniques. The performance of the proposed DCNN was calculated using different performance metrics such as classification accuracy, precision, recall, and F1-Score. The experimental results show that the proposed DCNN model achieves an average classification accuracy of 98.41% on the test dataset. Moreover, the overall performance of the proposed DCNN model was better than that of advanced transfer learning and machine learning techniques. The proposed DCNN model is useful in the identification of plant leaf diseases.
1. Introduction
Identification of plant diseases is essential to improving the farmer’s profit from the yield and growth of plants. Manual monitoring of plant diseases will not give accurate outcomes regularly. Moreover, finding domain experts for monitoring plant diseases is highly complicated and expensive for farmers. Currently, many artificial intelligence techniques are proposed for automatic plant disease detection and diagnosis with fewer human efforts [1]. A Deep Convolutional Neural Network (DCNN) is a recent artificial intelligence technique for solving computer vision and natural language processing challenges. The DCNN has been effectively applied in numerous fields, such as agriculture, healthcare, finance, security, and production. The DCNN uses a sequence of several layers to learn from the training data. The most common DCNN layers are convolution layers, pooling layers, and fully connected layers. The convolution layer extracts features from every input image [2]. The convolutional layer is a significant element of DCNN, and it understands the connection among pixels by learning features using input image data. Convolution is a mathematical operation that takes three inputs, such as an image matrix, filter matrix, and stride value, and produces feature maps. The filters are supported to extract the features from the input images. Moreover, the stride is a numerical value referred to as the number of pixels that move the filters over the input matrix [3]. After the convolutional layer, the pooling layer reduces the dimensionality of the future map. Max pooling and average pooling are the most common pooling techniques. The max-pooling takes the maximum value, and the average pooling takes the average value from the feature map. In this research, the performance of the different numbers of convolutional layers and different pooling functions was compared in terms of the most critical performance metrics.
In addition, the DCNN is required a large volume of training data, optimized hyperparameters value, and high-performance computing to improve the classification performance. Data augmentation is the most popular approach to dealing with the insufficient training data problem for image classification tasks [4]. Data augmentation is producing new images by different transformation techniques [5]. The five most advanced image augmentation techniques were used to produce new input images for the training process in this research. The augmentation techniques are Generative Adversarial Network (GAN), Neural Style Transfer (NST), Principal Component Analysis (PCA), four different color Augmentation, and seven different Position Augmentation. The position augmentation techniques are affine transformation, scaling, cropping, flipping, padding, rotation, and translation. Moreover, the color augmentation techniques are brightness, contrast, saturation, and hue.
The GAN is one of the unsupervised neural networks used to create a new set of realistic images. The GAN comprises two deep neural networks, such as a generator and a discriminator. The generator network creates new images similar to the training data, while the discriminator network classifies the original and newly created images [6]. GAN is one of the most successful image augmentation techniques in medical image processing applications. The NST is an image transformation technique to produce new images using three different images such as content images, style reference images, and input images. Hyperparameters are the most significant parameters that can influence the training process of machine learning techniques. The most common hyper-parameters in DCNN include learning rate, training epochs, filter size, batch size, loss function, activation function, and dropout value [7]. The selection of hyperparameter values is one of the most challenging tasks in machine learning algorithm design. Hyperparameter tuning is the technique of finding the value of the most suitable hyperparameters to achieve maximum performance [8]. The random search technique is a popular hyperparameter tuning technique to optimize the value of the hyperparameter of the machine learning algorithm [9]. In this research, random search techniques were used to optimize the values of the hyperparameters of all proposed DCNNs.
Transfer learning is an advanced learning technique to reuse a pre-trained model on a new similar task. It helps to accelerate the training process of the new model and achieve better performance [10]. The standard transfer learning techniques are AlexNet, VGG16, Inception-v3, and ResNet. The performance of the proposed DCNN is compared with the standard transfer learning and most common machine learning techniques. The machine learning techniques are Artificial Neural Network (ANN), Naive Bayes (NB), and Support Vector Machine (SVM). Moreover, DCNN requires a huge number of data and iterations to train the model. In order to train the model with better efficiency and less time consumption, the machine needs sufficient processing power. The GPU-accelerated deep learning framework is used in this work to train DCNN with a large dataset. The GPU-accelerated deep learning framework delivers high-performance and low-latency inference for the DCNN.
