Abstract
The main goal of this work was the use of the powerful solid-phase microextraction-gas chromatography-mass spectrometry (SPME-GC-MS) technique to unequivocally identify microbial volatile organic compounds (MVOCs) derived from the enzymatic activity produced during metabolic processes using analytical profile index (API) biochemical tests. Three bacteria were selected for this study: Escherichia coli, Proteus mirabilis, and Pseudomonas aeruginosa. They were inoculated and incubated to both API components and real cosmetics, as well as to a mixture of them. Specific MVOCs were successfully identified as biomarkers for each one of the studied microorganisms: Indole and 2-nitrophenol as Escherichia coli markers, 2-undecanone and phenylethyl alcohol as Proteus mirabilis-specific markers, and 1-undecene and 2′-aminoacetophenone as Pseudomonas aeruginosa ones. In addition, a high number of MVOCs were identified as general markers of bacterial presence. The results revealed that the MVOCs’ formation is highly subtract dependent. Therefore, the ultimate and most challenging objective is to establish a relationship between the identified MVOCs and the original compound present in the substrate. This work establishes the design and development of this original approach, and its practical application to the control of microbial contamination in real cosmetic samples.
1. Introduction
All cosmetic products placed in the European (EU) market are produced according to the Good Manufacturing Practices (GMPs) [1], which means that cosmetics are tested for microbiological, physical, and chemical properties to ensure their safety and quality. European Regulation EC No 1223/2009 [2] does not establish sterile conditions for cosmetic products but requires that they must be safe for consumers. Since bacteria contamination supposes an important concern for users, the European Scientific Committee of Consumer Products (SCCP) establishes specifications in the finished cosmetic products. In this way, products intended for children and to be used in the eye area and on mucous membranes should have not more than 100 CFU (colony-forming units) per gram or milliliter of aerobic mesophilic microorganisms. For all other products, the limit is higher, 1000 CFU per gram or milliliter. In addition, the absence of pathogens, such as Staphylococcus aureus, Pseudomonas aeruginosa, and Candida albicans, must be proven in 1 mL or 1 g of products intended for children or to be used in the eye area and on mucous membranes, or in 0.1 mL or 0.1 g of other finished products, whereas the presence of Escherichia coli and other Enterobacteriaceae is completely forbidden in all types of cosmetic products [3]. The appropriate microbiological control of cosmetic products is regulated by ISO (International Organization for Standardization) standards, and the path to follow is summarized in Figure 1, implying the preparation of multiple and laborious steps [4].
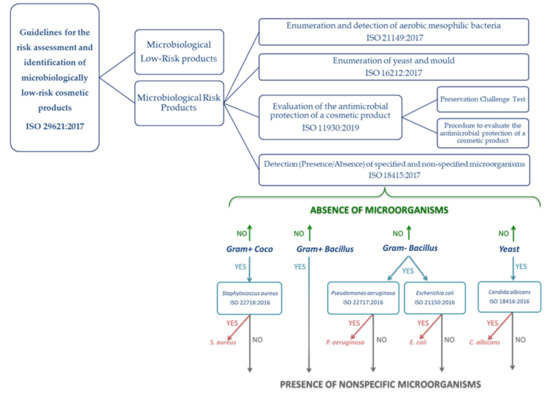
Figure 1.
Recommended path in carrying out the microbiological control of cosmetics products with the correspondent ISO standards to be followed and the conclusions that may be drawn.
Although preservatives are added to cosmetic formulations to avoid bacterial contamination, these products are recognized to be suitable substrates for the growth and survival of a large variety of microorganisms, since they posse some of the nutrients that facilitate growth, such as water, lipids, polysaccharides, alcohols, proteins, amino acids, glycosides, peptides, and vitamins [5,6]. Besides, inadequate storage conditions, such as high temperature and/or humidity exposition, and the use of expired products (the lifetime of the product is limited by the protection afforded by the preservation), can favor undesired microbial growth in the formulations. To assure consumers’ health, and to inform about unsafe products, the European Commission has an early warning system for safety management, called the Rapid Alert System (RAPEX) [7]. Figure 2 shows the evolution of the cosmetics withdrawals in the EU market due to microbiological contamination in the last decade. Only in the last year (2019 until April 2020), 27 products were withdrawn from the market due to microbial spoilage, with Pseudomonas aeruginosa, being the most frequently found microorganism [7,8]. A recent study evaluated the presence of bacteria in 467 cosmetics donated by product users from the United Kingdom [9]. The results revealed that 90% of the analyzed products contained microorganisms (up to 48 different bacteria species were identified), including the strictly forbidden Escherichia coli. This study also evaluated the CFU of beauty blenders, employed to homogenize the cosmetics, especially on the face. These products showed a worrying level higher than 106 CFU per g.
