Abstract
Skin microbiota, and its diversity and balance, play a key role in skin health and beauty, influencing skin moisture, barrier function, and radiance. A healthy skin microbiota limits the growth of detrimental species, protecting the skin from pathologies. Prebiotics can support beneficial populations in outcompeting detrimental ones. Dragon fruit (Hylocereus undatus) contains prebiotic polysaccharides effective on gut bacteria. Its extract was tested in vitro, in a coculture model including representative beneficial and detrimental species, and in double-blind, placebo-controlled clinical trials. Effects on the skin microbiota were measured via 16S rDNA sequencing, and skin health and beauty benefits were evaluated through image analysis, TEWL measurement, and chromametry. Doppler flowmetry measured skin resilience. The extract supported S. epidermidis and S. hominis (beneficial species), while limiting S. aureus and C. acnes (representing pathogens) in vitro. Clinical results demonstrated its beneficial effects on skin microbiota diversity, especially in older volunteers (Faith’s index up to +20% vs. placebo). Improvements were shown in skin sensitivity and resilience (by ca. 30% vs. placebo), skin redness (reflecting inflammation status), pigmentation and radiance (+11% ITA), barrier function (−13% TEWL), and wrinkling. This demonstrates this extract’s positive effects on the beauty, health, and microbiota balance of the skin.
Keywords:
Hylocereus; dragon fruit; pitaya; microbiota; microbiome; in vitro; in vivo; prebiotic; anti-inflammation; brightening 1. Introduction
The skin microbiota includes trillions of bacterial, viral, and fungal species, interacting with each other and the host organism to form a complex multifaceted ecosystem [1]. A balanced, diverse skin microbiota supports healthy skin homeostasis and strengthens the skin barrier through microbial, physical, and immune effects [2,3,4].
Beneficial bacteria form a protective microbial barrier limiting the growth of potentially pathogenic strains, such as Staphylococcus aureus, through competition, pH regulation, and antibiotics production [3,5]. For example, coagulase-negative Staphylococcus such as S. lugdunensis produce lugdunin, a thiazolidine-containing cyclic antibiotic peptide directly inhibiting S. aureus growth [6]. S. epidermidis deploys glutamyl endopeptidase (Esp), a serine protease which dismantles proteins key to S. aureus biofilm formation [7,8]. S. epidermidis can also trigger keratinocytes to produce antimicrobial peptides, including β-defensin [5]. Staphylococcus species, including S. epidermidis and S. hominis, were shown to produce lantibiotics synergistic with human LL-37 cathelicidin antimicrobial peptide, together significantly reducing S. aureus colonization [9]. Conversely, some skin microbes can promote S. aureus biofilm formation, such as Propionibacterium acnes (through coproporphyrin III production [10]).
The skin microbiota influences the physical skin barrier through the regulation of epidermis renewal and barrier structural component production [4]. Activation of the aryl hydrocarbon receptor (AHR) in keratinocytes by microbial metabolites (in a wound healing context) can promote epithelial differentiation and barrier integrity reinforcement [11]. S. epidermidis, for its part, secretes a sphingomyelinase enzyme, both serving the bacterium’s nutrient acquisition and facilitating host tissue production of ceramides (critical components of the epithelial barrier, contributing to skin hydration) [12]. The skin microbiota also plays a complex role in wound development and healing, the full comprehension of which is an ongoing endeavor [13,14].
The skin microbiota can influence innate immune responses and even form symbioses. Influencing immune cell recruitment, differentiation, and more, commensals can contribute to defense against harmful species, especially where the physical skin barrier is compromised [4]. S. epidermidis stimulates innate immunity by activating γδ T cells and upregulating antimicrobial peptide perforin-2 [15].
The microbiota profoundly influences the skin’s susceptibility to external conditions and its overall appearance. However, a definitive understanding of “balanced” skin microbiota remains elusive. Diverse microbial lineages exist in complex, dynamic equilibria, involving specific skin niche microenvironments and interwoven host immune–microbe and microbe–microbe interactions. These equilibria are highly individualized, and subject to continuous short- and long-term fluctuations, responding to changing environmental factors, aging processes, perturbations like changing hygiene practices, and even psychological stresses [16,17,18,19,20,21].
Available evidence suggests a strong positive correlation between skin health and microbial diversity, where healthy skin harbors more diverse and complex microbial communities compared to pathological states. This matches accumulating research positively correlating bacterial diversity with host protection, involving increased immune system adaptability and flexibility [2,22]. A “balanced”, diverse skin microbiota is thus associated with optimal skin barrier function and well-moisturized, calm, even, and radiant skin. Conversely, imbalances (dysbiosis) are associated with skin conditions like eczema, acne, allergies, and sensitive/irritated and dry skin [23,24]. Recent work shows that microbiota disruption can affect communication between the skin and immune system, leading to the emergence of uneven pigmentation [25].
“Biotic” strategies are used in cosmetic products to support a healthy and youthful skin appearance. “Probiotics” refers to the use of live beneficial bacteria—however, current regulations proscribing the presence of such in most cosmetic formulations make this impractical [26]. Microbial metabolites (“postbiotics”), such as Lactobacillus fermentation products and yeast ferment extracts, are sometimes used instead.
Cosmetic formulations also use “prebiotics”, nutrients including various oligosaccharides [27], to (selectively) encourage the growth of desirable commensal species [27,28]. Prebiotics can contribute to improved skin barrier function, reduced inflammation, and enhanced resilience against environmental stressors, and downregulate pro-inflammatory pathways, reducing redness and irritation associated with conditions like eczema and rosacea [29]. In our opinion, the cosmetic benefits of topical prebiotics remain under-studied.
Pitaya (Hylocereus spp.) is an epiphytic cactus, originating from Central and South America. At present, it is very widely cultivated, mainly in sub-tropical/tropical climates, for food, ornamental, medicinal, and other applications [30]. White-fleshed pitaya (Hylocerus undatus) is a naturally rich source of prebiotic oligosaccharides. H. undatus fruit flesh ethanol-extracted oligosaccharides trigger the growth of gut Bifidobacteria and Lactobacilli [31]. H. undatus oligosaccharides include raffinose, stachyose, maltopentaose, and maltotriose [32]. H. undatus also has anti-inflammatory and anti-oxidative potential, possibly due to bioactive compounds including betacyanin, p-coumaric acid, vanillic acid, and gallic acid [33]. Topical application of an H. undatus extract stimulated collagen synthesis (evidenced by elevated hydroxyproline and DNA content) and enhanced tissue integrity (reflected in increased tensile strength and epithelialization) in streptozotocin-induced diabetic rats, promoting wound healing [34]. Pitaya has also been used in traditional medicine to promote wound healing [30]. In light of the fruit’s composition and its demonstrated effects on microbial growth, and the relationships between wound healing and the skin microbiota, it is conceivable that some of the fruit’s efficacy rests on its effects on that microbiota.
On this background, we present here our investigations into the potential of an H. undatus extract to promote skin microbiota diversity and support skin health and appearance.
2. Materials and Methods
2.1. H. undatus Fruit Extract
H. undatus fruit was grown in Israel for Lucas Meyer Cosmetics Ltd. (Kfar Buli Agriculture Ltd., Kfar Bilu, Israel), based on original material identified by Pr. Yosef Mizrahi (Ben-Gurion University of the Negev) and using a sustainable disconnected substrate technique in which the vines grow disconnected from the ground, submerged in inert medium and fed through a micro-irrigation system using recycled wastewater. This allows for a highly sustainable crop with minimal effects on the local land, and optimal use of fertilizers and pest control. Pollination and harvest were performed by hand.
The fruits, harvested in early Fall 2017 (coculture study), 2015 (clinical trial 1), and 2013 (clinical trial 2), were peeled and blended to allow extraction of active components in their own water, thereby reducing additional use of water resources. Solids were separated by centrifugation, after which the supernatant was heated above 100 °C for a minimum of 30 min, to remove protein residues. The resulting mixture was centrifuged again and filtered down to 0.22 μm, followed by the addition of preservatives to prevent microbial contamination. The resulting finished aqueous extract (IBR-Dragon™, Lucas Meyer Cosmetics, Yavne, Israel) was used in the studies described herein.
2.2. In Vitro Competitive Coculture Assay
This study aimed at evaluating the effect of the H. undatus fruit extract on the population balance of a model coculture of four bacterial species: Staphylococcus epidermidis (ATCC #12228, Manassas, VA, USA); Staphylococcus hominis (ATCC #27844); Staphylococcus aureus (ATCC #6538); and Propionibacterium acnes (ATCC #11827). For the purposes of this study, the former two species represent commensal/beneficial bacterial populations, while the latter two represent detrimental/pathogenic populations. Each strain was initially grown alone in specific media: Tryptone soya broth (Oxoïd CM0129, Thermo Fisher Scientific, Waltham, MA, USA) for Staphylococcus species and Brucella broth (Condalab 1223, Madrid, Spain) for Cutibacterium acnes.
H. undatus fruit extract was placed in microplates in minimal culture medium at 0.1, 0.5, and 1.0%. Each sample was then contaminated with the four strains, in initial proportions intended to produce an equally balanced population distribution, at about 106 CFU/mL for each species after 3 to 6 h. The plates were incubated at 32 °C for 48 h.
Culture medium samples were collected at 24 and 48 h. Each sample was placed on specific agar for each selected strains (C. acnes: Columbia with 5% sheep blood, Oxoid PB5039A; for Staphylococcus spp., trypsone soya, Oxoid PO5012A), and incubated for 48 h at 37 °C. After incubation, a CFU count was carried out for each strain. Populations at T24h and T48h and in the absence of product were compared. The analysis was performed in triplicate for each sample. Two controls were included in the study: a minimal medium control (untreated condition, expected to yield slower growth), and an optimal medium control (expected to lead to optimal growth).
2.3. Clinical Trial 1
2.3.1. General Design
The H. undatus fruit extract described above was tested in a clinical trial, with the aim of evaluating its effect on the skin microbiota in vivo, together with is benefits on skin resilience and skin pigmentation spots, as well as the skin’s inflammation status.
An appropriate gel-cream model formulation containing 1% of the extract and a corresponding placebo product were designed for this study (Table 1).

Table 1.
Clinical trial formulations.
The design and execution of this study followed the principles of the Declaration of Helsinki and its amendments, as well as the spirit of Good Clinical Practice Guidelines, and complied with Portugal Law 46/2004 of 19 August 2004. The protocol was approved by an Internal Review Board (opinions 6882/2021 and 6883/2021), as well as by the Ethical Commission of PhD Trials (4 January 2021). Written informed consent was obtained from study participants, who also consented to the consequent use of study photographs. RNEC registration number 178,566 was attributed to this study, following local regulations (EMA/INFARMED, Portugal) for cosmetic clinical trials.
The protocol was built on the basis of a double-blind, placebo-controlled, randomized split-face/body design, where participants applied each product to a particular half-face (left/right) and the forearm on the same side. Application took place twice a day for 28 days, with measurements (including microbiota sampling) at Days 0 (D0), 7 (D7), and 28 (D28).