This research proposed a novel DCNN model for the diagnosis of plant leaf diseases using leaf images. The performance of the model was improved using data augmentation and hyperparameter optimization techniques. The remaining sections of the paper were organized as follows: Section 2 provides the literature survey of the plant leaf disease classification-related works. Section 3 describes the materials and methodologies of the proposed DCNN model. Section 4 contains results and related discussions. Section 5 gives the conclusions and future directions of the research.
2. Related Works
Detection of diseases in plant leaves is a significant challenge for precision crop protection and smart farming. Automatic plant disease detection can facilitate the control of diseases through suitable control approaches, for instance, chemical applications, pesticides, fungicides, and bactericide applications, and can increase production. Some earlier efforts to use machine learning techniques for plant disease detection are reviewed in this section. The authors in [2] proposed an SVM with a radial basis function and hyperspectral data for detecting three different sugar plant leaf diseases, such as leaf spot, leaf rust, and powdery mildew. On average, the classification accuracy using this technique was between 65% and 90%, subject to disease stage and category. The author in [11] proposed the SVM algorithm for solving the agriculture data classification problems. The SVM is an extensively used machine learning algorithm. It has been used in various fields, such as image processing and information retrieval. The F1 measure of the algorithm is higher than the typical machine learning algorithms, such as NB and ANN algorithms. The author in [12] proposed another approach based on SVM for the identification of Huanglongbing (HLB) or citrus greening diseases using multi-band imaging sensor data.
The authors of [13,14] discussed several methods for the detection of plant diseases using their leaf images that were based on various image processing and feature extraction techniques. Identification of tomato yellow leaf curl disease can be achieved using SVM with a quadratic kernel function as proposed by the author in [15]. Overall, the classification accuracy using this algorithm was 92%. The authors of [16] proposed a Huanglongbing disease identification model for citrus plants. The SVM and ANN were used to design the Huanglongbing detection technique. The testing accuracy of the SVM and ANN models was 92.8% and 92.2%, respectively. The accuracy of the SVM models is higher than that of the ANN model. The authors of [17] proposed an apple leaf spot disease forecasting model using a K-Nearest Neighbors (KNN) classifier. The forecasting model achieved 88% of accuracy, which is higher than other machine learning models.
The authors of [3,18] developed convolutional neural networks (CNN), based models for recognizing different plant species. Moreover, their experiment shows that performance improvements in plant leaf disease detection can be made using CNN rather than traditional machine learning techniques. The authors of [19,20] proposed a CNN-based model for solving crop disease identification problems using different datasets. In [21], the authors proposed the AlexNet and VGG16 models for disease detection in tomato crops with optimized hyperparameters. In [22], the authors proposed a leaf disease detection model using the AlexNet pre-trained model. The AlexNet based model achieved a test accuracy of 98%, which is better than machine learning techniques. The author of [23] developed the GoogLeNet and Cifar10 models based on deep learning for identifying maize leaf diseases. Overall, the top-1 accuracy using this GoogLeNet and Cifar10 technique was 98.9% and 98.8%, respectively.
The authors of [24] studied numerous deep learning approaches to solve many agriculture challenges. Likewise, plant disease detection and diagnosis can be achieved using convolutional neural network models. This technique was applied to 87,848 images of 25 different plants and presented in [25]. This model achieves an average accuracy of 99.53%. The authors of [26] presented a deep learning-based model for identifying ten different plant diseases from individual lesions and spots. A nine-layer DCNN model was developed in [27] for identifying different plant leaf diseases using optimized hyperparameters, six data augmentation methods, and the Graphics Processing Unit (GPU), and it achieved a classification accuracy of 96.46% in the test data. The authors of [28] proposed a leaf disease detection model for various plants using a transfer learning technique called TL-ResNet50. They achieved a classification accuracy of 98.20% on disease detection. Most recently, the authors of [29] proposed attention-based convolutional neural networks for tomato leaf disease classification. The model achieved a classification accuracy of 99.69% on the test dataset. The performance of the model was compared with standard transfer learning techniques. In [5], the authors proposed the apple leaf disease detection model using ResNet-50 and Mask R-CNN techniques. Moreover, they compared the performance of the MobileNetV3-Large and MobileNetV3-Large-Mobile models. The extensive survey shows the importance of developing a plant disease detection model with high detection accuracy. A DCNN with different convolutional layers and hyperparameters was trained and tested to develop an image-based plant leaf disease identification model in this research. The following section presents information about the dataset and the implemented models.