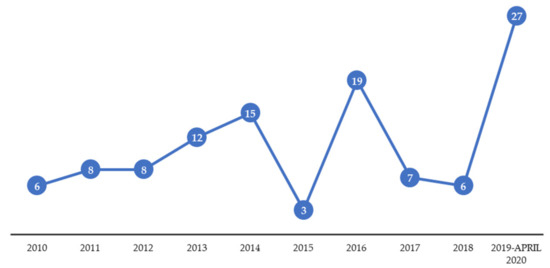
Figure 2.
Cosmetic recalls in the EU market due to microbiological contamination in the last decade.
Most of the employed techniques for detecting bacteria in cosmetics, including the official methodology, are based on the CFU count [4,6,10] and although they employ low-cost materials, they are laborious and require several steps, being very time consuming (up to 72 h). In addition, due to the broad variety of cosmetics available in the current market, including some ‘extreme formats’ and borderline products [11], such control methods are often not suitable for several types of products, like water-immiscible cosmetics. The use of molecular detection of bacterial indicators in cosmetics based on adenosine triphosphate (ATP) bioluminescence and polymerase chain reaction (PCR)-based assays has also been explored [12,13], and although they slightly reduce the analysis time up to 27 h, it is still too long. In addition, these molecular techniques employ expensive reagents and materials. Therefore, the current trend in microbial determination is towards biochemical tests, such as those based on the Analytical Profile Index (API®), a miniaturized version of the existing techniques. Briefly, it is based on wells containing dehydrated substrates to detect enzymatic activity, usually related to the fermentation of carbohydrates or catabolism of proteins or amino acids by the inoculated organisms. A bacterial suspension is used to rehydrate each of the wells, and they are incubated. During incubation, metabolism produces color changes that are either spontaneous or revealed by the addition of reagents [14]. However, this approach allows the identification of a limited number of Gram-negative Enterobacteriaceae or non-Enterobacteriaceae. Additional tests are sometimes necessary to distinguished between two species, since the coloration may not be clear, thus being able to give rise to false negatives.
On the other hand, it is well known that bacteria produce microbial volatile organic compounds (MVOCs), which are formed during bacterial metabolic processes. Although their formation is greatly affected by the substrate, pH, humidity, and temperature, approximately 2000 volatile organic compounds from microorganisms have been already identified as MVOCs [15]. For their monitoring, the combination of solid-phase microextraction (SPME) with gas chromatography-mass spectrometry (GC-MS) is a very suitable option, since SPME allows an in-situ extraction and preconcentration of the MVOCs in a single step, whereas GC-MS provides the sensitivity and selectivity required for an unequivocal identification of the extracted MVOCs. The determination of MVOCs by SPME-GC-MS has been successfully used in the diagnosis of respiratory and gastrointestinal infections, and to investigate microbial growth in food [16,17,18,19]. However, to the best of our knowledge, only a previous study of the authors reported the use of SPME-GC-MS to identify MVOCs produced by bacterial growth in cosmetics [20], work that provided the basis for the present study by being combined with API® technology.
The main goal of this work was, therefore, the use of the powerful SPME-GC-MS technique to unequivocally identify MVOCs derived from the enzymatic activity produced using an API® biochemical test. Besides, real cosmetics will be inoculated with bacteria to identify MVOCs coming from the cosmetic substrate, as general markers of bacterial presence. The ultimate and most challenging goal is to establish a link between the identified MVOCs and the presence of a specific species of bacteria in the contaminated cosmetic.
2. Materials and Methods
2.1. Chemicals and Materials
Methanol and ethyl acetate were provided by Scharlab (Barcelona, Spain). API® 10 S system, containing as substrates: 2-nitrophenyl-β-d-galactopyranoside, d-glucose, l-arabinose, l-lysine, l-ornithine, trisodium citrate, sodium thiosulfate, urea, and L-tryptophan was provided by BioMerieux (Marcy-l’Étoile, France). The standards N,N-dipropyl-1-propanamine, 1,2-hexanediol, 1,2-octanediol, ethylhexanol, 1-octanol, phenylethyl alcohol, 2-nitrophenol, 1-nonanol, indole, 2-undecanone, and 2-tridecanone were supplied by Sigma Aldrich (Steinheim, Germany). Individual stock solutions of each compound were prepared in methanol. Further dilutions and mixtures were prepared in ethyl acetate. Stock solutions were stored in glass vials protected from light at −20 °C.
A 50/30 µm divinylbenzene-carboxen-polydimethylsiloxane (DVB/CAR/PDMS) fiber housed in a manual SPME holder was obtained from Supelco (Bellefonte, PA, USA). Prior to the first use, the fiber was conditioned as recommended by the manufacturer, in the GC injection port with carrier flow (He) at 270 °C for 1 h.
2.2. Cosmetic Samples
A moisturizing hand cream free of silicone, allergens, and dyes (Stokolan®) was supplied by Deb group (Denby, UK). It was employed as a cosmetic matrix model to be inoculated with the studied microorganisms. Since, in the last decade, more than 56% of the cosmetics recalled in the EU market were leave-on products [7], this matrix was selected as a model to carry out the experiments. The sample was kept in its original container at room temperature and protected from the light until its inoculation, incubation, and further analysis.
