Adenosine Improves Mitochondrial Function and Biogenesis in Friedreich's Ataxia Fibroblasts Following L-Buthionine Sulfoximine-Induced Oxidative Stress
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Dermal Fibroblasts Culture
2.3. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Viability Assay
2.4. Mitochondrial Membrane Potential (MMP) Assay
2.5. Aconitase Assay
2.6. Adenosine Triphosphate (ATP) Assay
2.7. Mitochondrial Biogenesis Assay
2.8. Reverse Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
2.9. Statistical Analysis
3. Results
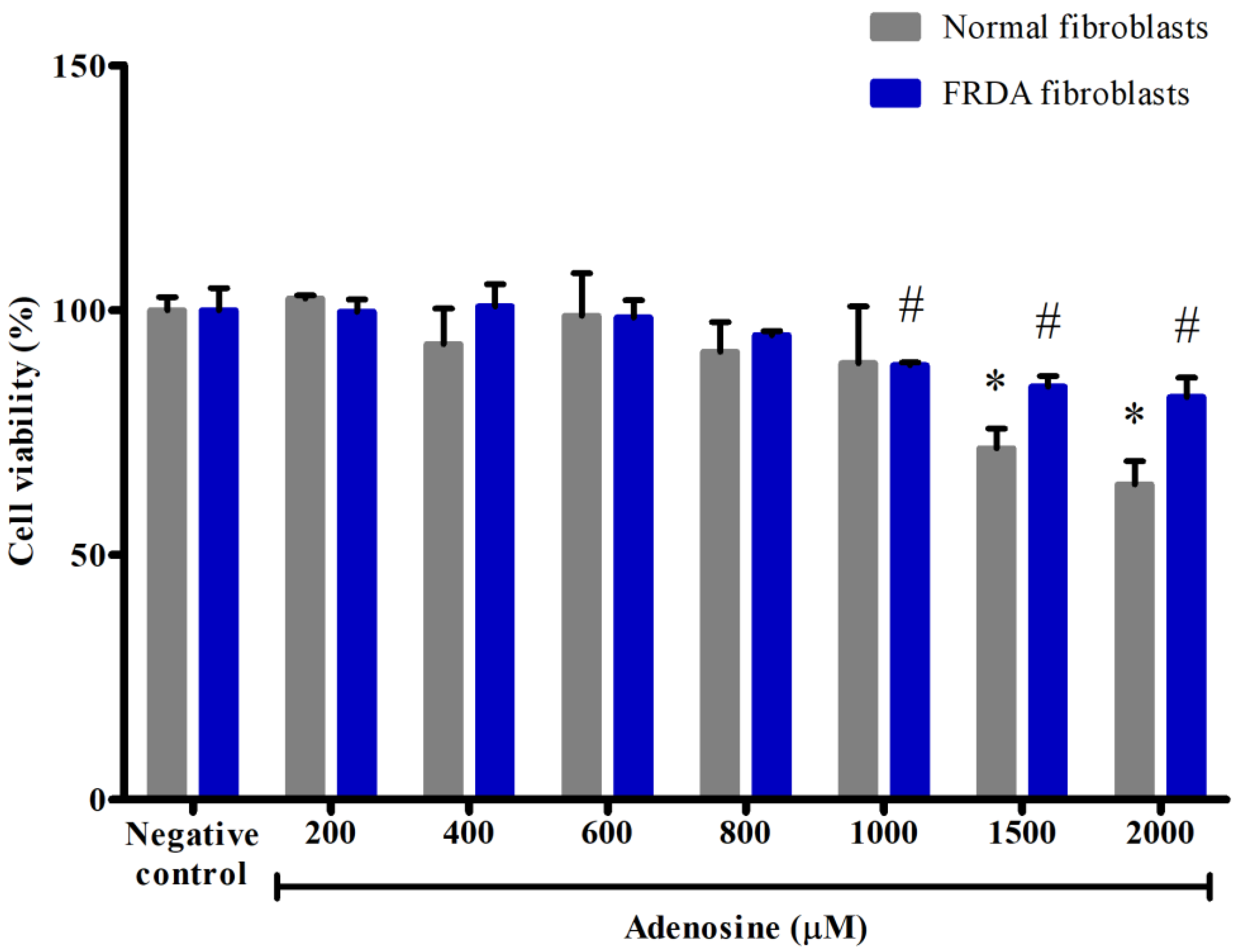
3.1. Effects of Adenosine on the Viability of Normal and FRDA Fibroblasts
3.2. Effects of Adenosine on the Mitochondrial Membrane Potential (MMP) in FRDA Fibroblasts Treated with BSO
3.3. Effects of Adenosine on the Aconitase Activity in FRDA Fibroblasts Treated with BSO
3.4. Effects of Adenosine on the Adenosine Triphosphate (ATP) Level in FRDA Fibroblasts Treated with BSO
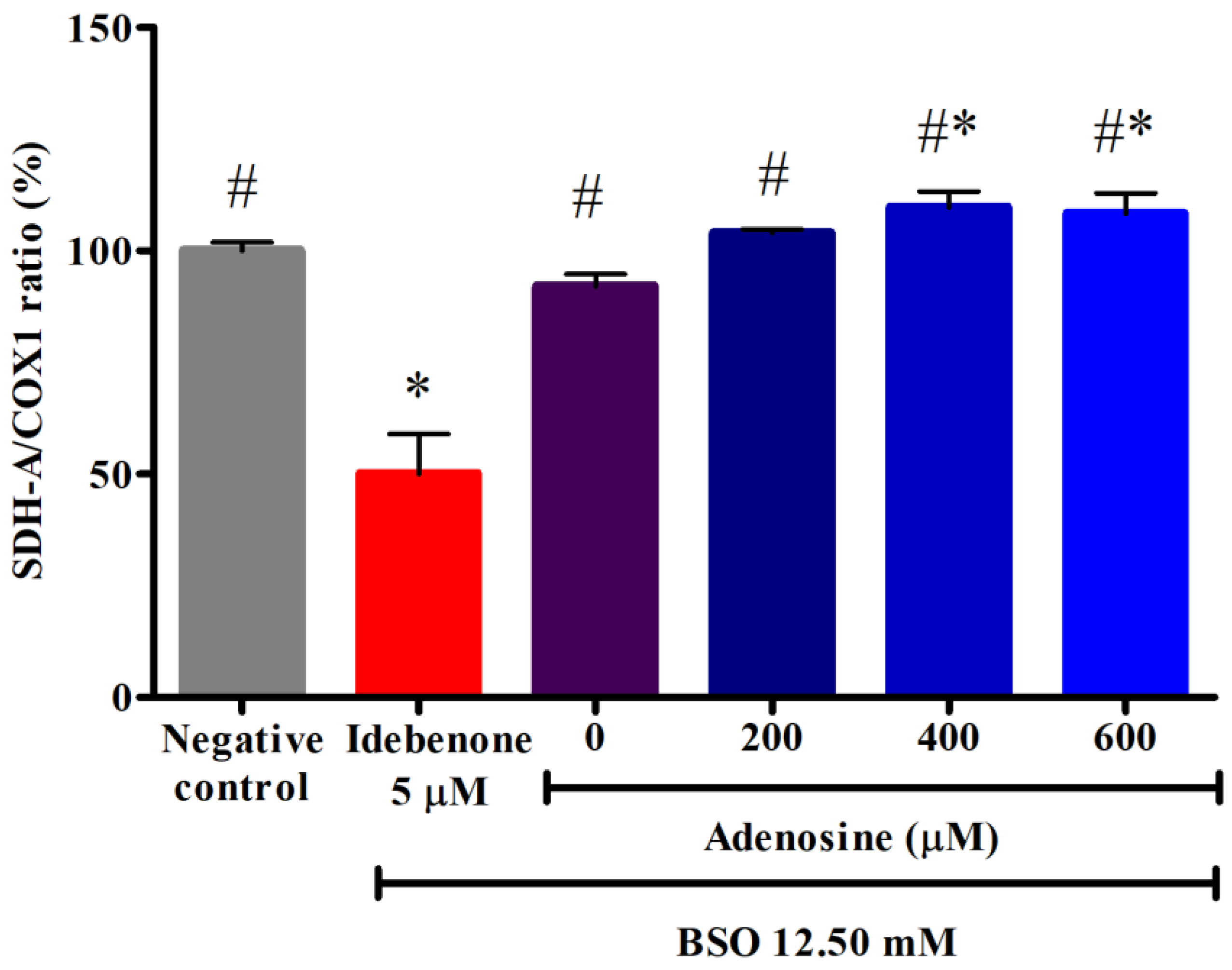
3.5. Effects of Adenosine on the Mitochondrial Biogenesis in FRDA Fibroblasts Treated with BSO
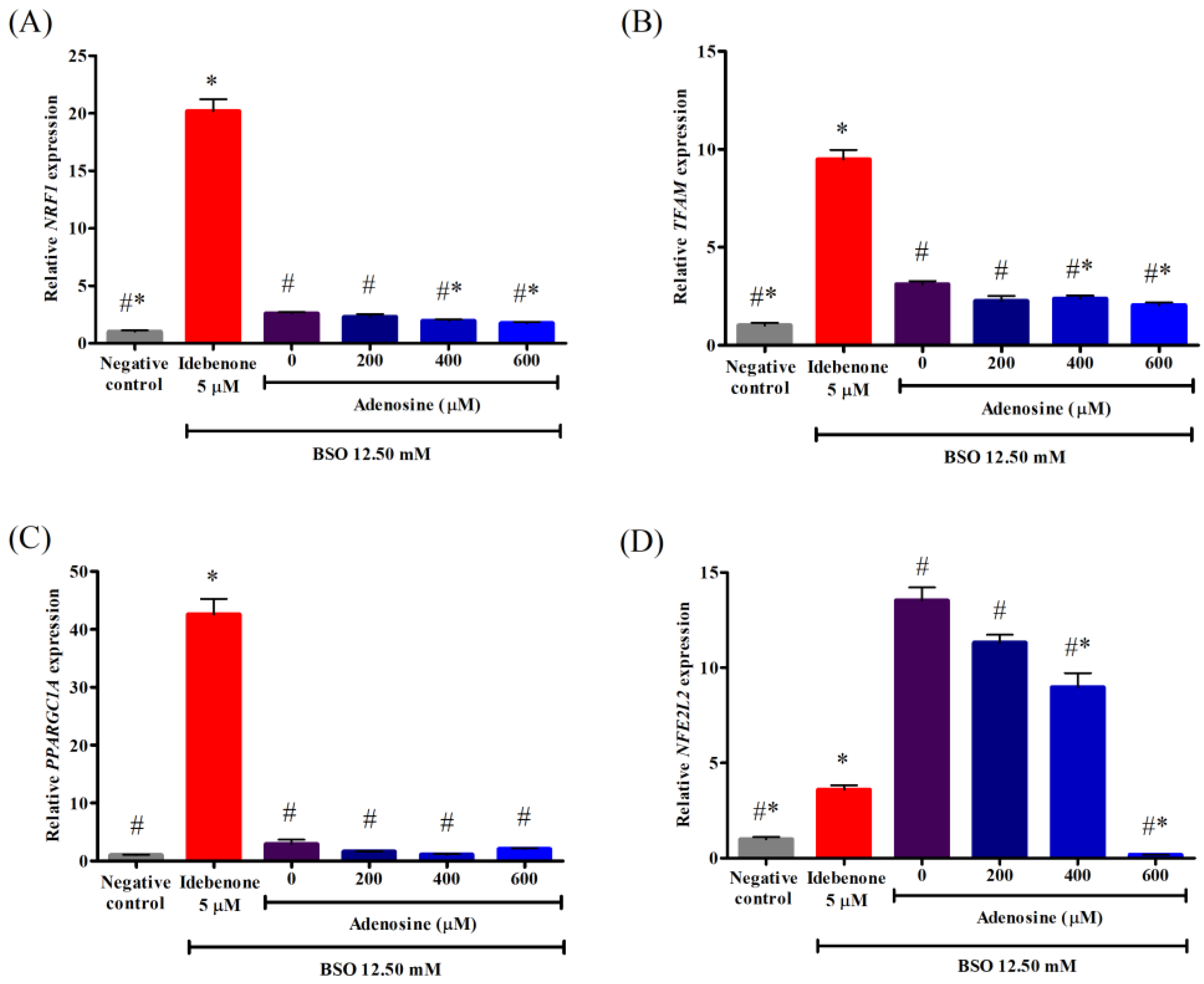
3.6. Effects of Adenosine on the Gene Expression Associated with Mitochondrial Biogenesis in FRDA Fibroblasts Treated with BSO