3. Materials and Methods
This section provides a complete procedure for creating and preprocessing the dataset and designing and developing the proposed DCNN for plant leaf disease identification. Initially, a novel CNN model was proposed with a different number of convolutional layers between 3 and 5. A hyperparameter optimization technique was used to select the most appropriate hyperparameters. The models were trained using optimized hyperparameters on an augmented dataset. The training and testing of the DCNN and pre-trained models were performed using an NVIDIA DGX-1 GPU deep learning server and an HP Z600 workstation. The Python programming language and advanced libraries were used to implement the proposed and existing models. The essential Python libraries are Keras, OpenCV, NumPy, Pillow, SciPy, and TensorFlow.
3.1. Data Collection and Preprocessing
The original dataset was downloaded from an open research data repository [27]. The original dataset contained 39 different classes of 55,448 healthy and diseased plant leaves and background images. Figure 1 shows the four sample images from random classes of the original dataset.

Figure 1.
Sample images from random classes of the original dataset.
Data augmentation techniques can increase the dataset size and reduce overfitting during the training state of models by adding a small number of distorted images to the training set. There are five types of data augmentation techniques used in this research to create new training images. The augmentation techniques are Neural Style Transfer (NST), Deep Convolutional Generative Adversarial Network (DCGAN), Principal Component Analysis (PCA), Color Augmentation, and Position Augmentation. The augmentation techniques increased the dataset size from 55,448 to 234,008 images. It also increased the size of all the classes to 6000 original and augmented images.
3.1.1. Neural Style Transfer
Neural Style Transfer is a convolutional network to create augmented images using content images and style reference images [30]. The NST performs the image transformation, converting the style properties of the style image into a content image for generating a new output image. The sample content, style reference, and output images of NST are shown in Figure 2.
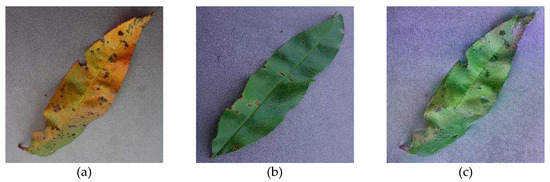
Figure 2.
NST technique: (a) Content image; (b) style reference image; (c) output image.
The VGG-19 based NST technique was used to generate 22,000 different plant leaf images in this research. The augmented images were generated using 5000 iterations and GPU techniques. Figure 3 shows some random augmented image samples of the different classes using the NST technique.

Figure 3.
Sample augmented images using NST technique.
3.1.2. Deep Convolutional Generative Adversarial Networks
Generative Adversarial Networks (GANs) are one of the exciting applications of deep neural networks. It creates new images from the input images using random noise and maps them into the input images. Two models are trained simultaneously by an adversarial process such as generator and discriminator. The generator network learns to create new images similar to the input image. Furthermore, the discriminator learns to classify the real dataset images and generator-created images. A Deep Convolutional Generative Adversarial Network (DCGAN) was used to create augmented images in this research [6]. The layered architecture of the generator and discriminator of the DCGAN are shown in Figure 4 and Figure 5, respectively. The DCGAN creates the generator and discriminator networks, mainly using convolution layers without a pooling function. Transposed convolution and stride are used to upsampling and downsample the features of convolution layers.
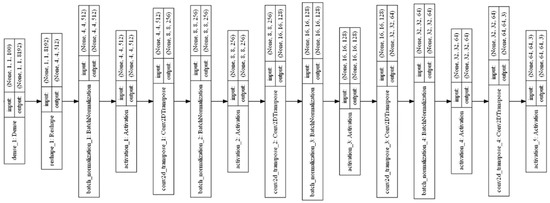
Figure 4.