2.3. Bacterial Cultures and Inoculation Procedure
Three different Gram-negative bacterial species of different genera were included in this study: Escherichia coli, Proteus mirabilis, and Pseudomonas aeruginosa.
Escherichia coli is probably the most studied and experimentally used bacteria. It is considered as an indicator of fecal contamination, and its presence in cosmetics is associated with poor hygiene conditions during manufacturing and/or product packaging. Proteus mirabilis is one of the most resistant to antibiotics since it can transform urea in NH3 to alkalize the medium staying alive. Its presence in a cosmetic would indicate either contamination of a raw material, such as surfactant or the use of water containing high levels of pollution. Finally, Pseudomonas aeruginosa was included in this work since it is the most detected in cosmetics, especially in eye products, being the cause of most cosmetic recalls, and its presence is attributed to both manufacturing processes and consumer manipulation [21].
From a pure culture of each bacterium, a bacterial solution of 0.5 McFarland was prepared in saline medium (0.85%) and measured by densitometry (DENSIMAT, Biomerieux, France). This solution was equivalent to approximately 1.5 × 108 CFU of each of the bacteria species, and it was the standard dilution for conducting identification with API tests. The appropriate volume of the bacterial solution for leveling the API® 10 S test substrates (see components in Section 2.1) was added. Then, the mixture was transferred to a 10-mL sterile glass vial that was sealed with an aluminum cap furnished with polytetrafluoroethylene (PTFE) (Figure 3a). For experiments carried out with the cosmetic sample, 100 mg of cosmetic were placed in a sterile 10-mL glass vial and inoculated with the corresponding bacteria culture suspension (Figure 3b), or with the inoculated API® 10 S substrate (Figure 3c). In all cases, the vials were incubated in a stove at 37 °C for 24 h prior to SPME-GC-MS analysis.
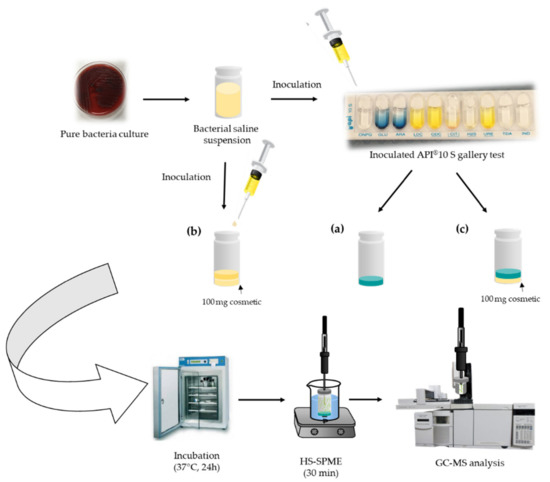
Figure 3.
API-SPME-GC-MS experimental procedure for the analysis of inoculated (a) API® 10 S gallery, (b) cosmetic, and (c) API® 10 S + cosmetic.
2.4. Solid-Phase Microextraction Procedure
The 10-mL glass vials containing the incubated API® 10 S substrates or the incubated cosmetic, depending on the experiment, were immersed in a water bath maintained at 60 °C. Samples were left to equilibrate for 5 min. Then, the PDMS/DVB/CAR SPME fiber was introduced into the vial and exposed to the headspace over the sample for 30 min. Afterwards, the fiber was retracted into the needle of the holder syringe, desorbed in the GC injector for 5 min at 250 °C, and GC-MS analysis was carried out.
2.5. Gas Chromatography-Mass Spectrometry Analysis
Analyses were performed using an Agilent 7890A (GC) coupled to an Agilent 5975C inert mass spectral detector (MSD) with a triple axis detector and an Agilent 7693 autosampler from Agilent Technologies (Palo Alto, CA, USA). Separation was carried out on a ZB-Semivolatiles (30 m × 0.25 mm i.d., × 0.25 µm) obtained from Phenomenex (Torrance, CA, USA). Helium (99.999%) was employed as the carrier gas at a flow rate of 1.0 mL min−1. The GC oven temperature was programmed from 50 °C (3 min) to 200 °C (2 min) at 4 °C min−1, and to 290 °C (3 min) at 20 °C min−1. The total run time was 50 min. The injection volume was 1 µL (for standards injection). Splitless mode was used for injection (1 min, 75 mL min−1). The injection temperature was set at 250 °C. The temperatures of the transfer line, quadrupole, and ion source were set to 290, 150, and 230 °C, respectively. The MSD was operated in the full scan (FS) acquisition mode, monitoring the mass/charge (m/z) fragments between 30 and 800. The MVOCs identification was performed by comparison (match > 80%) between the obtained experimental MS spectral and provided by the commercial spectral library database (NIST). For several compounds (see Section 2.1), the identification was also performed employing analytical standards.