In total, 33 subjects were included in the final analysis (36 subjects were recruited in total, and 3 dropped out for reasons unrelated to the products). Participants were 30–55 years old and female. From the standpoint of ethnicity, 23 participants who finished the study were described as Caucasian, and 10 as Brazilian (i.e., mixed Caucasian–African ancestry). Fitzpatrick phototypes were, respectively, II–III and IV. Exclusion criteria included the presence of cutaneous marks in the areas of interest, such as extreme pigmentation irregularities, scars, and excessive pilosity; allergy or reactivity to skin care products; history of malignant melanomas, bilateral mastectomy with lymph node removal, or bilateral axillary lymph node removal; immune deficiency or auto-immune disease; treatment for malignancy within 6 months prior to enrollment; current treatment for asthma or diabetes; treatments susceptible of interfering with the study measurements, including anti-aging, anti-wrinkle, or hormonal treatments; plastic surgery; intensive sunlight or UV exposure; systemic disorders (cardiovascular, pulmonary, digestive, neurologic, psychiatric, genital, urinary, hematological, or endocrine); anticipated vaccination during the study, or vaccination within 3 weeks prior; and pregnancy or breast-feeding.
2.3.2. Microbiota Analysis
The skin microbiota was sampled by swabbing. While awaiting analysis, swab samples were kept at −80 °C.
Extraction and purification of DNA were carried out using a DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany, 47016). Cells were lysed using a combination of mechanical (beads) and chemical (detergent) means as provided by the kit. Extracted DNA was then purified on a silica membrane.
16S sequencing: The 16S Ribosomal DNA (rDNA) variable region (V1 to V3) was amplified using primers AGAGTTTGATCCTGGCTCAG (27F) and ATTACCGCGGCTGCTGG (534R) as described in Human Microbiome Project sequencing protocols [35]. An Illumina MiSeq (illuina, San Diego, CA, USA) with paired-end technology was used for amplicon sequencing.
Sequence processing and taxonomic assignment: Fastq files from sequencing were processed to filter, denoise, and merge sequences, remove chimeras, and assign taxonomy by using the Qiime2 software [36] (sequence quality was evaluated with the FASTQC software [37]); sequences were then trimmed and filtered according to sequence quality, before denoising and chimera removal by the DADA2 algorithm [38]; each sequence was then assigned to its Operational Taxonomic Unit (OTU) classification using the Qiime2 classify-sklearn classifier, trained on the SILVA release 138 database [39].
The abundance of each OTU corresponds to the number of associated reads. Taxa abundance was estimated by summing OTU abundances associated with each taxon (from Phylum to Species). The differential abundance of each taxon was analyzed using the Deseq2 R package [40]. A taxon was considered as differentially abundant if a. it was detected in at least three samples in one out of the two compared groups; b. the log2 Fold Change was <−0.5 or >0.5; and c. the non-adjusted p-value was <0.05.
Diversity analysis was performed on denoised and classified Operational Taxonomic Units (OTUs) with Qiime2 diversity core analysis to estimate alpha-diversity indexes for each sample and beta-diversity distances between the different samples.
Faith’s phylogenetic diversity index [41,42] (which takes into account OTU phylogenetic proximity) was selected as our index for alpha diversity. Values were compared between groups using the Wilcoxon paired test; the diversity index ratio between two groups was calculated as the median of pair (subject) ratios.
2.3.3. Other Measurements
Red spots and visible spots were evaluated through image analysis (VISIA-CR, Canfield, Parsippany, NJ, USA), respectively, serving as indicators of the extract’s effect on the skin’s irritation status and pigmentation. Standardized photographic images were obtained under cross-polarized lighting for both hemifaces at D0, D7, and D28. A mask was created for the defined measurement area and applied in subsequent images in order to calculate differences vs. D0. Visia images were also used as the source for general macrophotography records.
Skin redness/irritation states were further evaluated through clinical evaluation by a dermatologist. Grading was carried out on the whole hemiface, along a 5-point scale from Score 0: very red or irritated skin to Score 4: No skin redness/irritation.
Skin sensitivity (or conversely, resilience) was evaluated by Laser Doppler Flowmetry (via skin microcirculation) with histamine stimulation. Histamine is known to induce itching and to increase capillary permeability and vasodilation. This is a useful model to assess cutaneous susceptibility to inflammation and the resilience of the cutaneous barrier, through simultaneous measurement of skin microcirculation. Histamine was dispensed by iontophoresis (30 s at 200 μA, Perilont iontophoresis system, Perimed, Jakobsberg, Sweden). Laser Doppler Flowmetry (LDF) was used to assess skin microcirculation, using a monochromatic low-energy (780 nm) laser (Perilont LDPM PF750/PF5000 + PF 5010 Periflux laser channel, Perimed, Jakobsberg, Sweden), to detect the movement of red blood cells in skin microcirculation (average depth of 1.5 mm, corresponding to the superficial dermal plexus). The results are expressed in arbitrary units (AUs) or BPUs (blood perfusion units). Time elapsed between histamine application and blood flow increase was defined as the reaction onset time. We also evaluated the maximal amplitude of the increase in microcirculation.
2.4. Clinical Trial 2
2.4.1. Clinical Design
A second clinical trial aimed at evaluating the effects of the H. undatus fruit extract on indicators of skin health and beauty, including skin barrier function, skin redness, skin radiance, and wrinkling. This trial used the same formulations as described above (Table 1) for clinical trial 1.
The design and execution of the study followed the principles of the Declaration of Helsinki and its amendments, as well as the spirit of Good Clinical Practice Guidelines and Portugal Law 46/2004 (9 August 2004). The protocol received Internal Review Board approval (opinions 1015/15 and 1016/15, 5 January 2015) and that of an Internal Ethical Commission (PhD Trials, opinion nº 028/2014, 29 December 2014). Written informed consent was obtained from study participants, including for subsequent study photograph use.
The study was designed based on a double-blind, placebo-controlled, randomized split-face design, where participants applied each product to a defined half-face (left/right). Application took place twice a day for 28 days, with measurements taken at D0 and D28.
Twenty-six subjects were recruited (distinct from the group recruited for Trial 1 above) and included in the final analysis. Participants were 35–55 years old, female, of Caucasian ethnic background, with Fitzpatrick phototypes II–III. All skin types were represented. Participants were selected for the presence of ageing signs (wrinkling) around the periocular area. Exclusion criteria included the presence of cutaneous marks in the areas of interest, such as extreme pigmentation irregularities, scars, and excessive pilosity; allergy or reactivity to skin care products; any history of malignant melanomas; treatments susceptible of interfering with the study measurements, including anti-aging, anti-wrinkle, or hormonal treatments; plastic surgery; intensive sunlight or UV exposure; and pregnancy or breast-feeding.
2.4.2. Measurements
Trans-Epidermal Water Loss (TEWL), measured using a Tewameter® TM 300 (Courage & Khazaka, Koln, Germany) with a cylindrical probe 10 mm in diameter and 20 mm in height, was used to evaluate skin barrier function.
The effect on skin complexion, radiance, and redness was evaluated through the expression of skin tone as Hunter L,a,b color space coordinates, as measured using a CR-400 Chromameter (Minolta, Tokyo, Japan). Skin redness was evaluated through the a* (red-green axis) coordinate, while skin radiance was evaluated through the L* (luminescence/light–dark axis), and the Individual Typological Angle (ITA) index [43,44], a one-number indicator of skin tone calculated from the L* and b* (blue–yellow axis) values as ITA = tan−1[(L8 − 50)/b*]. A higher ITA index indicates lighter skin.
Wrinkling was evaluated in the crow’s feet area using a PRIMOS 3D (Phase Shifting Rapid In-vivo Measurement of Skin) instrument (GFMesstechnik, Berlin, Germany) with a 40 × 30-mm evaluation area. Using this instrument, 3D images of the skin topography were obtained through a digital fringe projection using micro mirror displays, and quantified based on observed fringe deflection.
Illustrative photographs were produced using standardized photographic images obtained under normal and cross-polarized lighting at D0 and D28 using a VISIA-CA system (Canfield, Parsippany, NJ, USA) with software v6.
2.5. Statistics
Prism 9 (GraphPad, Boston, MA, USA) was used to analyze the numerical data obtained in our in-vivo studies, using p = 0.05 (per generally accepted practice in this field) as the statistical significance threshold. Normality was used to select specific statistical significance tests: when the data showed positive normality, we used the paired Student t-test, a parametric test; and when the data showed negative normality, we applied the Wilcoxon test, a non-parametric test.
3. Results
3.1. In-Vitro Competitive Coculture Assay
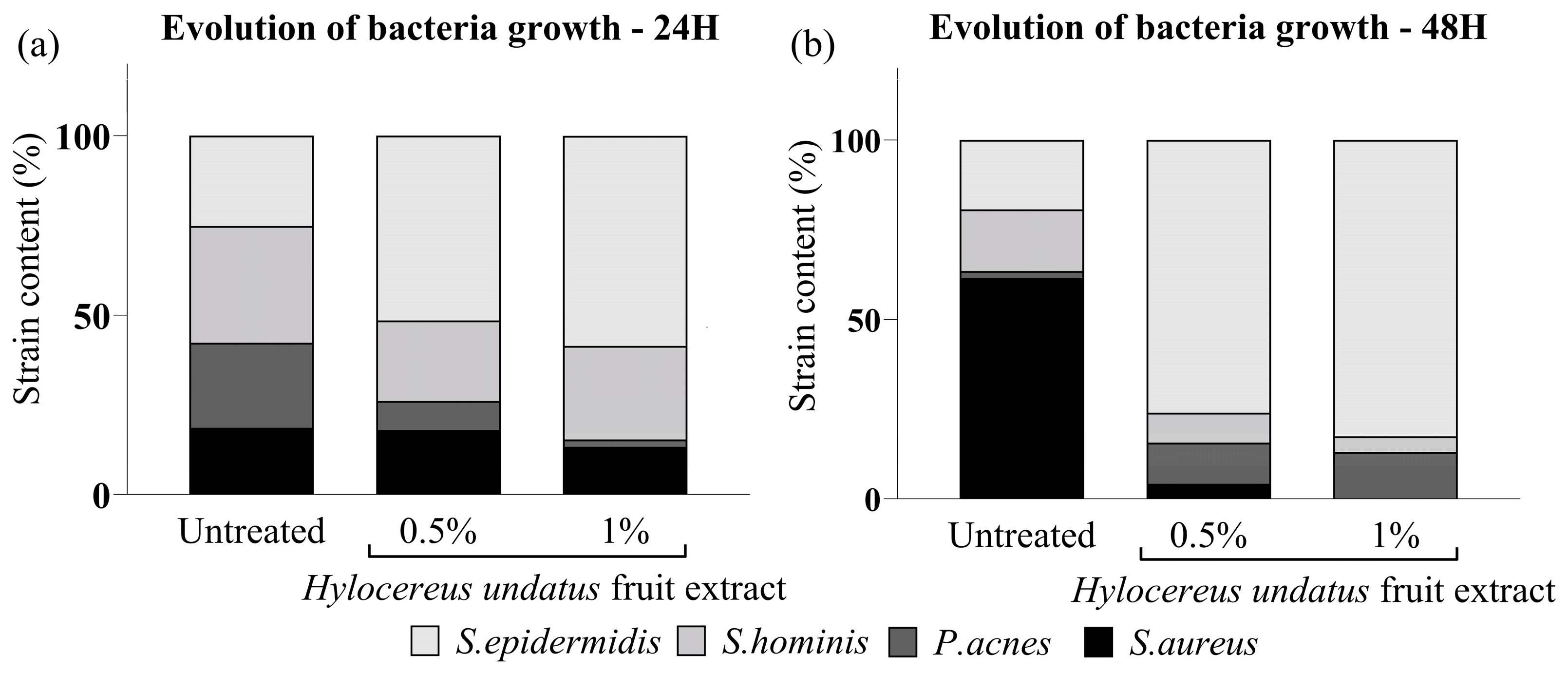
The H. undatus fruit extract was shown to increase bacterial growth overall, to levels comparable to optimal conditions (full medium). S. hominis and S. epidermidis (beneficial strains) exhibited optimal growth under treatment with the extract. On the other hand, the harmful strains P. acnes and S. aureus showed strongly reduced preponderance under the same treatment (Figure 1). This indicates that the extract could possess a prebiotic effect, leading to a beneficial rebalancing of the skin microbiota.