Layered structure of DCGAN generator.
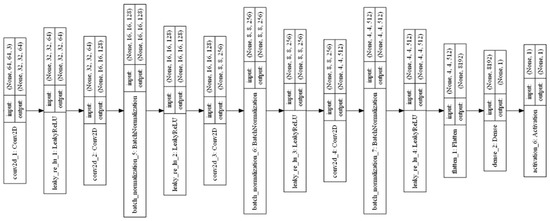
Figure 5.
Layered structure of DCGAN discriminator.
The DCGAN network was trained using the training epoch of 30,000, the stride dimension of 2 × 2, and the mini-batch size of 64. It generates 35,700 augmented images in different classes. Four samples of augmented images from different classes using DCGAN are shown in Figure 6.

Figure 6.
Sample augmented images using DCGAN.
3.1.3. Principal Component Analysis Color Augmentation
Principal Component Analysis (PCA) color augmentation is one of the advanced image augmentation techniques that shifts the values of the red, green, and blue pixels in the input image based on which values are the maximum existing in the image by calculating eigenvectors and eigenvalues of the image matrix. The PCA algorithm helps to discover the relative color balance of the input image. This research performed PCA on the set of RGB pixel values of original plant leaf disease images and generated 26,800 new images in various classes. Figure 7 shows the random sample augmented images using the PCA technique.

Figure 7.
Sample augmented images using the PCA color augmentation technique.
3.1.4. Color Augmentation
Color augmentation techniques adjust the color properties of the original images to create augmented images. In this research, a combination of four different color properties is used to generate new images, such as brightness, contrast, saturation, and hue. The color augmentation techniques create over 47,000 augmented images in different classes. Figure 8 illustrates the sample images of color augmentation from random classes.

Figure 8.
Sample augmented images using color augmentation techniques.
3.1.5. Position Augmentation
Position augmentation techniques affect the position of pixel values of the input original image to create augmented images. A combination of seven-position augmentation techniques was used in this research, such as affine transformation, scaling, cropping, flipping, padding, rotation, and translation. It created 47,052 images in all the classes of the dataset. The sample position augmented images of random classes in the dataset are shown in Figure 9.

Figure 9.
Sample augmented images using position augmentation techniques.
3.2. Building the DCNN Model
DCNN achieved great success in solving image classification problems. Three different DCNN configurations were proposed for making plant leaf disease image classification from the augmented dataset: three convolutional layer DCNN (Conv-3 DCNN), four convolutional layer DCNN (Conv-4 DCNN), and five convolutional layers DCNN (Conv-5 DCNN). A random search hyperparameter tuning technique was used to optimize the hyperparameter value of each proposed DCNN. The augmented dataset was divided into the training and testing processes. The training and testing datasets contain 224,552 and 9448 images, respectively. Table 1 shows the hyperparameter options for the hyperparameter optimization process. The random search optimization technique generates the random combination of parameters for training the model. The generated combinations were used to train the model for a few epochs. The best form generated combination was identified from the training performance of the model. The designing and training processes of each model were explained in the following subsections, starting with the Conv-3 DCNN model.

Table 1.
Options for hyperparameters optimization.
3.2.1. Three Convolutional Layer DCNN
Training the three convolutional layer DCNN model using the augmented image dataset that is described in Section 3.2 was proposed. Three convolutional layers and three max-pooling layers were used in this Conv-3 DCNN model. The max-pooling and 2 × 2 stride were used to downsample the feature dimensions. The layered structure of the Conv-3 DCNN model is shown in Figure 10.
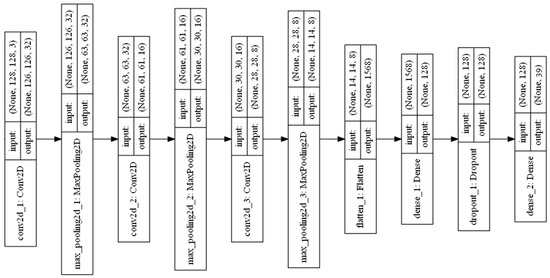
Figure 10.