3. Results and Discussion
3.1. Preliminary Experiments
3.1.1. SPME Optimization
Based on previous studies, some parameters, such as the SPME fiber coating and extraction time, were kept constant [20]. One of the most critical experimental parameters affecting the SPME efficiency is the extraction temperature. It is common that temperature fluctuation affects the microbial growth and their enzymatic reaction kinetics [21]. Considering MVOCs can be thermolabile, a wide range of SPME temperatures, 25, 60, and 100 °C, were tested. These experiments were performed in triplicate (see detailed procedure in Section 2.4) for the API® 10 S substrates (blank), and for the API® 10 S inoculated with each one of the three considered bacteria. Figure 4 shows the obtained chromatograms for Escherichia coli. Although the chromatographic abundance was clearly higher performing the SPME at 100 °C, this temperature was discarded since several fatty acids were observed at the end of the chromatogram, probably coming from the bacterial cellular membrane decomposition. This behavior was also observed for the other studied microorganisms. Although the same chromatographic profile was obtained between 25 °C and 60 °C, finally 60 °C was selected as the SPME temperature since the chromatographic abundance was slightly higher in comparison with those obtained performing SPME at 25 °C.
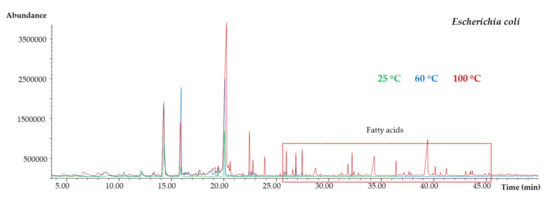
Figure 4.
Comparison between the chromatographic abundance performing SPME at three different temperatures.
3.1.2. Blanks and Bacterial Cultures Characterization
The identification of MVOCs’ origin in complex matrices, such as cosmetics, supposes a great challenge, since it is difficult to evaluate if the detected MVOCs are generated by the metabolism of microorganisms (reaction with different components of the substrate), if they are present in the culture media, or if they come from the cosmetic matrix. For this reason, blank analyses are necessary and must be the first step. In this way, API® 10 S components were incubated according to specifications (37 °C, 24 h) without the presence of bacteria and SPME-GC-MS analysis was carried out. The analysis of the selected moisturizing hand cream sample used as a matrix model without further incubation was also performed. In addition, to evaluate MVOCs produced by bacteria themselves in the absence of substrate, individual bacteria cultures (Escherichia coli, Proteus mirabilis, and Pseudomonas aeruginosa) were analyzed by SPME-GC-MS. Results are depicted in Figure 5, and the identified compounds are summarized in Table 1.
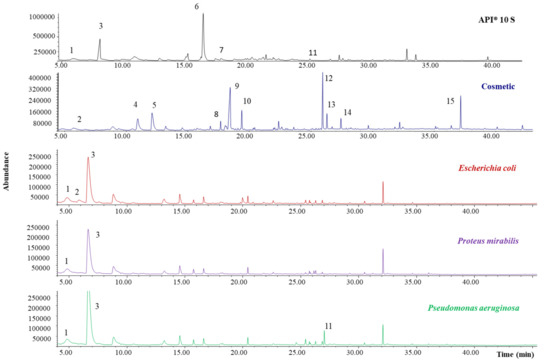
Figure 5.
Obtained SPME-GC-MS chromatograms for the blanks: API® 10 S components, cosmetic, and pure culture media of the 3 studied microorganisms.

Table 1.
Identified compounds by SPME-GC-MS in the blanks: API® 10 S components, cosmetic, and pure culture media of Escherichia coli, Proteus mirabilis, and Pseudomonas aeruginosa.
A total of 15 compounds were identified. The API® 10 S analysis revealed the presence of six MVOCs, with four of them also being detected in the bacteria culture analysis. It is important to note that the chromatographic abundance of the identified compounds in both API® 10 S and individual bacteria culture analysis is very low. It is also important to highlight that, although the API® 10 S chemical composition is well known (see Section 2.1), most of the compounds, excluding 2-nitrophenyl-β-d-galactopyranoside, were not detected by SPME-GC-MS due to their low volatility. In any case, this fact is not required, since the objective of the work was the tentative establishment of a relationship between the formed MVOCs and the bacteria species. Regarding the cosmetic analysis, up to 11 compounds were identified, being an important source of volatile compounds, which bacteria can use as a substrate.
3.2. Formation of MVOCs from API® 10 S
Since the API® 10 S components are known, the formation of MVOCs derived from API® 10 S inoculated with the target microorganisms was evaluated. The experimental procedure was previously detailed in Section 2.3 and Section 2.4. Figure 6 shows the TIC chromatograms obtained after SPME-GC-MS analysis for Escherichia coli, Proteus mirabilis, and Pseudomonas aeruginosa. As can be seen, the chromatographic profile was completely different in the three cases. The identified MVOCs are summarized in Table 2. Up to 10 different compounds were successfully identified for the different microorganisms, with three of them being common for more than one species. Most of the detected MVOCs have been previously reported [15,20] as bacterial presence indicators. Two MVOCs at least were exclusively formed by any of the target bacteria: 2-nitrophenol, and indole by Escherichia coli, phenylethyl alcohol and 2-undecanone by Proteus mirabilis, and 1-undecene and 2′-aminoacetophenone by Pseudomonas aeruginosa.