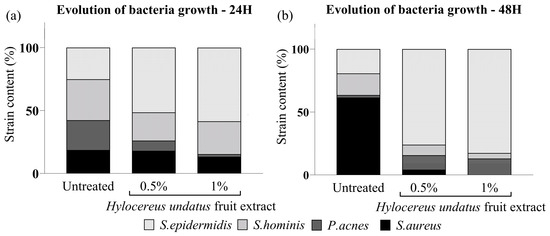
Figure 1.
In-vitro competitive coculture assay showing the effect of the H. undatus fruit extract on representative commensal and pathogenic bacteria, as mean relative proportion of total population for each strain, at 24 h (a) and 48 h (b).
3.2. Clinical Trial 1
3.2.1. Microbiota Analysis Results
A total of 11,658 OTUs were identified in the dataset; 6236 of them (53%) were assigned down to the genus level and 4620 (40%) to the species level. Those OTUs were, respectively, assigned to 602 different genera and 998 different species. In all sample groups, the three dominant Phyla were Actinobacteriota, with Cutibacterium and Corynebacterium as the main genera, and Firmicutes, with Staphylococcus as the main genus. This is considered typical for sebaceous areas of the skin, such as the face [1].
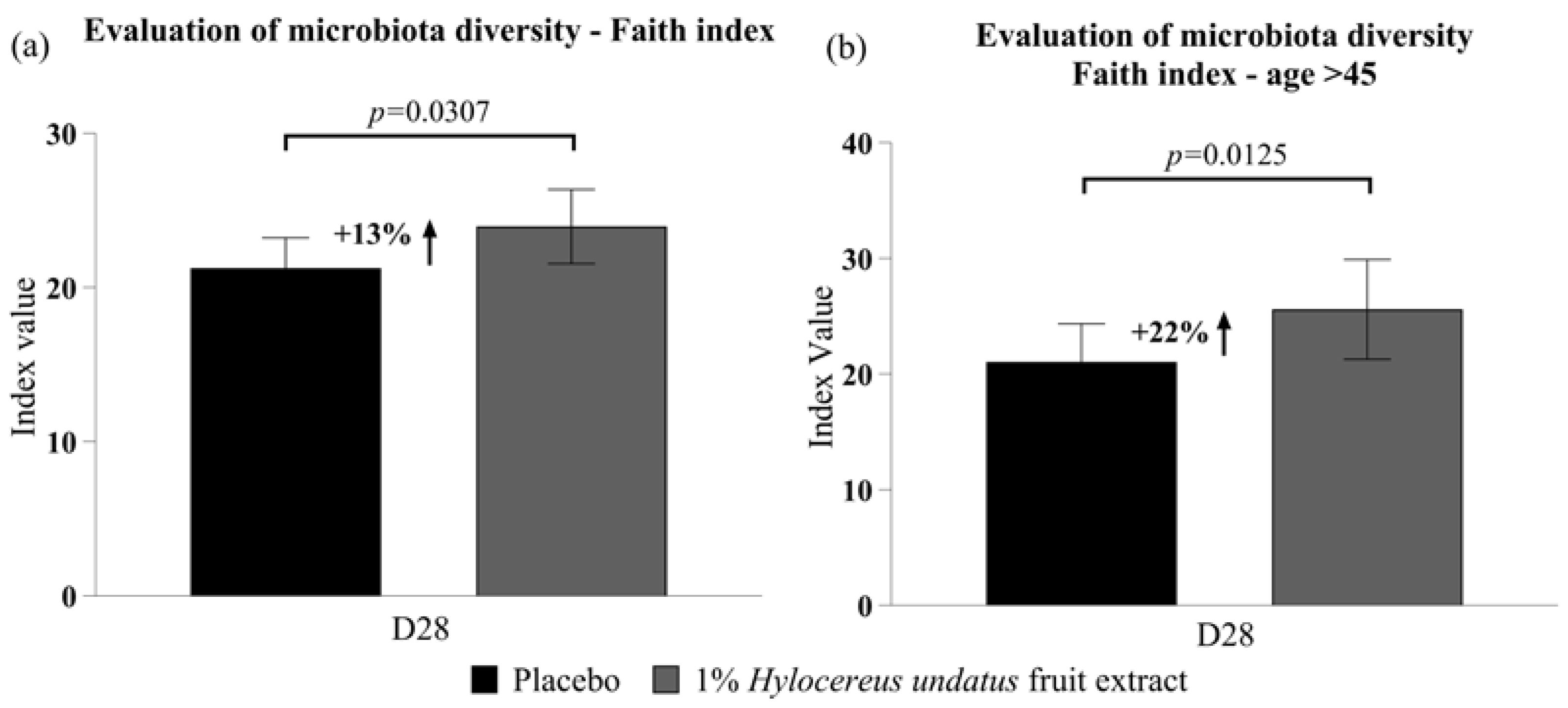
After 28 days, compared to placebo, 1% H. undatus fruit extract in cream showed, in the global trial participant population, a 13% improvement in Faith’s phylogenetic diversity index (p < 0.05) (Figure 2a; this improvement was observed in 67% of volunteers). In the subgroup of volunteers aged 45+ (n = 13), Faith’s phylogenetic diversity was improved by 22% (p < 0.01) with the active vs. the placebo (Figure 2b; an improvement was observed in 85% of volunteers over 45).
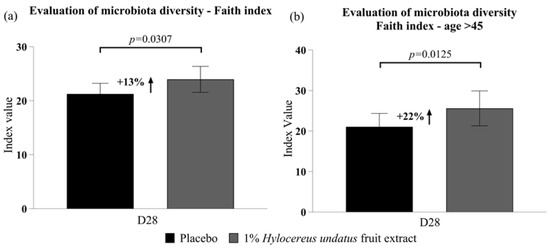
Figure 2.
Evaluation of microbiota diversity (mean +/− SEM) according to Faith’s index (a) in the whole participant group; (b) in participants aged over 45 years—showing improvements in phylodiversity with the product containing 1% H. undatus fruit extract.
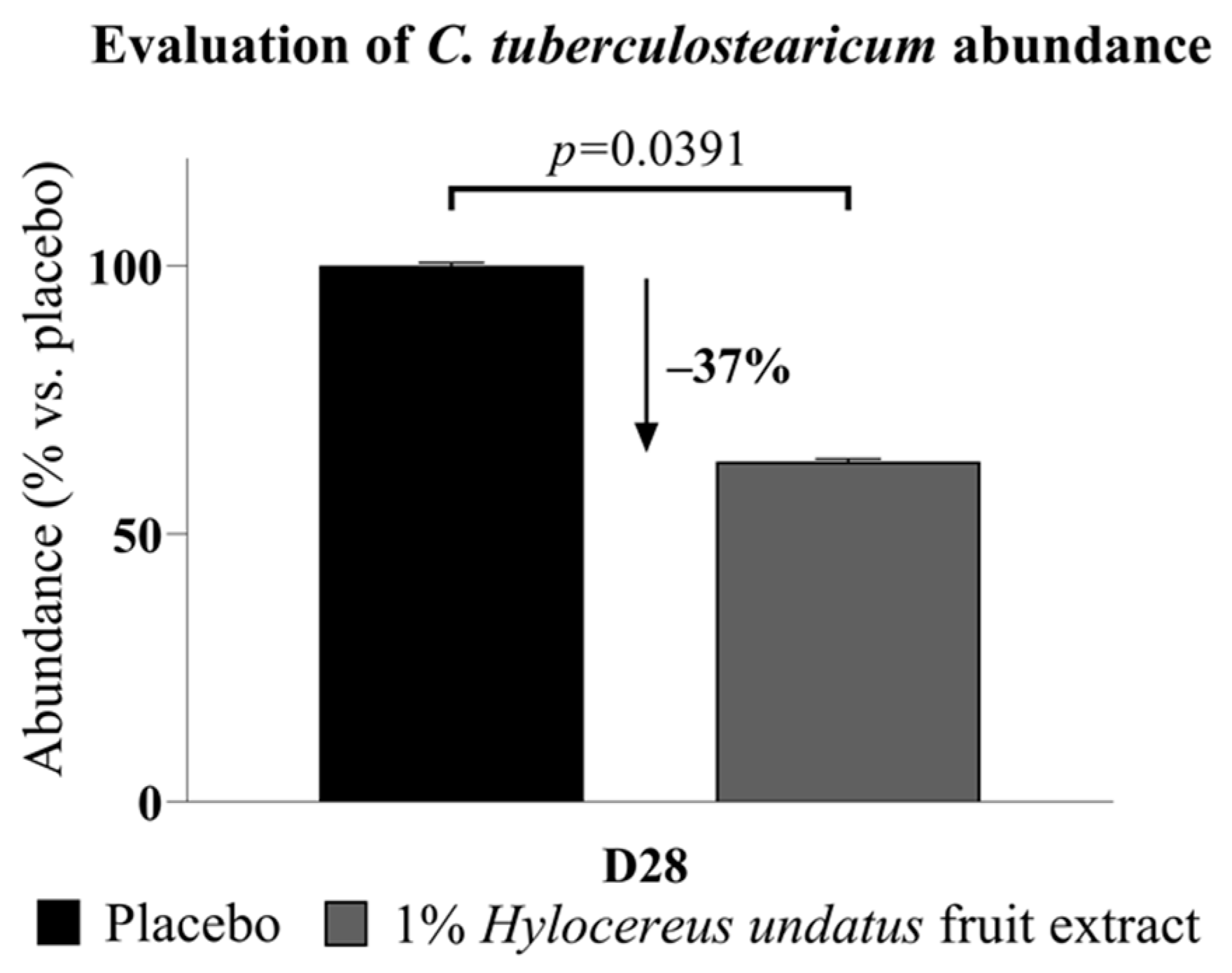
Of particular interest among the sequencing data, Corynebacterium tuberculostearicum, a major skin bacterium and pathogen identified in almost all samples (97%), showed significantly reduced abundance (−37%) after 28 days (Figure 3).
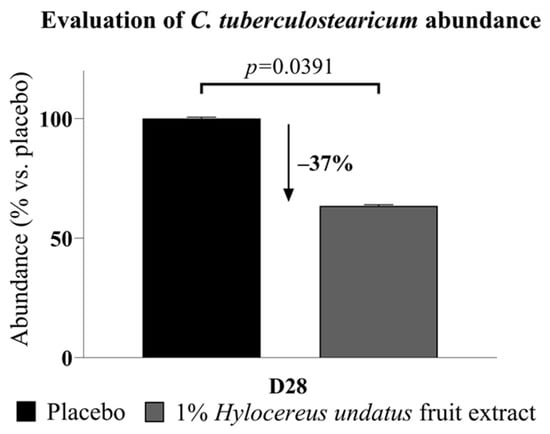
Figure 3.
Corynebacterium tuberculostearicum abundance, reduced after 28 days’ treatment with the product containing 1% H. undatus fruit extract (mean +/− SEM).