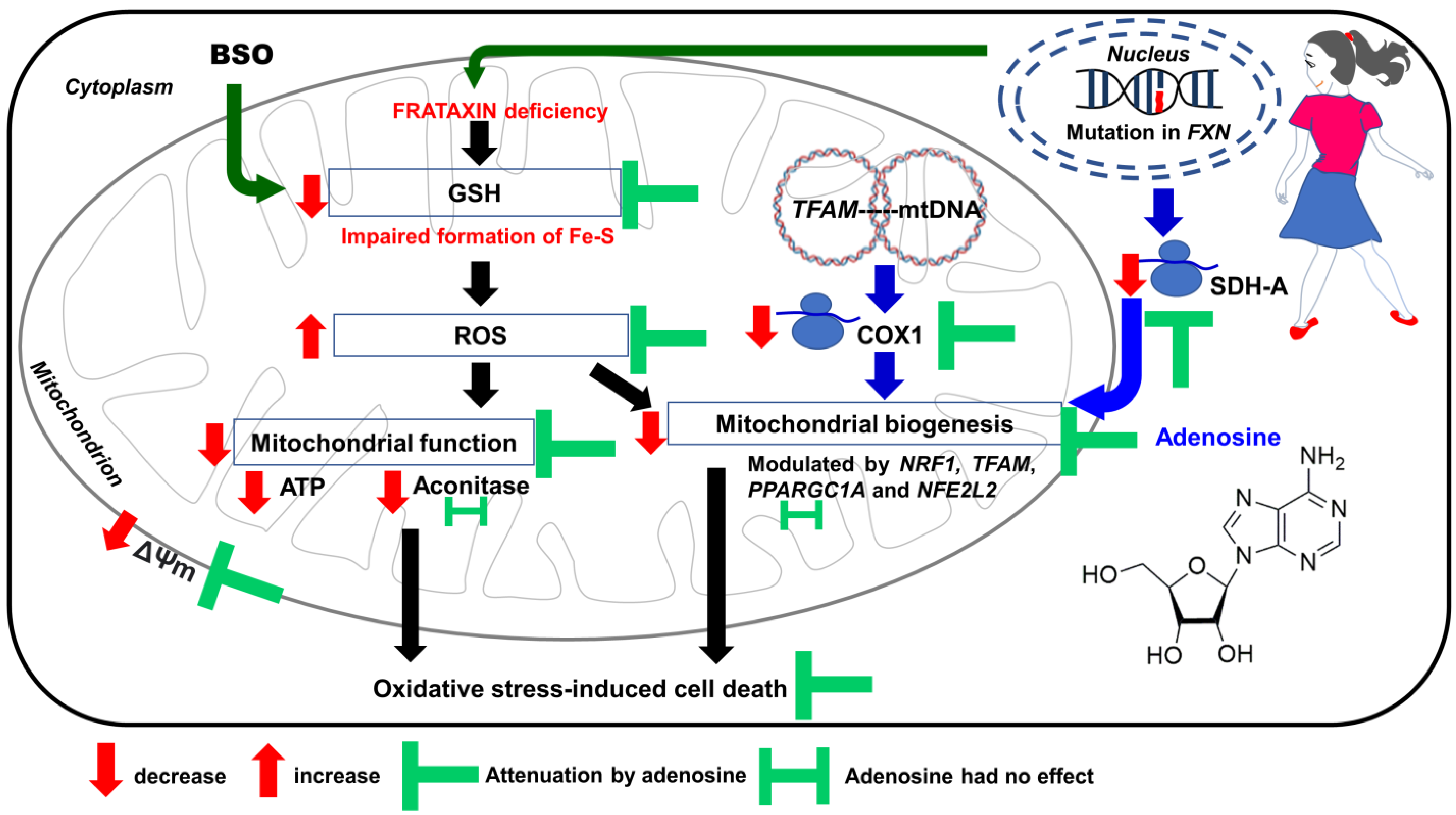
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACTB | actin beta |
| ANOVA | one-way analysis of variance |
| A1R | adenosine A1 receptor |
| A2AR | adenosine A2A receptor |
| A2BR | adenosine A2B receptor |
| A3R | adenosine A3 receptor |
| ARE | antioxidant response element |
| ATP | adenosine triphosphate |
| BSO | L-buthionine sulfoximine |
| cDNA | complementary DNA |
| CNS | central nervous system |
| COX1 | cytochrome c oxidase subunit 1 |
| CO2 | carbon dioxide |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DMSO | dimethyl sulfoxide |
| FBS | fetal bovine serum |
| FDA | Food Drug and Administration |
| Fe-S | Iron–sulfur |
| FRDA | Friedreich’s ataxia |
| FXN | frataxin |
| GAA | guanine–adenine–adenine |
| h | hour |
| HMEC-1 | human microvascular endothelial cells |
| H2O2 HSD LHON | hydrogen peroxide Tukey’s honestly significant difference Leber’s hereditary optic neuropathy |
| mM | millimolar |
| MMP/ΔΨm | mitochondrial membrane potential |
| mtDNA | mitochondrial DNA |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| nDNA | nuclear DNA |
| NFE2L2 NIGMS | NFE2-like bZIP transcription factor 2 National Institute of General Medical Sciences |