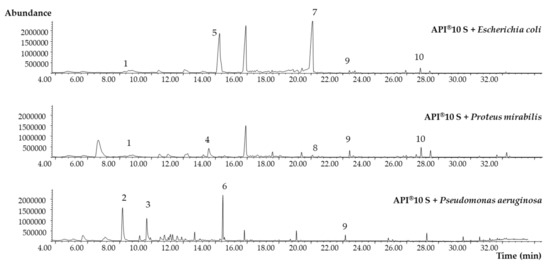
Figure 6.
Identified MVOCs in the obtained SPME-GC-MS chromatograms after API® 10 S inoculation. See the numbers’ correspondence in Table 2.

Table 2.
Identified MVOCs by SPME-GC-MS generated by the studied microorganisms from inoculated API® 10 S, cosmetic, and both substrates.
Indole (peak 7) is a well-known MVOC produced by Escherichia coli specimens through the cytoplasmic enzyme tryptophanase, which hydrolyses tryptophan, generating indole, pyruvate, and ammonia. However, when employing the API® 10 S test, it is necessary to add complementary reagents to reveal the results. Thus, to evaluate the formation of indole, the Kovacs reagent is added, and after 2 min, a red circle is formed, indicating a positive result. In addition, to observe the indole-pyruvic acid that produces a dark-brown color, it is necessary to add ferric chloride after incubation. However, the coloration may be not clear in many cases, resulting in false negatives. The other specific MVOC formed by Escherichia coli, 2-nitrophenol (peak 5), derives from 2-nitrophenyl-β-d-galactopyranoside, and its formation is due to the enzyme β-galactosidase, which allows the metabolization of the disaccharide lactose, producing two reaction products, galactose and 2-nitrophenol. In the API® 10 S test, the presence of 2-nitrophenol is visualized since the test solution turns yellow in the case of positive samples. However, in some cases, the reaction is weak, complicating the conclusion. One of the advantages of using SPME-GC-MS is that the addition of other reagents to unequivocally identify the formed MVOC is not necessary. The formation of phenylethyl alcohol (peak 4) has been reported by several authors, being mainly synthetized by the genus Enterobacter fungi, such as Aspergillus niger, and yeasts, through the shikimate pathway from simple sugars [22]. Recently, Liu et al. used Escherichia coli to mimic a phenylethyl alcohol production pathway found in Proteus mirabilis [23], confirming the obtained results. The presence of 2′-aminoacetophenone (peak 6) identified as specific MVOC synthetized by Pseudomonas aeruginosa was previously reported as a biomarker of its presence in food. This grape-like odorant facilitates attraction to food for several fly species, and constant feeding on 2′-aminoacetophenone improves the level of long-term colonization of the flies’ intestine [24,25].
3.3. Formation of MVOCs from Inoculated Cosmetic Samples
The selected leave-on cosmetic, a moisturizing hands cream, was inoculated with the bacteria cultures in four independent experiments. First, the cosmetic sample was inoculated with each one of the three studied microorganisms, and afterwards, it was inoculated with a mixture of the three species to evaluate possible synergistic effects. Figure 7 shows the obtained chromatograms for the inoculated cosmetic sample, and the identified compounds are also summarized in Table 2.
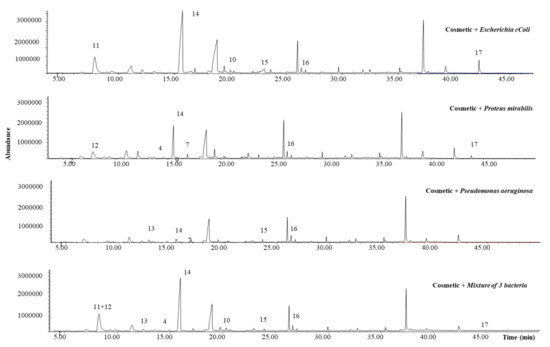
Figure 7.
Identified MVOCs in the obtained SPME-GC-MS chromatograms after cosmetic inoculation.