Layered structure of Conv-3 DCNN model.
Table 2 shows the optimized hyperparameters using the random search hyperparameters tuning technique of the Conv-3 DCNN for the identification of plant leaf diseases. The random search technique was used to address the parametric uncertainty challenge of the model development. The training and validation accuracy of the Conv-3 DCNN were 98.32 and 97.87, respectively. The training progress of the Conv-3 DCNN using the augmented dataset and optimized hyperparameters is shown in Figure 11.

Table 2.
Optimized hyperparameters values of the Conv-3 DCNN.
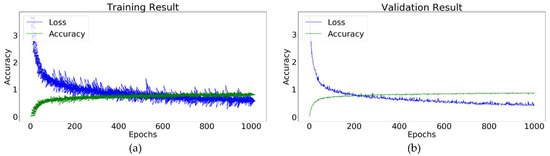
Figure 11.
Conv-3 DCNN model: (a) Training result; (b) validation result.
3.2.2. Four Convolutional Layer DCNN
The four-convolutional layer DCNN (Conv-4 DCNN) model using the augmented image dataset and optimized hyperparameters values was proposed. Four convolutional layers and four max-pooling layers are used in this Conv-4 DCNN model. The layered structure of the Conv-4 DCNN model is shown in Figure 12.
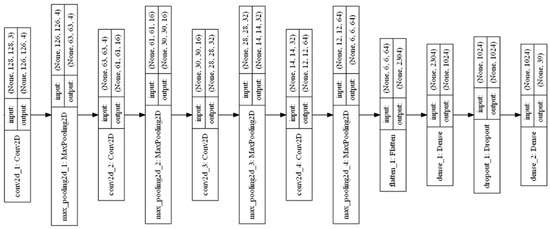
Figure 12.
Layered structure of Conv-4 DCNN model.
The training accuracy of the Conv-4 DCNN was 99.12, and the validation accuracy was 98.74. Table 3 shows the optimized hyperparameters of the conv-4 DCNN for the identification of plant leaf diseases. Figure 13 illustrates the training performance of the Conv-4 DCNN model for the identification of plant leaf diseases.

Table 3.
Optimized hyperparameters of the Conv-4 DCNN.
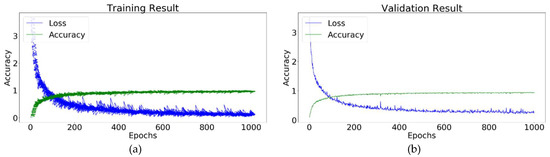
Figure 13.
Conv-4 DCNN model: (a) Training result; (b) validation result.
3.2.3. Five Convolutional Layer DCNN
The plant leaf disease identification model using a five convolutional layer DCNN (Conv-5 DCNN) was proposed. Five convolutional layers and five max-pooling layers were used in this Conv-5 DCNN model. The layered structure of the Conv-5 DCNN model is shown in Figure 14.
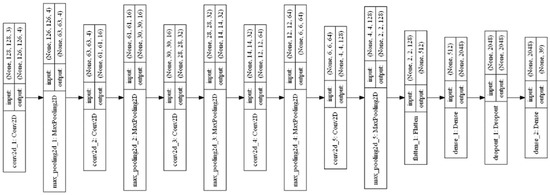
Figure 14.
Layered structure of Conv-5 DCNN model.
The random search hyperparameter tuning technique was used to discover the value of the hyperparameter of the conv-5 DCNN. Table 4 shows the optimized hyperparameters for the plant leaf disease identification model using the conv-5 DCNN. The conv-5 DCNN achieves a training accuracy of 9923 and a validation accuracy of 98.93. These accuracies are much higher than the other proposed DCNN algorithms. Figure 15 illustrates the training performance of the Conv-5 DCNN model for the identification of plant leaf diseases.

Table 4.
Optimized hyperparameters of the conv-5 DCNN.