3.2.2. Effects on Skin Health and Beauty
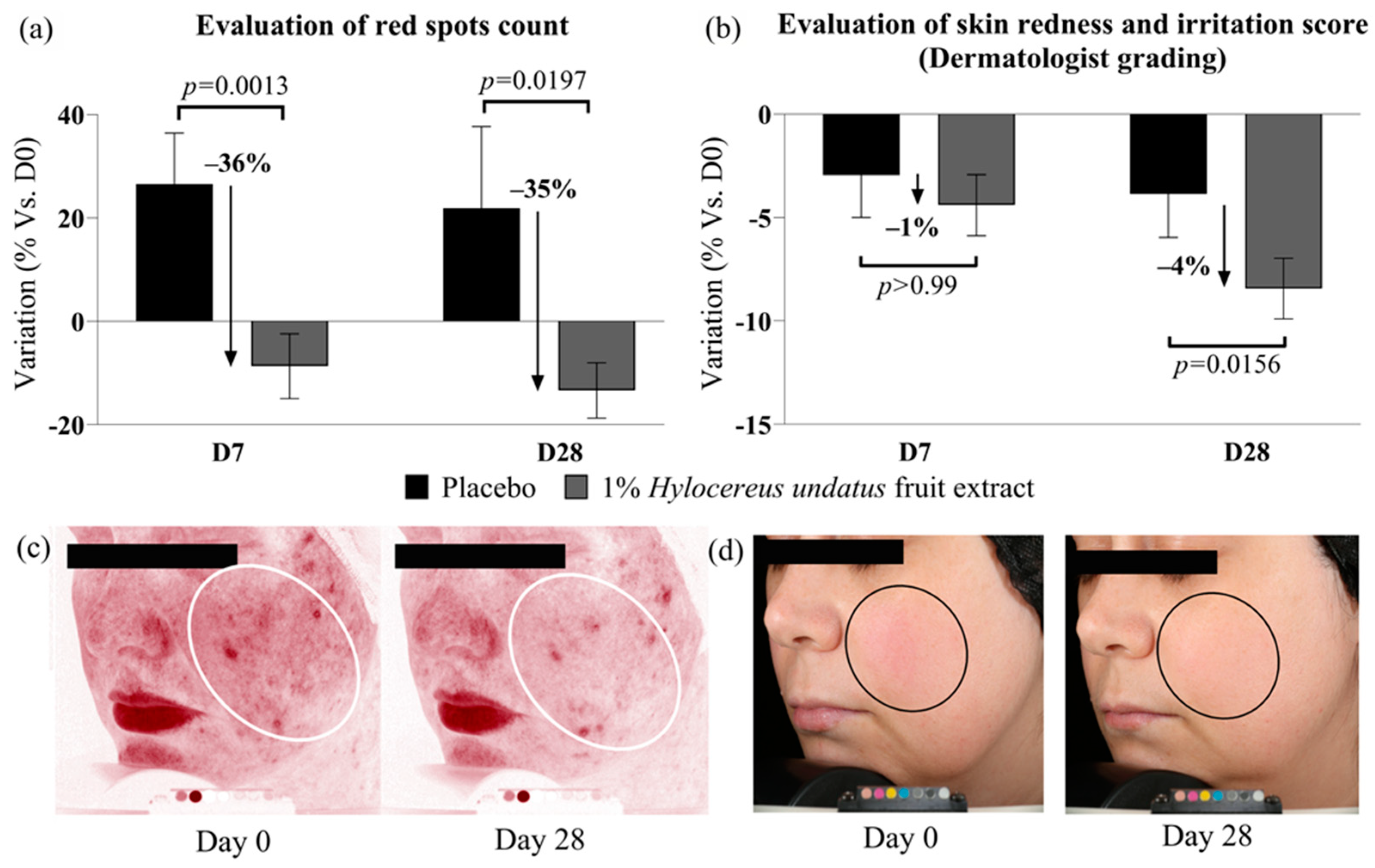
1% H. undatus fruit extract in formula showed strong efficacy on red spot counts when compared to placebo: −35% at D28, and −36% at D7, with statistical significance vs. placebo in both cases (Figure 4a). Compared to D0, the reduction in red spot counts was statistically significant at D28 only (variations with placebo were not statistically significant vs. D0). These data indicate reduced skin inflammation with the active product. This is borne out by the results of clinician grading, which show a small (4%) but statistically significant improvement in redness/irritation grades at D28 vs. placebo and D0 with 1% H. undatus fruit extract in formulation (Figure 4b). Figure 4c,d show illustrative Visia photographs, demonstrating the effect on red spots and skin redness.
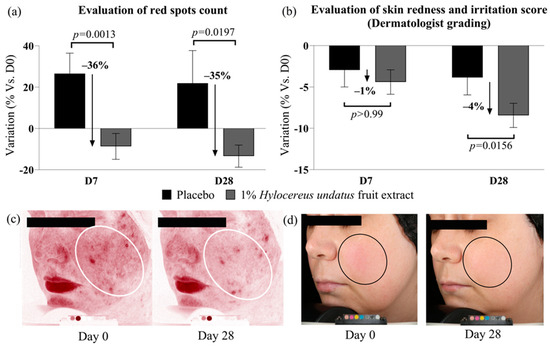
Figure 4.
Evaluation of skin inflammation status indicators: red spot count (a) and clinical scoring of skin redness and irritation (b), both indicating reduced skin inflammation status with the product containing 1% H. undatus fruit extract (shown as mean +/− SEM). Illustrative Visia images: (c) red spots, volunteer #14 (age 35); (d) visible redness, volunteer #36 (age 42). Ovals indicate regions of interest.
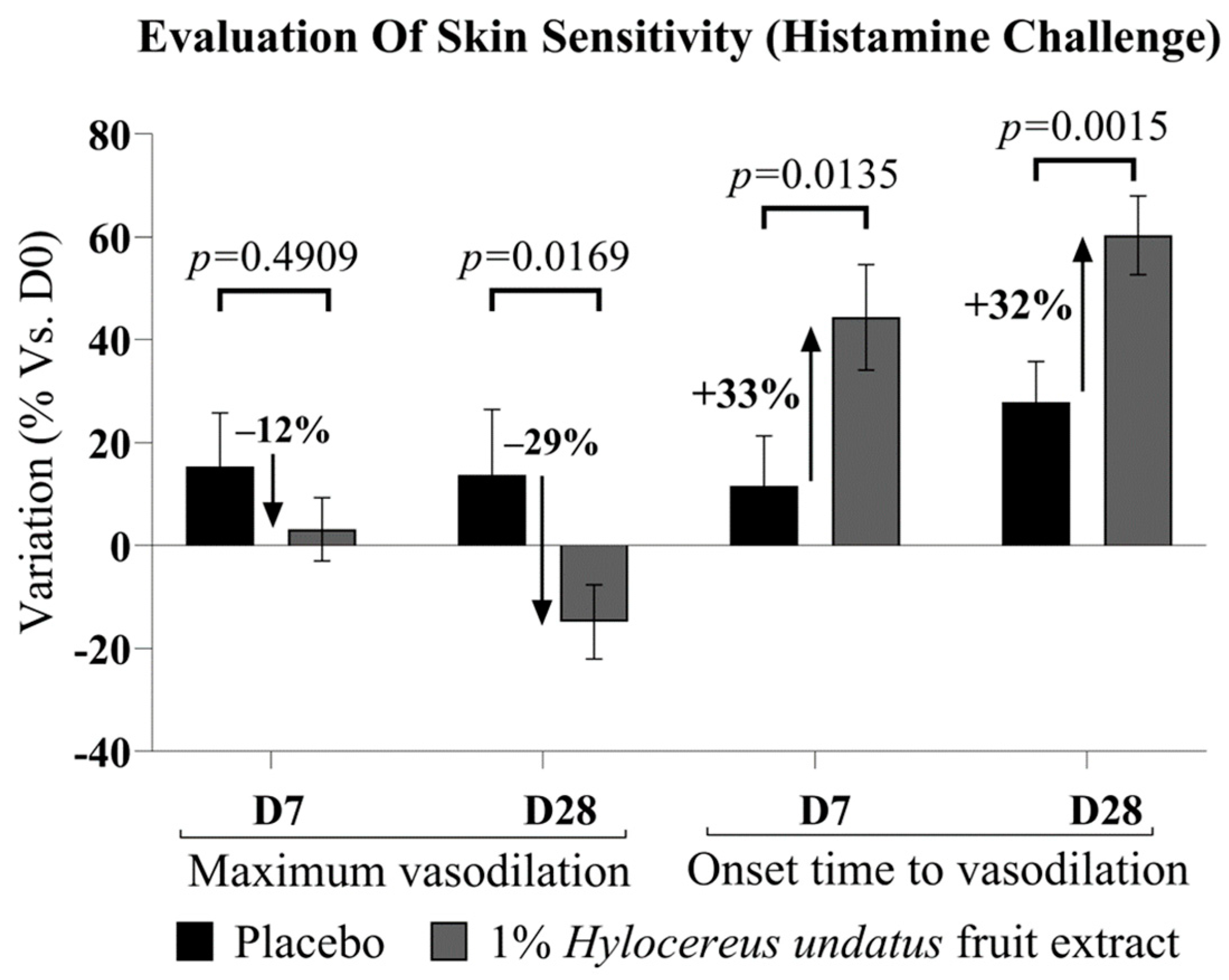
Upon histamine challenge, microcirculation data (Figure 5) showed a strong advantage for 1% H. undatus fruit extract in formula over placebo at D28, both in terms of onset time (time required to reach maximum irritation) and in terms of peak irritation—exhibiting, respectively, a 32% delay in onset time and a 29% drop in maximum amplitude. This effect is statistically significant vs. placebo and D0. At D7, an advantage for the active formulation’s effect on onset time is already observable and significant vs. placebo and D0. The active formulation’s advantage in terms of maximum irritation is observable as a trend at D7, but is not yet statistically significant. In this test, irritation was observed as an increase in microcirculation; therefore, a reduction in the peak value and an increase in onset time indicate anti-inflammatory effects.
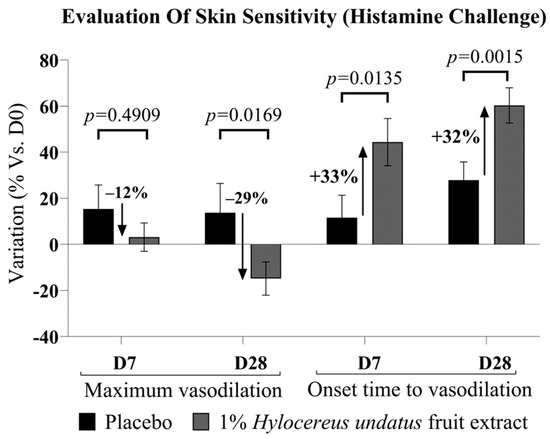
Figure 5.
Evaluation of skin sensitivity, as resilience vs. histamine challenge, showing maximum vasodilation amplitude (left) and onset time (right) (mean +/− SEM). In all cases, improvements are observed, indicating greater skin resilience, with the product containing 1% H. undatus fruit extract.