As can be seen, up to nine MVOCs were successfully identified. Two of them, phenylethyl alcohol (peak 4) and indole (peak 7), both identified in the previous experiments as Proteus mirabilis and Escherichia coli-derived MVOcs, were also detected when the cosmetic was inoculated. Although their chromatographic abundance is lower than the observed in the previous experiments, these two MVOCs could be proposed, a priori, as specific biomarkers for Proteus mirabilis and Escherichia coli cosmetic contamination, respectively. Another identified MVOC, coming exclusively from one of the studied microorganisms, Pseudomonas aeruginosa, was 2-methyl-1-undecanol (peak 13). To the best of our knowledge, it was only identified in soil bacteria and associated to Carnobacterium divergens [15,26], being reported for the first time in this work as a metabolite from Pseudomonas aeruginosa. Regarding MVOCs generated by more than one species, it highlights the chromatographic abundance of 2-oxooctanoic acid (peak 14) for all the studied microorganisms. This branched-chain alpha-keto acid is usually the intermediate metabolite of branched-chain amino acids, which lead to keto acids that release CO2 to yield, in turn, aldehydes, which can be easily oxidized to acid forms [18]. In fact, an aldehyde, 3,3-dimethyl-2-oxobutanal (peak 11), was also identified as Escherichia coli-derived MVOC. Other compounds, such as n-decanoic acid, pentadecane, and 1-octadecanol (peaks 15, 16, and 17, respectively), were detected in more than one species. All of them are common MVOCs generated by several microorganisms [15], and as well as 2-oxooctanoic acid, could be employed as general biomarkers of bacterial contamination in leave-on cosmetics with similar matrices.
3.4. Formation of MVOCs from Inoculated Mixed Substrate
Individual cultures of each studied species and a mixture of them were inoculated to a mixed substrate formed by API® 10 S components and the cosmetic sample to evaluate potential bacteria preferences for one or another substrate. The identified MVOCs after SPME-GC-MS analysis are summarized in Table 2. As can be seen, the 10 MVOCs previously identified coming from the metabolism of the API® 10 S components (numbers 1–10) were also detected when these components are alongside the cosmetic matrix. However, not all the MVOCs previously identified as coming from the bacterial metabolism of the cosmetic matrix (numbers 11–17) were detected. The compounds 3,3-dimethyl-2-oxobutanal, 2,2-dimethylpropanoicaldehyde, and 2-methyl-1-undecanal (numbers 11, 12, and 13, respectively) were not found when the mixed substrate was inoculated, whereas other compounds not previously formed were now identified, such as ethyl octanoate, ethyl decanoate, and 2-nonanone. The two esters were observed when Escherichia coli and the mixture of the three species were inoculated, whereas the ketone was only observed after the inoculation of the three species individually. This behavior could confirm that in the presence of several substrates, the bacteria could select between different components to metabolize.
In view of the obtained results, when SPME-GC-MS analysis is combined with the API® 10 S test, the MVOCs indole and 2-nitrophenol can be considered as Escherichia coli-specific markers, 2-undecanone and phenylethyl alcohol as Proteus mirabilis-specific markers, and 1-undecene and 2′-aminoacetophenone as indicators of bacterial contamination by Pseudomonas aeruginosa. These results were confirmed for indole and phenylethyl alcohol, since they were also identified in the inoculated cosmetic sample. It is undoubted that the MVOCs’ formation is highly dependent on the substrate. To easily visualize this behavior, the differences, expressed in chromatographic abundance, for the specific biomarkers indole, 2-nitrophenol, 2-undecanone, phenylethyl alcohol, 1-undecene, and 2′-aminoacetophenone in the function of the substrate are depicted in Figure 8. As can be seen, when bacteria growth is produced in the presence of a wide range of components that can metabolize (API® 10 S + cosmetic), the MVOCs abundance is lower than in the case of employing only API® 10 S as the substrate, which contains a limited number of nutrients.
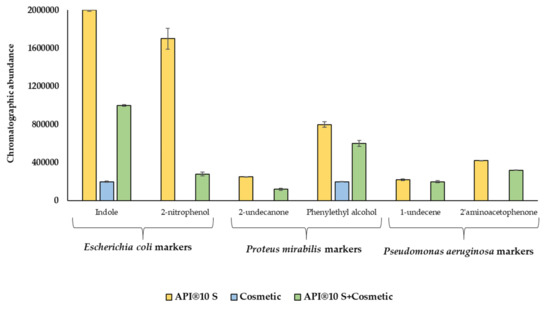
Figure 8.
Influence of substrate in the MVOCs’ formation.
The ultimate challenge of this proposal is to establish the metabolic pathway between the identified MVOCs and the original compound. For several MVOCs, such as indole, the metabolic route is well defined. This compound is produced by Escherichia coli species through the cytoplasmic enzyme tryptophanase, which hydrolyses tryptophan, generating indole, pyruvate, and ammonia, as it is shown in Figure 9a. However, since cosmetics formulations contain a wide range of ingredients that can act as substrates, further experiments with different cosmetic matrices will be necessary to establish a relationship between the formed MVOC and the ingredient present in the substrate. In this work, the formation of 2-oxooctanoic acid when the model cosmetic sample is inoculated with the three studied microorganisms stands out. One of the most abundant organic compounds present in the formulation is 1,2-octanediol (see peak 9 in Figure 5). Taking into account the theoretical approaches and experimental results reported for the degradation of a similar compound, 1,2-propanediol by Escherichia coli [27,28], Figure 9b elucidates a tentative pathway for the formation of 2-oxooctanoic acid from 1,2-octanediol.