| NRF1 | nuclear respiratory factor 1 |
| PC-12 | rat pheochromocytoma |
| PPARGC1A | PPARG coactivator 1 alpha |
| RNA | ribonucleic acid |
| ROS | reactive oxygen species |
| RT-qPCR | reverse transcription quantitative real-time polymerase chain reaction |
| SD | standard deviation |
| SDH-A | succinate dehydrogenase subunit A |
| TFAM | transcription factor A, mitochondrial |
| TNF-α | tumor necrosis factor alpha |
| v/v | volume/volume |
| µM | micromolar |
| °C | degree Celsius |
References
- Dürr, A.; Cossee, M.; Agid, Y.; Campuzano, V.; Mignard, C.; Penet, C.; Mandel, J.-L.; Brice, A.; Koenig, M. Clinical and genetic abnormalities in patients with Friedreich’s ataxia. N. Engl. J. Med. 1996, 335, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Rummey, C.; Farmer, J.M.; Lynch, D.R. Predictors of loss of ambulation in Friedreich’s ataxia. Eclinicalmedicine 2020, 18, 100213. [Google Scholar] [CrossRef]
- Hanson, E.; Sheldon, M.; Pacheco, B.; Alkubeysi, M.; Raizada, V. Heart disease in Friedreich’s ataxia. World J. Cardiol. 