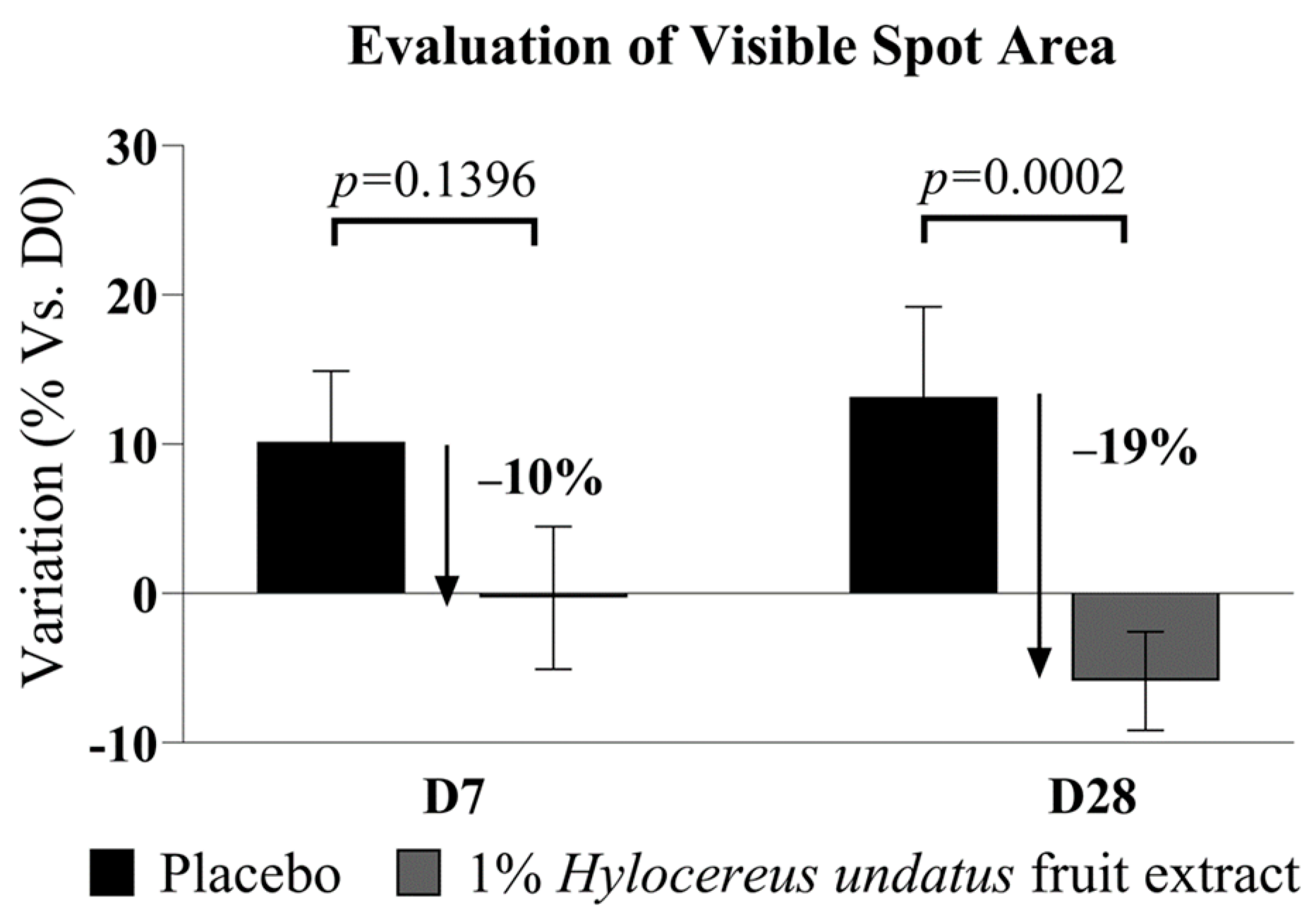
Finally, 1% H. undatus fruit extract in formula shows an advantage vs. placebo in visible spot areas at D28 (−19% vs. placebo, with statistical significance vs. placebo) (Figure 6). At D7, a somewhat smaller advantage is already observable, but not yet statistically significant. This result indicates that treatment with the extract reduces uneven skin pigmentation.
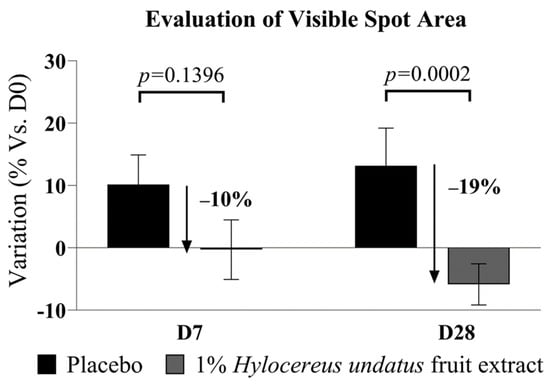
Figure 6.
Evaluation of visible spot areas (Visia image analysis, mean +/− SEM), showing reduced visible spotting with the product containing 1% H. undatus fruit extract.
3.3. Clinical Trial 2
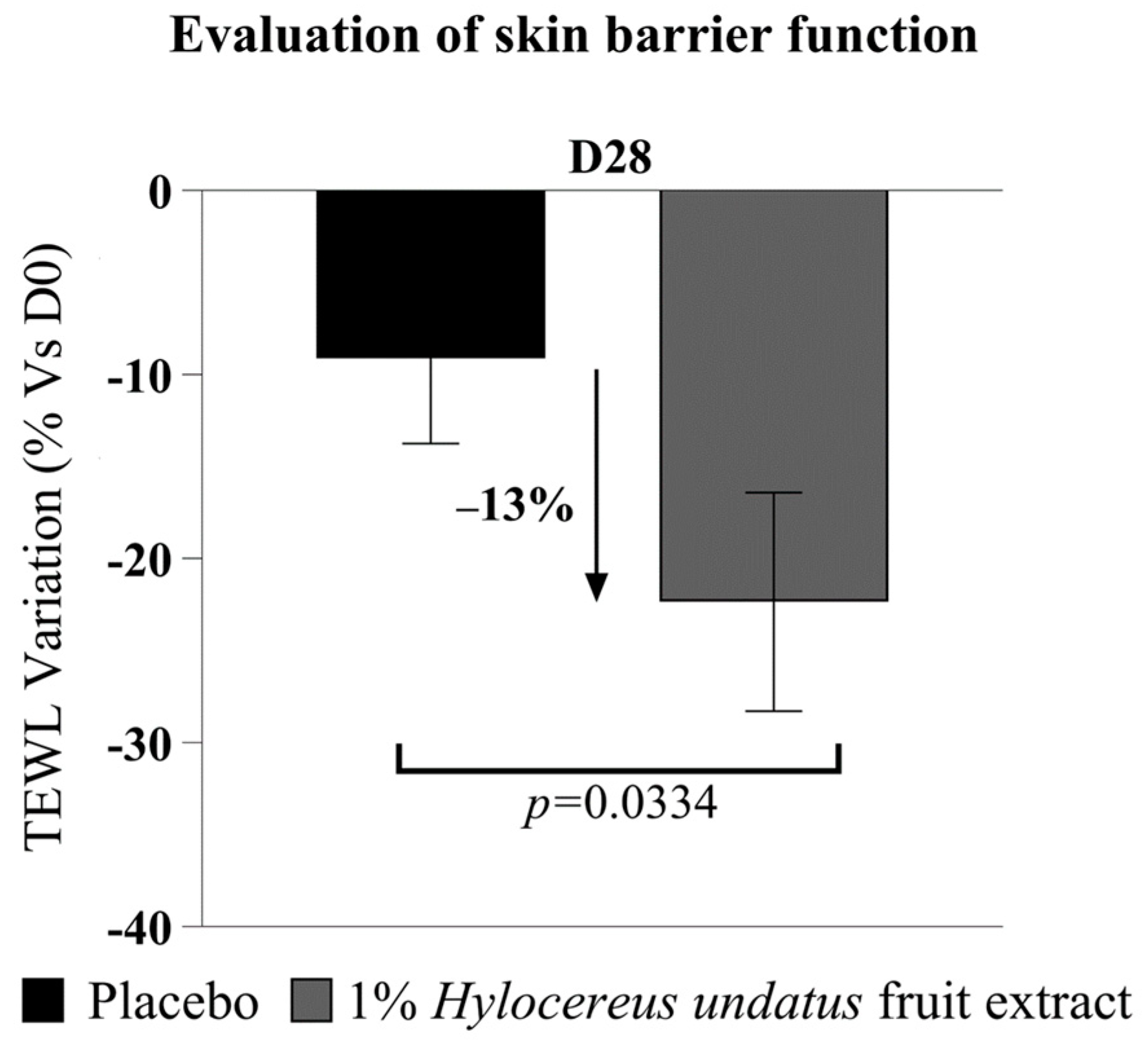
The Tewameter data (Figure 7) show a significant improvement in skin barrier with 1% H. undatus fruit extract in formula, as a reduction in TEWL at D28 (−13% vs. placebo), with statistical significance vs. D0 and vs. placebo—indicating that treatment with the extract reinforces the skin barrier.
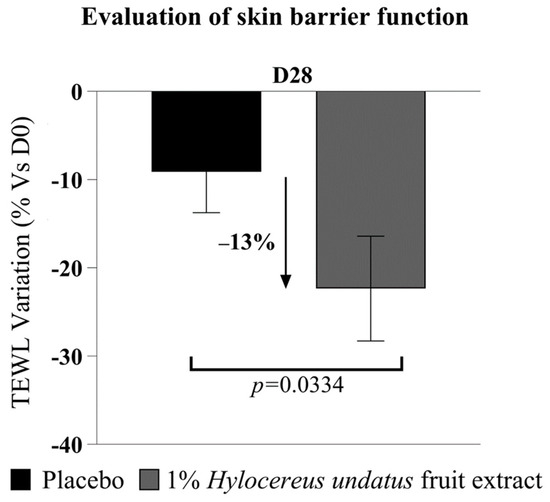
Figure 7.
Evaluation of transepidermal water loss (mean +/− SEM), showing enhanced skin barrier function with the product containing 1% H. undatus fruit extract.
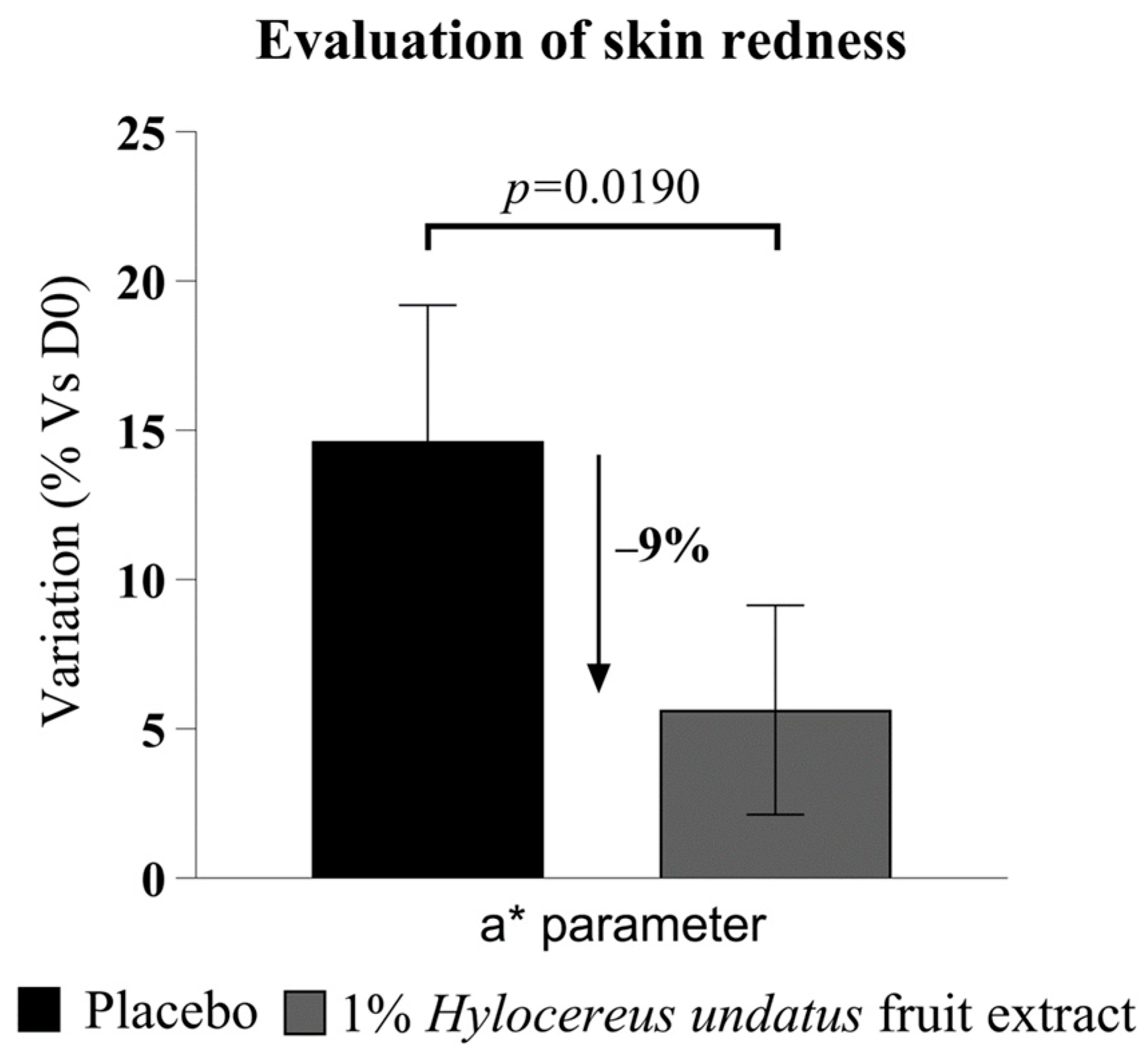
The chromameter data (Figure 8) show a statistically significant advantage in redness for 1% H. undatus fruit extract in formula vs. placebo at 28 days. Both products seem to increase redness slightly, but the increase is smaller with the active product. This indicates that treatment with the extract leads to reduced skin inflammation, reinforcing the indications derived from the results of the first clinical trial described above.