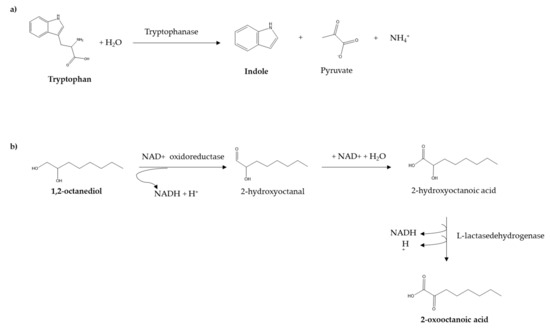
Figure 9.
Proposed metabolic pathways for the formation of (a) indole from API® 10 S substrate, and (b) 2-oxooctanoic acid from a cosmetic ingredient.
4. Conclusions and Future Trends
The combination API-SPME-GC-MS was demonstrated to be a promising technique for the identification of microbial presence in cosmetics. It allowed the identification of several MVOCs as specific markers for each one of the studied bacteria: Indole and 2-nitrophenol for Escherichia coli, 2-undecanone and phenylethyl alcohol for Proteus mirabilis, and 1-undecene and 2′-aminoacetophenone for Pseudomonas aeruginosa, and at least another three MVOCs as general indicators of bacterial presence. The results for indole and phenylethyl alcohol were successfully confirmed when a leave-on cosmetic model was employed as a substrate, demonstrating also that the MVOCs formation is highly dependent on the substrate. In this way, to establish a relationship between the formed MVOC and the original compound present in the substrate, further analysis will be required. These experiments will be carried out considering cosmetic matrixes with common ingredients in a wide range of formulations, so that the scope of this methodology can be extended. Still, the use of SPME-GC-MS was demonstrated to be a fast, robust, and very valuable tool to complement API biochemical tests to evaluate the presence of microbial contamination in cosmetics, offering high extraction and concentration capacities, as well as an unequivocal identification of MVOCs.
In further experiments, the proposed approach can be complemented by other microextraction environmentally friendly techniques, such as micro-matrix solid-phase dispersion (µ-MSPD), to detect other non-volatile metabolites generated by bacteria in cosmetics. Besides, the use of devices, such as ‘Nanodrop’, allowing cell size quantification, could establish a relationship between the bacteria concentration and chromatographic abundance, allowing also the ability to distinguish bacteria if similar types of microorganisms are being studied. The overall objective is to contribute to the development of methodologies for the early detection of bacterial contamination in cosmetics to guarantee product safety and, by extension, consumer safety.
Author Contributions
Conceptualization, M.L., E.V. and M.P.; methodology, M.C., M.L. and M.P.; formal analysis, M.C., E.V., M.P. and E.V.; investigation, M.C. and R.R.; resources, M.L., E.V. and M.P.; data curation, M.C. and M.L.; writing—original draft preparation, M.C.; writing—review and editing, M.C., E.V. and M.L.; supervision, M.L.; project administration, M.L.; funding acquisition, M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by project QFashion (IN852A 2016/157, CONECTA-PEME, Xunta de Galicia). The authors belong to the CRETUS Strategic Partnership (ED431E 2018/01). All these programmes are co-funded by FEDER (UE).
Acknowledgments
Authors would like to thank Pablo Caramés-Méndez (University of Leeds) and Eugenia Guerra (University of Santiago de Compostela) for their valuable support with the KEGG database.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Good Manufacturing Practices (GMP). Guidelines on Good Manufacturing Practices; ISO: Geneva, Switzerland, 2007; p. 22716. [Google Scholar]
- UNION; PEAN. Regulation (EC) No 1223/2009 of the European Parliament and of the Council. Off. J. Eur. Union L 2009, 342, 59. [Google Scholar]
- Scientific Committee on Consumer Products (SCCP). The SCCP’s Notes of Guidance for the Testing of Cosmetic Ingredients and Their Safety Evaluation; SCCP: Brussels, Belgium, 2018. [Google Scholar]
- ISO. Cosmetics-Microbiology-Detection of Specified and Non-Specified Microorganisms; ISO: Geneva, Switzerland, 2017; p. 18415. [Google Scholar]
- Herrera, A.G. Microbiological analysis of cosmetics. In Public Health Microbiology; Springer: Berlin/Heidelberg, Germany, 2004; pp. 293–295. [Google Scholar]
- March, G.A.; Garcia-Loygorri, M.C.; Eiros, J.M.; Bratos, M.A.; Ortiz de Lejarazu, R.l.; Salvador, A.; Chisvert, A. Chapter 18—Microbiological Quality in Cosmetics. In Analysis of Cosmetic Products, 2nd ed.; Elsevier: Boston, MA, USA, 2018; pp. 585–597. [Google Scholar] [CrossRef]
- Commission, E. Safety Gate: The Rapid Alert System for Dangerous Non-Food Products. Available online: https://ec.europa.eu/consumers/consumers_safety/safety_products/rapex/alerts/repository/content/pages/rapex/index_en.htm (accessed on 15 April 2020).