2019, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bürk, K. Friedreich Ataxia: Current status and future prospects. Cerebellum Ataxias 2017, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, V.; Montermini, L.; Moltò, M.D.; Pianese, L.; Cossée, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996, 271, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Lefevre, S.D.; Sliwa, D.; Seguin, A.; Camadro, J.-M.; Lesuisse, E. Friedreich ataxia: Molecular mechanisms, redox considerations, and therapeutic opportunities. Antioxid. Redox Signal. 2010, 13, 651–690. [Google Scholar] [CrossRef]
- Carletti, B.; Piemonte, F. Friedreich’s Ataxia: A neuronal point of view on the oxidative stress hypothesis. Antioxidants 2014, 3, 592–603. [Google Scholar] [CrossRef]
- Lupoli, F.; Vannocci, T.; Longo, G.; Niccolai, N.; Pastore, A. The role of oxidative stress in Friedreich’s ataxia. FEBS Lett. 2018, 592, 718–727. [Google Scholar] [CrossRef]
- Llorens, J.V.; Soriano, S.; Calap-Quintana, P.; Gonzalez-Cabo, P.; Moltó, M.D. The role of iron in Friedreich’s ataxia: Insights from studies in human tissues and cellular and animal models. Front. Neurosci. 2019, 13, 75. [Google Scholar] [CrossRef]
- Pandolfo, M.; Pastore, A. The pathogenesis of Friedreich ataxia and the structure and function of frataxin. J. Neurol. 2009, 256, 9–17. [Google Scholar] [CrossRef]