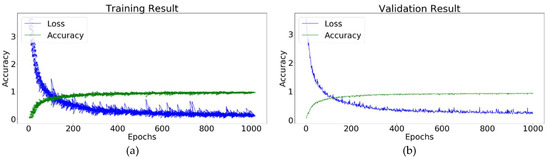
Figure 15.
Conv-5 DCNN model: (a) Training result; (b) validation result.
The performance of all the proposed models was calculated using the different performance metrics that were discussed in the subsequent section. New input images were used to compare the performance of the proposed models with advanced classification techniques.
4. Results and Discussions
The proposed Conv-3 DCNN, Conv-4 DCNN, and Conv-5 DCNN models were compared to advanced classification techniques using the most critical performance metrics and 9448 original input images. Classification accuracy, precision, recall, and F1-Score are the most critical performance metrics. The following subdivision calculates the classification accuracy of all the models.
4.1. Classification Accuracy
Classification accuracy is one of the critical performance metrics, which refers to the percentage of correct predictions made by the classification model. Figure 16 illustrates the individual class testing accuracy of the proposed Conv-3 DCNN, Conv-4 DCNN, and Conv-5 DCNN models.
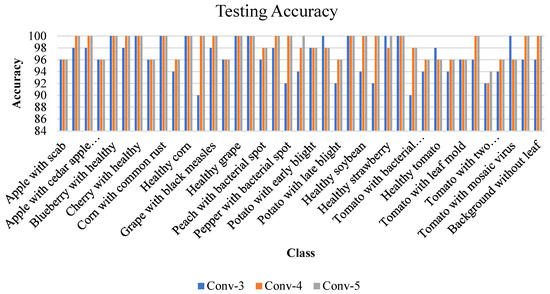
Figure 16.
Testing accuracy of individual classes for proposed Conv-3 DCNN, Conv-4 DCNN, and Conv-5 DCNN.
Figure 17 shows that the classification accuracy of the proposed Conv-5 DCNN model is higher than the Conv-3 DCNN, Conv-4 DCNN, and advanced machine learning and transfer learning techniques. The range of the classification accuracy is between 0 and 100%.

Figure 17.
Testing accuracy of all the advanced and proposed techniques.
4.2. Precision
Precision is defined as the ratio of correctly predicted values to the total predicted values. Figure 18 represents that the proposed Conv-5 DCNN model achieved a precision value of 0.94, much higher than the other techniques.
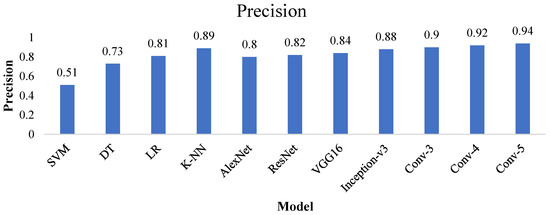
Figure 18.
Precision value of all the advanced and proposed techniques.
4.3. Recall
The recall is also known as sensitivity and is a ratio of correctly predicted values to all available values in a particular individual class. Figure 19 represents the recall value of the Conv-4 DCNN, and the Conv-5 DCNN is more significant than all other techniques.
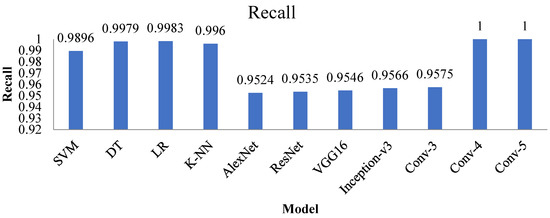
Figure 19.
Recall value of all the advanced and proposed techniques.
4.4. F1-Score
The F1 score is the most crucial performance metric and is defined as the weighted average of precision and recall. The F1-Score of the proposed models and other advanced techniques are shown in Figure 20. The F1-score of the Conv-4 DCNN and Conv-5 DCNN is higher than other techniques.
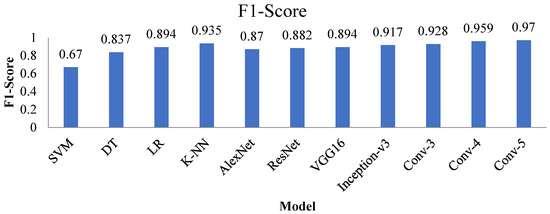
Figure 20.