- Neza, E.; Centini, M. Microbiologically contaminated and over-preserved cosmetic products according Rapex 2008–2014. Cosmetics 2016, 3, 3. [Google Scholar] [CrossRef]
- Bashir, A.; Lambert, P. Microbiological study of used cosmetic products: Highlighting possible impact on consumer health. J. Appl. Microbiol. 2020, 128, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Dadashi, L.; Dehghanzadeh, R. Investigating incidence of bacterial and fungal contamination in shared cosmetic kits available in the women beauty salons. Health Promot. Perspect. 2016, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Lores, M.; Celeiro, M.; Rubio, L.; Llompart, M.; Garcia-Jares, C. Extreme cosmetics and borderline products: An analytical-based survey of European regulation compliance. Anal. Bioanal. Chem. 2018, 410, 7085–7102. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, L. Molecular diagnosis of microbial contamination in cosmetic and pharmaceutical products: A review. J. Aoac Int. 2001, 84, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, L.; Ignar, R.; Smalls, S.; Grech, P.; Hamilton, J.; Bosko, Y.; English, D. Molecular detection of bacterial indicators in cosmetic/pharmaceuticals and raw materials. J. Ind. Microbiol. Biotechnol. 1999, 22, 93–95. [Google Scholar] [CrossRef]
- API. Be first to know. Available online: https://www.biomerieux-usa.com/clinical/api (accessed on 25 March 2020).
- Lemfack, M.C.; Gohlke, B.-O.; Toguem, S.M.T.; Preissner, S.; Piechulla, B.; Preissner, R. mVOC 2.0: A database of microbial volatiles. Nucleic Acids Res. 2018, 46, D1261–D1265. [Google Scholar] [CrossRef]
- Saptalena, L.G.; Kerpen, K.; Kuklya, A.; Telgheder, U. Rapid detection of synthetic biomarkers of Escherichia coli in water using microAnalyzer: A field dependence study. Int. J. Ion Mobil. Spectrom. 2012, 15, 47–53. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Pu, Q.; Li, Y.; Wang, X.; Jiang, Y.; Yang, D.; Yang, Y.; Yang, J.; Sun, C. Rapid identification of Staphylococcus aureus, Vibrio parahaemolyticus and Shigella sonnei in foods by solid phase microextraction coupled with gas chromatography–mass spectrometry. Food Chem. 2018, 262, 7–13. [Google Scholar] [CrossRef]
- Elmassry, M.M.; Piechulla, B. Volatilomes of Bacterial Infections in Humans. Front. Neurosci. 2020, 14, 257. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Yang, J.; Ruan, J.; Sun, C. Microbial volatile organic compounds and their application in microorganism identification in foodstuff. Trac Trends Anal. Chem. 2016, 78, 1–16. [Google Scholar] [CrossRef]
- Alvarez-Rivera, G.; De Miguel, T.; Llompart, M.; Garcia-Jares, C.; Villa, T.G.; Lores, M. A novel outlook on detecting microbial contamination in cosmetic products: Analysis of biomarker volatile compounds by solid-phase microextraction gas chromatography-mass spectrometry. Anal. Methods 2013, 5, 384–393. [Google Scholar] [CrossRef]
- Brannan, D.K. Biology of microbes. In Cosmetics Microbiology. A Practical Approach; Geis, P.A., Ed.; Taylor & Francis: Oxfordshire, UK, 2006; p. 50. [Google Scholar]
- Martà nez-Avila, O.; Sánchez, A.; Font, X.; Barrena, R. Bioprocesses for 2-phenylethanol and 2-phenylethyl acetate production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2018, 102, 9991–10004. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, J.; Bai, Y.; Fan, T.-P.; Zhao, Y.; Zheng, X.; Cai, Y. Mimicking a new 2-phenylethanol production pathway from Proteus mirabilis JN458 in Escherichia coli. J. Agric. Food Chem. 2018, 66, 3498–3504. [Google Scholar] [CrossRef] [PubMed]
- Kapsetaki, S.-E.; Tzelepis, I.; Avgousti, K.; Livadaras, I.; Garantonakis, N.; Varikou, K.; Apidianakis, Y. The bacterial metabolite 2-aminoacetophenone promotes association of pathogenic bacteria with flies. Nat. Commun. 2014, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hilton, M.D.; Cain, W.J. Bioconversion of cinnamic acid to acetophenone by a pseudomonad: Microbial production of a natural flavor compound. Appl. Env. Microbiol. 1990, 56, 623–627. [Google Scholar] [CrossRef]
- Effmert, U.; Kalderás, J.; Warnke, R.; Piechulla, B. Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 2012, 38, 665–703. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-H.; Palsson, B.Ã. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1, 2-propanediol. Appl. Env. Microbiol. 2010, 76, 4158–4168. [Google Scholar] [CrossRef] [PubMed]
- KEGG: Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.genome.jp/kegg/ (accessed on 28 April 2020).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).