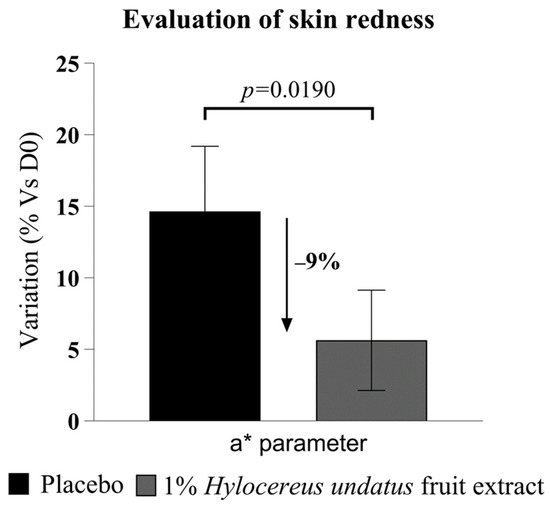
Figure 8.
Evaluation of skin redness, as Hunter color space parameter a* (mean +/− SEM), showing reduced skin redness with the product containing 1% H. undatus fruit extract.
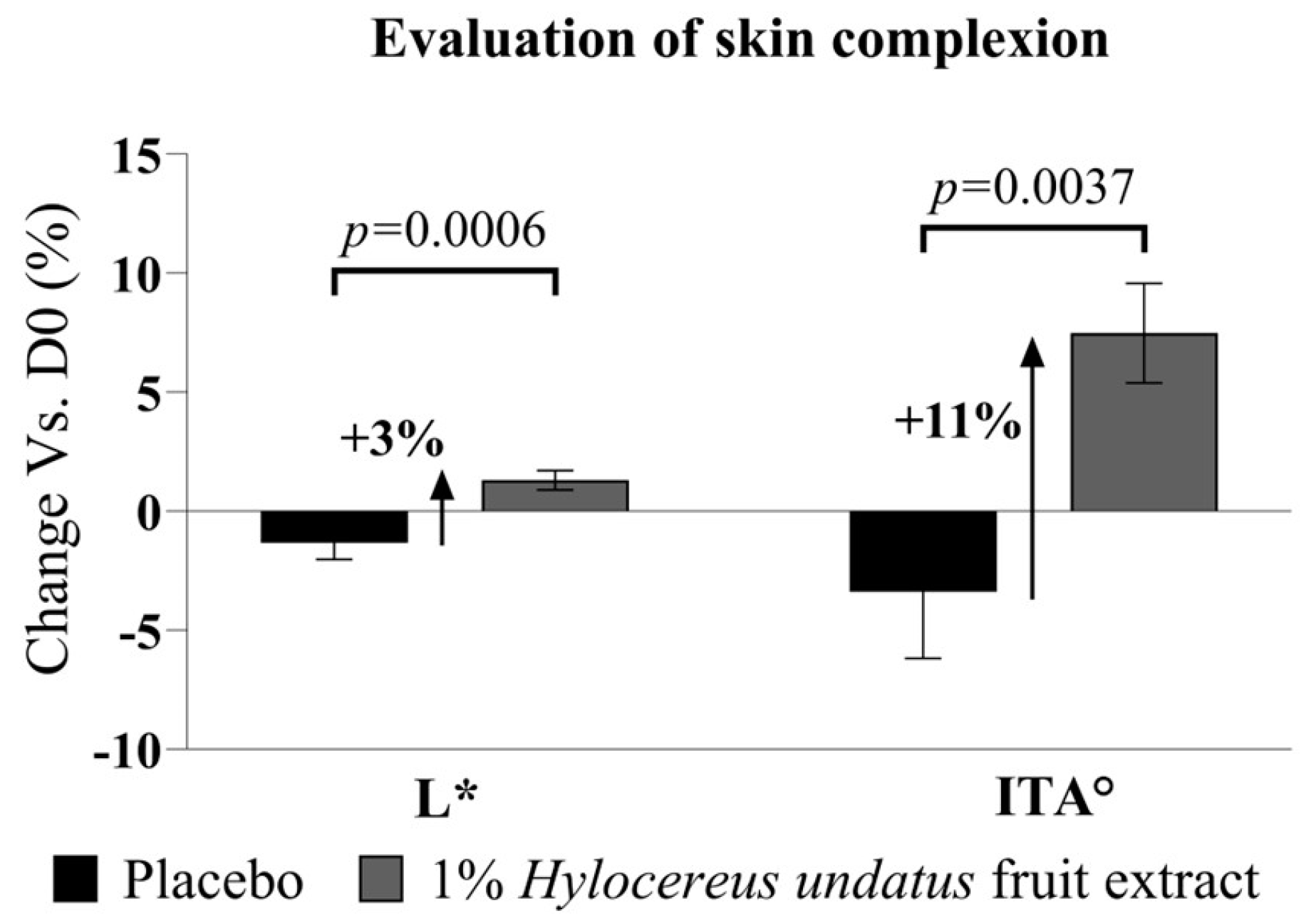
Additionally, the chromameter data at D28 also show modest but significant improvements in L* (luminance) with 1% H. undatus fruit extract in formula (+3%, with statistical significance vs. placebo and D0 baseline) (Figure 9, left). The ITA (skin tone index calculated based on L* and b* values) similarly shows a statically significant improvement (+11%, where higher ITA values indicate lighter skin) vs. placebo (statistically significant vs. placebo and D0) (Figure 9, right). This indicates that treatment with the extract has a skin-brightening effect.
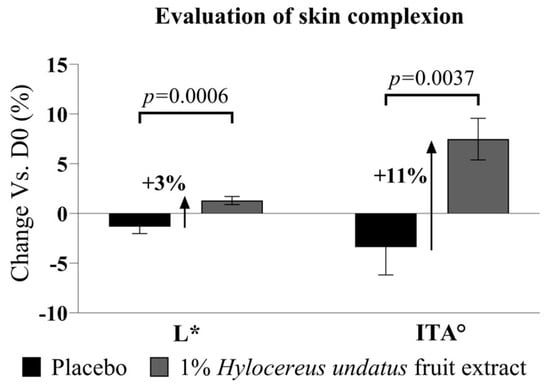
Figure 9.
Evaluation of skin complexion, as Hunter color space parameter L* (left) and ITA skin tone index (right) (both as mean +/− SEM), showing a skin-brightening effect with the product containing 1% H. undatus fruit extract.
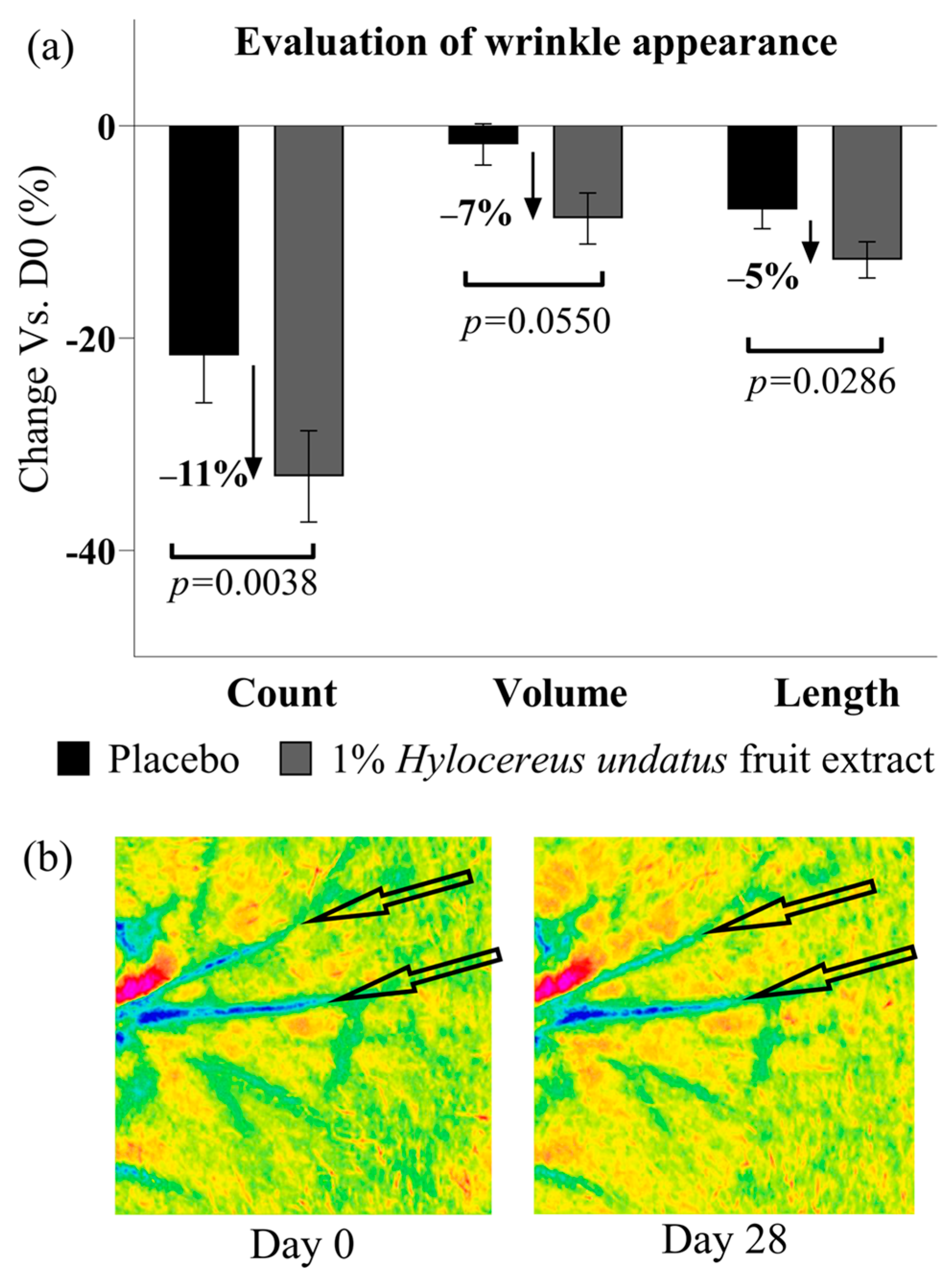
Finally, PRIMOS data show that after 28 days of treatment with 1% H. undatus fruit extract in formula, an anti-aging/anti-wrinkle effect can be observed as a significant reduction in wrinkle count (−11% vs. placebo), as well as lower wrinkle volume and length values (−7% and −5%, respectively vs. placebo), all with statistical significance vs. D0 and placebo (except for wrinkle volume, achieving borderline significance with p = 0.055) (Figure 10).