F1-Score value of all the advanced and proposed techniques.
Outcomes of all performance metrics show that the Conv-5 DCNN model produced much higher performance than the other proposed Conv-3 DCNN, Conv-4 DCNN models, and state-of-the-art transfer learning and machine learning techniques. The proposed Conv-5 DCNN model achieved superior results using optimized hyperparameters and an augmented dataset. Moreover, the proposed Conv-5 DCNN model uses the optimized number of convolutional layers for extracting the feature information from the dataset.
5. Conclusions
In this research, a DCNN based approach was proposed to identify plant leaf diseases from leaf images. This model can successfully identify 26 different plant diseases through leaf images. Five augmentation techniques were used to enhance the dataset size from 55,448 to 234,000 images. The augmentation techniques are Neural Style Transfer (NST), Deep Convolutional Generative Adversarial Network (DCGAN), Principal Component Analysis (PCA), Color Augmentation, and Position Augmentation. The individual class size of the dataset was 6000 original and augmented images. Training of the most successful Conv-5 DCNN model was completed with the use of an augmented dataset of 224,552 images and optimized hyperparameter values. The random search hyperparameter tuning technique was used to optimize the value of the hyperparameters. The proposed Conv-5 DCNN model achieved a classification accuracy of 98.41%, a precision value of 0.94, a recall value of 1.0, and an F1-Score of 0.97 using an augmented plant leaf image dataset. The optimized hyperparameters and the data augmentation process had a more considerable influence on the result. Compared with other proposed approaches and advanced machine learning and transfer learning techniques, the proposed Conv-5 DCNN model has superior classification performance. An extension of this study will add new classes of plant diseases and an increasing number of training images to the dataset and modify the architecture of the DCNN model.
Author Contributions
Conceptualization, J.A.P. and K.K.; methodology, V.D.K.; software, J.A.P. and K.K.; validation, R.G., M.J. and Z.L.; formal analysis, E.J.; investigation, J.A.P. and K.K.; resources, V.D.K.; writing—original draft preparation, J.A.P., K.K. and V.D.K.; writing—review and editing, M.J.; visualization, J.A.P. and K.K.; supervision, V.D.K., E.J., R.G. and Z.L.; funding acquisition, R.G. and Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
SGS Grant from VSB—Technical University of Ostrava under grant number SP2022/21.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early detection and classification of plant diseases with Support Vector Machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Lee, S.H.; Chan, C.S.; Wilkin, P.; Remagnino, P. Deep-plant: Plant identification with convolutional neural networks. In Proceedings of the 2015 IEEE International Conference on Image Processing (ICIP), Quebec City, QC, Canada, 27–30 September 2015; pp. 452–456. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-D.; Dong, Z.; Chen, X.; Jia, W.; Du, S.; Muhammad, K.; Wang, S.H. Image based fruit category classification by 13-layer deep convolutional neural network and data augmentation. Multimed. Tools Appl. 2019, 78, 3613–3632. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Sezer, A.; Altan, A. Detection of solder paste defects with an optimization-based deep learning model using image processing techniques. Solder. Surf. Mt. Technol 2021, 33, 291–298. [Google Scholar] [CrossRef]
- Sezer, A.; Altan, A. Optimization of Deep Learning Model Parameters in Classification of Solder Paste Defects. In Proceedings of the 2021 3rd International Congress on Human-Computer Interaction, Optimization and Robotic Applications (HORA), Ankara, Turkey, 11–13 June 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Altan, A.; Karasu, S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos Solitons Fractals 2020, 140, 110071. [Google Scholar] [CrossRef]