- Armstrong, J.S.; Khdour, O.; Hecht, S.M. Does oxidative stress contribute to the pathology of Friedreich’s ataxia? A radical question. FASEB J. 2010, 24, 2152–2163. [Google Scholar] [CrossRef]
- Bolotta, A.; Pini, A.; Abruzzo, P.M.; Ghezzo, A.; Modesti, A.; Gamberi, T.; Ferreri, C.; Bugamelli, F.; Fortuna, F.; Vertuani, S.; et al. Effects of tocotrienol supplementation in Friedreich’s ataxia: A model of oxidative stress pathology. Exp. Biol. Med. 2020, 245, 201–212. [Google Scholar] [CrossRef]
- Lynch, D.R.; Farmer, G. Mitochondrial and metabolic dysfunction in Friedreich ataxia: Update on pathophysiological relevance and clinical interventions. Neuronal Signal. 2021, 5, NS20200093. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, J.; Li, X.; Zhou, X.; Hu, Y.; Chu, S.; Peng, Y.; Chen, N. Research progress on adenosine in central nervous system diseases. CNS Neurosci. Ther. 2019, 25, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; van Aerde, K.; Abel, T.; Feldmeyer, D. Adenosine differentially modulates synaptic transmission of excitatory and inhibitory microcircuits in layer 4 of rat barrel cortex. Cereb. Cortex 2017, 27, 4411–4422. [Google Scholar] [CrossRef] [PubMed]
- Ziganshin, A.; Khairullin, A.; Hoyle, C.; Grishin, S. Modulatory roles of ATP and adenosine in cholinergic neuromuscular transmission. Int. J. Mol. Sci. 2020, 21, 6423. [Google Scholar] [CrossRef]