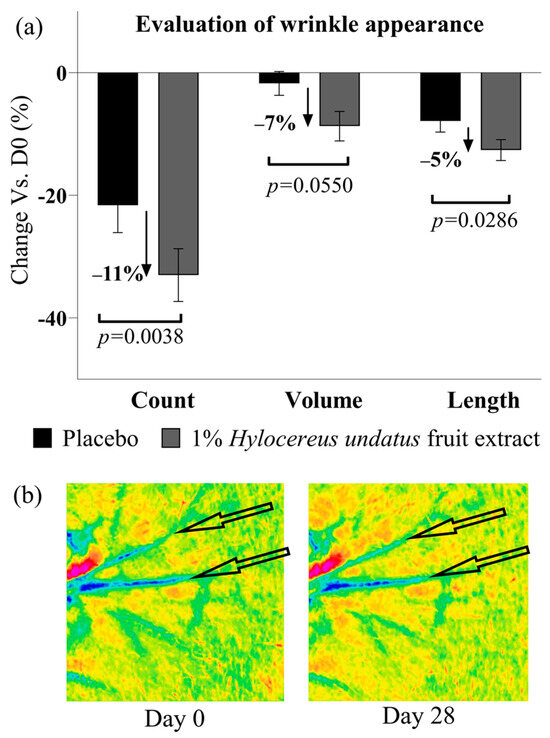
Figure 10.
(a) Evaluation of skin wrinkles in the crow’s feet area (next to the corner of the eyelids) by PRIMOS: wrinkle count (left), volume (center), and length (right) (all shown as mean +/− SEM), showing an ani-wrinkle effect with the product containing 1% H. undatus fruit extract. (b) Illustrative Primos false-color depth map images, volunteer #16 (age 42). Arrows indicate features of interest.
4. Discussion
As shown above, introduction of the H. undatus fruit extract described here into a model bacterial coculture resulted in enhanced growth for the two strains representing common beneficial commensal species (S. hominis, S. Epidermidis) while severely restricting the relative proportion of the two strains representing detrimental species (S. aureus, C. acnes). This indicated potential prebiotic activity on the extract’s part. In a double-blind, placebo-controlled clinical study, this potential appeared borne out by the enhancement of microbiota diversity observed after 28 days of treatment with a formulation containing 1% of the extract, as evaluated through the Faith biodiversity index. This index is considered of special interest due to the fact that, where other commonly used indices may consider more limited factors, for example only the number of taxons recorded, the Faith index takes into account the phylogenetic distance between the various taxons. In other words, instead of simply recording how many different bacterial species were observed, Faith’s index reflects how different the species recorded are from one another.
Of special interest was the observation that this enhancement was greater in older volunteers. While the correlations between aging and microbiota diversity and balance are not fully understood, recent research shows, perhaps counterintuitively, higher diversity in older age [18,45,46]. This could be due to accumulated exposure (seeding) to a consequentially accumulating variety of different microbial species over a lifetime. Age-related changes in host immune function could also play a part. Regardless, it is clear we do not yet understand everything there is to understand about the relationship between age/aging and the skin microbiota. It is even conceivable that the effects of the microbiota on health and beauty could be more strongly related to specific elements in the microbiota balance than to diversity itself. In light of this state of affairs, it may be particularly valuable to focus more future research on correlations between skin health or beauty elements and particular microbial populations or balance components, in order to shed more specific light on the relationships between skin condition and changes in microbiota composition driven, inter alia, by aging—as well as on the ultimate value of diversity assessments themselves, or conversely perhaps to enable the definition of a more discriminating, weighted diversity index geared towards specifically evaluating the microbiota for health- and beauty-enabling microbiota diversity and balance elements. Still, the correlation with the end-product skin beauty benefits demonstrated here remains interesting. If nothing else, this suggests that the extract described here could at the least represent a valuable tool to answer specific needs relating to skin health and beauty as well as microbiota health in aging or older populations.
We also showed a direct effect in vivo against at least one skin pathogen, C. tuberculostearicum, further reinforcing the indication that the extract exerted an overall positive effect on the skin microbiota balance. Corynebacterium is a major genus of skin microbiota that is reported as predominant on moist skin [1]. Some Corynebacterium species, including C. tuberculostearicum, are reported as causal agents of axillary odor [47]. It is also reported in the literature as involved in human skin diseases, including tinea pedis [48]. This species is also a major bacterium of the nasal microbiota, and is involved in pathologies there [49,50]. In keratinocytes and cutaneous squamous carcinoma cell cultures, C. tuberculostearicum is also reported as triggering inflammation through canonical NF-κB pathway activation, and thus inducing pro-inflammatory cytokine production. Mechanistically, pathogen-associated molecular patterns of C. tuberculostearicum could interact with host cells’ Toll-Like-Receptors (TLRs), leading to NF-κB translocation in the nucleus and consequent inflammatory gene expression modulation. Furthermore, C. tuberculostearicum could upregulate the expression of Toll-Like-Receptor 2, hence amplifying inflammation [51]. This modulation of the abundance of Corynebacterium tuberculostearicum with the active vs. placebo could thus correlate with the active’s anti-inflammatory effects.
In our clinical studies, 1% of H. undatus fruit extract also delivered significant skin health and beauty benefits, compared with placebo, including in terms of skin inflammation status (as shown by a reduction in red spots; improved grading by a trained clinician; and reduced redness as measured by chromametry), as well as resilience to chemical insult (as shown by reduced reactivity to histamine); an amelioration in uneven pigmentation (spots); strengthened skin barrier function (as evaluated via transepidermal water loss); improved skin radiance (also as evaluated by chromametry, expressed as ITA); and reduced signs of aging (wrinkling).
One main limitation of the research described here remains, of course, that it is difficult to establish a direct link between the two types of effects observed: the effects on the skin microbiota, on the one hand, and the effects on skin health and beauty measures on the other. The data indicate that treatment with the extract positively affects both the microbiota balance and the overall condition of the skin. It is possible that these effects are linked, or that they are parallel and unrelated to one another. The data presented here do not provide a definite answer, but they appear to at least suggest a correlation. This is especially so in light of the association of a disturbed microbiota balance with increased skin inflammation, compromised skin barrier, and dry skin inter alia. Our data show that the extract alleviates precisely such phenomena while improving microbiota diversity, which appears suggestive in this context. This is especially so considering the extract’s composition, which could provide favorable nutrition for some bacterial species, and the extract’s demonstrated prebiotic properties in other contexts. As pointed out by Boxberger et al. [52] in their review of the known effects of pre- and probiotics on the human skin microbiota, scientific data enabling clear conclusions remain “elusive”, but the results presented above may represent one more correlative element backing the current models for these effects.
In future studies, it could be valuable to extend the clinical investigations described here to cover more different types of skin and of volunteers, including wider ranges of age and ethnicities, and possibly attempting to address some specific conditions with a known microbiota-related component, such as acne or atopic dermatitis. It may be valuable to examine the extract’s effects on especially dry or oily skin. The effect of the extract on pruritus (itching) may also be interesting in light of recent research showing that S. aureus can drive skin itch, leading to skin damage from scratching [53]. It might also be interesting to examine the effect of the extract on wound healing and the formation of visible scar tissue, including parameters such as thickness and pigmentation, seeing as microbial populations can also strongly influence these processes and outcomes. In the context of the findings by Morvan and Vallee, who identified differences in the microbiota composition of highly stressed volunteers compared to a reference population, it may be interesting to see if this extract can correct the effects of high psychological stress in ways similar, or different, from what these authors reported with a different topical treatment [21]. There may also be valuable lessons to be learned from the consideration of other forms of microbial life present in the skin microbiota, such as fungi or viruses, especially phages, in any such future in vitro or in vivo studies with this extract. Finally, it may be interesting to examine synergies between this extract and other substances capable of influencing the skin microbiota, such as the Lysine dendrimer recently described by Leignadier et al. in the context of acne [54].
5. Conclusions
The natural extract of H. undatus fruit described here showed effects in preclinical and clinical studies, consistent with a prebiotic activity leading to a beneficial rebalancing of the skin microbiota. In vitro, the extract positively affected the balance of a model bacterial coculture. In vivo, the extract was shown to enhance microbiota diversity, especially in older volunteers, and reduce the prevalence of at least one pathogenic species, Corynebacterium tuberculostearicum. Further, beneficial effects of treatment with this extract were observed in vivo, including reduced skin inflammation; increased skin resilience to chemical insult; strengthened skin barrier; increased skin radiance; and reduced signs of aging (wrinkles). The available literature suggests that at least some of these effects are consistent with improved skin microbiota balance and diversity. These results indicate that this extract has prebiotic potential, potentially valuable in cosmetic applications aimed at delivering skin health and beauty benefits through the preservation and/or rebalancing of the skin microbiota.
Author Contributions
Conceptualization, F.H., S.K. and J.A.-V.; methodology, F.H., J.A.-V. and S.K.; formal analysis, M.C., S.K. and F.H.; investigation, F.H., M.C., S.K. and J.A.-V.; resources, J.A.-V.; data curation, M.C. and F.H.; writing—original draft preparation, F.H. and S.K.; writing—review and editing, F.H., J.A.-V. and S.K.; supervision, J.A.-V.; project administration, F.H. and S.K. All authors have read and agreed to the published version of the manuscript.
Funding
Lucas Meyer Cosmetics funded this research. No external funding was obtained.
Institutional Review Board Statement
Clinical studies described herein were carried out according to Declaration of Helsinki guidelines. The Clinica Dr. Carlos Ramos’s Ethics Committee (Portugal) approved the protocols (respectively, opinions 1015/15 and 1016/15 of 5 January 2015, and opinions 6882/2021 and 6883/2021 of 4 January 2021).
Informed Consent Statement
All subjects included in the clinical trials reported here provided written informed consent.
Data Availability Statement
Upon reasonable request, the corresponding author may make available the data presented in this study.
Conflicts of Interest
Fabien Havas, Shlomi Krispin, Moshe Cohen, and Joan Attia-Vigneau are employed by Lucas Meyer Cosmetics. The authors declare that these studies received funding directly from Lucas Meyer Cosmetics. The funder was not involved in study design, collection, analysis, or interpretation of data, or in the writing of this article or the decision to submit it for publication.