- Shi, L.; Duan, Q.; Ma, X.; Weng, M. The Research of Support Vector Machine in Agricultural Data Classification. In International Conference on Computer and Computing Technologies in Agriculture; Li, D., Chen, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 265–269. [Google Scholar]
- Garcia-Ruiz, F.; Sankaran, S.; Maja, J.M.; Lee, W.S.; Rasmussen, J.; Ehsani, R. Comparison of two aerial imaging platforms for identification of Huanglongbing-infected citrus trees. Comput. Electron. Agric. 2013, 91, 106–115. [Google Scholar] [CrossRef]
- Khirade, S.D.; Patil, A.B. Plant Disease Detection Using Image Processing. In Proceedings of the 2015 International Conference on Computing Communication Control and Automation, Pune, India, 26–27 February 2015; pp. 768–771. [Google Scholar]
- Bharate, A.A.; Shirdhonkar, M.S. A review on plant disease detection using image processing. In Proceedings of the 2017 International Conference on Intelligent Sustainable Systems (ICISS), Palladam, India, 7–8 December 2017; pp. 103–109. [Google Scholar]
- Mokhtar, U.; Ali, M.A.S.; Hassanien, A.E.; Hefny, H. Identifying Two of Tomatoes Leaf Viruses Using Support Vector Machine. In Information Systems Design and Intelligent Applications; Mandal, J.K., Satapathy, S.C., Kumar Sanyal, M., Sarkar, P.P., Mukhopadhyay, A., Eds.; Springer: New Delhi, India, 2015; pp. 771–782. [Google Scholar]
- Wetterich, C.B.; de Oliveira Neves, R.F.; Belasque, J.; Ehsani, R.; Marcassa, L.G. Detection of Huanglongbing in Florida using fluorescence imaging spectroscopy and machine-learning methods. Appl Opt. 2017, 56, 15–23. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, J.; Zhang, J.; Yuan, L.; Zhou, X.; Xu, X.; Yang, G. Forecasting Alternaria Leaf Spot in Apple with Spatial-Temporal Meteorological and Mobile Internet-Based Disease Survey Data. Agronomy 2022, 12, 679. [Google Scholar] [CrossRef]
- Grinblat, G.L.; Uzal, L.C.; Larese, M.G.; Granitto, P.M. Deep learning for plant identification using vein morphological patterns. Comput. Electron. Agric. 2016, 127, 418–424. [Google Scholar] [CrossRef] [Green Version]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johannes, A.; Picon, A.; Alvarez-Gila, A.; Echazarra, J.; Rodriguez-Vaamonde, S.; Navajas, A.D.; Ortiz-Barredo, A. Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case. Comput. Electron. Agric. 2017, 138, 200–209. [Google Scholar] [CrossRef]
- Rangarajan, A.K.; Purushothaman, R.; Ramesh, A. Tomato crop disease classification using pre-trained deep learning algorithm. Procedia Comput. Sci. 2018, 133, 1040–1047. [Google Scholar] [CrossRef]
- Chen, H.-C.; Widodo, A.M.; Wisnujati, A.; Rahaman, M.; Lin, J.C.; Chen, L.; Weng, C.-E. AlexNet Convolutional Neural Network for Disease Detection and Classification of Tomato Leaf. Electronics 2022, 11, 951. [Google Scholar] [CrossRef]
- Zhang, X.; Qiao, Y.; Meng, F.; Fan, C.; Zhang, M. Identification of Maize Leaf Diseases Using Improved Deep Convolutional Neural Networks. IEEE Access 2018, 6, 30370–30377. [Google Scholar] [CrossRef]
- Kamilaris, A. Prenafeta-Boldú FX. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Arnal Barbedo, J.G. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Geetharamani, G.; Pandian, J.A. Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput. Electr. Eng. 2019, 76, 323–338. [Google Scholar]
- Ali, S.; Hassan, M.; Kim, J.Y.; Farid, M.I.; Sanaullah, M.; Mufti, H. FF-PCA-LDA: Intelligent Feature Fusion Based PCA-LDA Classification System for Plant Leaf Diseases. Appl. Sci. 2022, 12, 3514. [Google Scholar] [CrossRef]
- Bhujel, A.; Kim, N.-E.; Arulmozhi, E.; Basak, J.K.; Kim, H.-T. A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification. Agriculture 2022, 12, 228. [Google Scholar] [CrossRef]
- Gatys, L.A.; Ecker, A.S. Matthias Bethge. A Neural Algorithm of Artistic Style. arXiv 2015, arXiv:1508.06576. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).