References
- Grice, E.; Segre, J. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Wallen-Russell, C.; Wallen-Russell, S. Meta Analysis of Skin Microbiome: New Link between Skin Microbiota Diversity and Skin Health with Proposal to Use This as a Future Mechanism to Determine Whether Cosmetic Products Damage the Skin. Cosmetics 2017, 4, 14. [Google Scholar] [CrossRef]
- Byrd, A.; Belkaid, Y.; Segre, J. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Kim, M. Skin Barrier Function and the Microbiome. Int. J. Mol. Sci. 2022, 23, 13071. [Google Scholar] [CrossRef]
- Scudiero, O.; Brancaccio, M.; Mennitti, C.; Laneri, S.; Lombardo, B.; De Biasi, M.G.; De Gregorio, E.; Pagliuca, C.; Colicchio, R.; Salvatore, P.; et al. Human Defensins: A Novel Approach in the Fight against Skin Colonizing Staphylococcus aureus. Antibiotics 2020, 9, 198. [Google Scholar] [CrossRef]
- Zipperer, A.; Konnerth, M.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, S.; Iwamoto, T.; Takada, K.; Okuda, K.; Tajima, A.; Iwase, T.; Mizunoe, Y. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 2013, 195, 1645–1655. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Wollenberg, M.S.; Claesen, J.; Escapa, I.F.; Aldridge, K.L.; Fischbach, M.A.; Lemon, K.P. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio 2014, 5, e01286-14. [Google Scholar] [CrossRef]
- Uberoi, A.; Bartow-McKenney, C.; Zheng, Q.; Flowers, L.; Campbell, A.; Knight, S.A.B.; Chan, N.; Wei, M.; Lovins, V.; Bugayev, J.; et al. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe 2021, 29, 1235–1248.e1238. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hunt, R.L.; Villaruz, A.E.; Fisher, E.L.; Liu, R.; Liu, Q.; Cheung, G.Y.C.; Li, M.; Otto, M. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe 2022, 30, 301–313.e9. [Google Scholar] [CrossRef] [PubMed]
- Tomic-Canic, M.; Burgess, J.L.; O’Neill, K.E.; Strbo, N.; Pastar, I. Skin Microbiota and its Interplay with Wound Healing. Am. J. Clin. Dermatol. 2020, 21, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Canchy, L.; Kerob, D.; Demessant, A.; Amici, J.M. Wound healing and microbiome, an unexpected relationship. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Pastar, I.; O’Neill, K.; Padula, L.; Head, C.R.; Burgess, J.L.; Chen, V.; Garcia, D.; Stojadinovic, O.; Hower, S.; Plano, G.V.; et al. Staphylococcus epidermidis Boosts Innate Immune Response by Activation of Gamma Delta T Cells and Induction of Perforin-2 in Human Skin. Front. Immunol. 2020, 11, 550946. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, J.J.; Myeong, N.R.; Kim, T.; Kim, D.; An, S.; Kim, H.; Park, T.; Jang, S.I.; Yeon, J.H.; et al. Segregation of age-related skin microbiome characteristics by functionality. Sci. Rep. 2019, 9, 16748. [Google Scholar] [CrossRef] [PubMed]
- Flowers, L.; Grice, E.A. The Skin Microbiota: Balancing Risk and Reward. Cell Host Microbe 2020, 28, 190–200. [Google Scholar] [CrossRef]
- Townsend, E.C.; Kalan, L.R. The dynamic balance of the skin microbiome across the lifespan. Biochem. Soc. Trans. 2023, 51, 71–86. [Google Scholar] [CrossRef]
- Ratanapokasatit, Y.; Laisuan, W.; Rattananukrom, T.; Petchlorlian, A.; Thaipisuttikul, I.; Sompornrattanaphan, M. How Microbiomes Affect Skin Aging: The Updated Evidence and Current Perspectives. Life 2022, 12, 936. [Google Scholar] [CrossRef]
- Li, Z.; Bai, X.; Peng, T.; Yi, X.; Luo, L.; Yang, J.; Liu, J.; Wang, Y.; He, T.; Wang, X.; et al. New Insights into the Skin Microbial Communities and Skin Aging. Front. Microbiol. 2020, 11, 565549. [Google Scholar] [CrossRef]
- Morvan, P.Y.; Vallee, R. Evaluation of the effects of stressful life on human skin microbiota. Appl. Microbiol. Open Access 2018, 4, 1000140. [Google Scholar]
- Swaney, M.H.; Kalan, L.R. Living in your skin: Microbes, molecules, and mechanisms. Infect. Immun. 2021, 89, 10–128. [Google Scholar] [CrossRef]
- Sfriso, R.; Egert, M.; Gempeler, M.; Voegeli, R.; Campiche, R. Revealing the secret life of skin-with the microbiome you never walk alone. Int. J. Cos. Sci. 2020, 42, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, H.E.; Bhatia, N.C.; Friedman, A.; Eng, R.M.; Martin, R.; Seite, S. The role of cutaneous microbiota harmony in maintaining a functional skin barrier. J. Drugs Dermatol. 2017, 16, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Zanchetta, C.; Vilanova, D.; Jarrin, C.; Scandolera, A.; Chapuis, E.; Auriol, D.; Robe, P.; Dupont, J.; Lapierre, L.; Reynaud, R. Bacterial taxa predictive of hyperpigmented skins. Health Sci. Rep. 2022, 5, e609. [Google Scholar] [CrossRef] [PubMed]
- Souak, D.; Barreau, M.; Courtois, A.; André, V.; Duclairoir Poc, C.; Feuilloley, M.G.J.; Gault, M. Challenging Cosmetic Innovation: The Skin Microbiota and Probiotics Protect the Skin from UV-Induced Damage. Microorganisms 2021, 9, 936. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Feng, N.; Guo, F.; Chen, Z.; Liang, J.; Wang, T.; Guo, X.; Xu, Z. Applications of Probiotic Constituents in Cosmetics. Molecules 2022, 28, 6765. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghazzewi, F.H.; Tester, R.F. Impact of prebiotics and probiotics on skin health. Benef. Microbes 2014, 5, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Lolou, V.; Panayiotidis, M.I. Functional Role of Probiotics and Prebiotics on Skin Health and Disease. Fermentation 2019, 5, 41. [Google Scholar] [CrossRef]
- Shah, K.; Chen, J.; Chen, J.; Qin, Y. Pitaya Nutrition, Biology, and Biotechnology: A Review. Int. J. Mol. Sci. 2023, 24, 13986. [Google Scholar] [CrossRef]
- Wichienchot, S.; Jatupornpipat, M.; Rastall, R. Oligosaccharides of pitaya (dragon fruit) flesh and their prebiotic properties. Food Chem. 2010, 120, 850–857. [Google Scholar] [CrossRef]
- Rohin, M.A.K.; Abu Bakar, C.A.; Ali, A.M. Isolation and characterization of oligosaccharides composition in organically grown red pitaya, white pitaya and papaya. Int. J. Pharm. Pharm. Sci. 2014, 6, 131–136. [Google Scholar]
- Nishikito, D.F.; Borges, A.C.A.; Laurindo, L.F.; Otoboni, A.M.M.B.; Direito, R.; Goulart, R.d.A.; Nicolau, C.C.T.; Fiorini, A.M.R.; Sinatora, R.V.; Barbalho, S.M. Anti-Inflammatory, Antioxidant, and Other Health Effects of Dragon Fruit and Potential Delivery Systems for Its Bioactive Compounds. Pharmaceutics 2023, 15, 159. [Google Scholar] [CrossRef]
- Perez, G.R.M.; Vargas, S.R.; Ortiz, H.Y.D. Wound healing properties of Hylocereus undatus on diabetic rats. Phytother. Res. 2005, 19, 665–668. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. 16S 454 Sequencing Protocol. 2010. Available online: https://hmpdacc.org/hmp/doc/16S_Sequencing_SOP_4.2.2.pdf (accessed on 25 December 2023).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 December 2023).
- Callahan, B.; McMurdie, P.; Rosen, M.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P.; Veron, S.; Pavoine, S.; Pellens, R. Indicators for the Expected Loss of Phylogenetic Diversity. In Phylogenetic Diversity; Scherson, R., Faith, D.P., Eds.; Springer: Cham, Switzerland; Berlin/Heidelberg, Germany, 2018; pp. 73–91. [Google Scholar]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Wu, Y.; Tanaka, T.; Akimoto, M. Utilization of individual typology angle (ITA) and hue angle in the measurement of skin color on images. Bioimages 2020, 28, 1–8. [Google Scholar]
- Del Bino, S.; Bernerd, F. Variations in skin colour and the biological consequences of ultraviolet radiation exposure. Br. J. Dermatol. 2013, 169, 33–40. [Google Scholar] [CrossRef]
- Kim, H.J.; Oh, H.N.; Park, T.; Kim, H.; Lee, H.G.; An, S.; Sul, W.J. Aged related human skin microbiome and mycobiome in Korean women. Sci. Rep. 2022, 12, 2351. [Google Scholar] [CrossRef] [PubMed]
- Myers, T.; Bouslimani, A.; Huang, S.; Hansen, S.T.; Clavaud, C.; Azouaoui, A.; Ott, A.; Gueniche, A.; Bouez, C.; Zheng, Q.; et al. A multi-study analysis enables identification of potential microbial features associated with skin aging signs. Front. Aging 2024, 4, 1304705. [Google Scholar] [CrossRef] [PubMed]
- Onwuliri, V.; Agbakoba, N.R.; Anukam, K.C. Topical cream containing live lactobacilli decreases malodor-producing bacteria and downregulates genes encoding PLP-dependent enzymes on the axillary skin microbiome of healthy adult Nigerians. J. Cosmet. Dermatol. 2021, 20, 2989–2998. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tan, J.; Yang, H.; Gao, Z.; Cai, Q.; Meng, L.; Yang, L. Characterization of Skin Microbiome in Tinea Pedis. Indian J. Microbiol. 2019, 59, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Abreu, N.A.; Nagalingam, N.A.; Song, Y.; Roediger, F.C.; Pletcher, S.D.; Goldberg, A.N.; Lynch, S.V. Sinus Microbiome Diversity Depletion and Corynebacterium tuberculostearicum Enrichment Mediates Rhinosinusitis. Sci. Transl. Med. 2012, 4, 151ra124. [Google Scholar] [CrossRef] [PubMed]
- Rhee, R.L.; Lu, J.; Bittinger, K.; Lee, J.J.; Mattei, L.M.; Sreih, A.G.; Chou, S.; Miner, J.J.; Cohen, N.A.; Kelly, B.J.; et al. Dynamic Changes in the Nasal Microbiome Associated with Disease Activity in Patients with Granulomatosis with Polyangiitis. Arthritis Rheumatol. 2021, 73, 1703–1712. [Google Scholar] [CrossRef]
- Altonsy, M.O.; Kurwa, H.A.; Lauzon, G.J.; Amrein, M.; Gerber, A.N.; Almishri, W.; Mydlarski, P.R. Corynebacterium tuberculostearicum, a human skin colonizer, induces the canonical nuclear factor-κB inflammatory signaling pathway in human skin cells. Immun. Inflamm. Dis. 2020, 8, 62–79. [Google Scholar] [CrossRef]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in exploring and manipulating the human skin microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Deng, L.; Costa, F.; Blake, K.J.; Choi, S.; Chandrabalan, A.; Yousuf, M.S.; Shiers, S.; Dubreuil, D.; Vega-Mendoza, D.; Rolland, C.; et al. S. aureus drives itch and scratch-induced skin damage through a V8 protease-PAR1 axis. Cell 2023, 186, 5375–5393. [Google Scholar] [CrossRef]
- Leignadier, J.; Drago, M.; Lesouhaitier, O.; Barreau, M.; Dashi, A.; Worsley, O.; Attia-Vigneau, J. Lysine-Dendrimer, a New Non-Aggressive Solution to Rebalance the Microbiota of Acne-Prone Skin. Pharmaceutics 2023, 15, 2083. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).