- Van Aerde, K.I.; Qi, G.; Feldmeyer, D. Cell type-specific effects of adenosine on cortical neurons. Cereb. Cortex 2015, 25, 772–787. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Vollmer, C.; Nurse, C.A. Adenosine and dopamine oppositely modulate a hyperpolarization-activated currentIhin chemosensory neurons of the rat carotid body in co-culture. J. Physiol. 2018, 596, 3101–3117. [Google Scholar] [CrossRef] [PubMed]
- Boison, D.; Singer, P.; Shen, H.-Y.; Feldon, J.; Yee, B.K. Adenosine hypothesis of schizophrenia—Opportunities for pharmacotherapy. Neuropharmacology 2012, 62, 1527–1543. [Google Scholar] [CrossRef]
- Fredholm, B.B. Adenosine, an endogenous distress signal, modulates tissue damage and repair. Cell Death Differ. 2007, 14, 1315–1323. [Google Scholar] [CrossRef]
- Van Calker, D.; Biber, K.; Domschke, K.; Serchov, T. The role of adenosine receptors in mood and anxiety disorders. J. Neurochem. 2019, 151, 11–27. [Google Scholar] [CrossRef]
- Melani, A.; Pugliese, A.M.; Pedata, F. Adenosine receptors in cerebral ischemia. Int. Rev. Neurobiol. 2014, 119, 309–348. [Google Scholar] [CrossRef] [PubMed]
- Coelho, J.E.; Alves, P.; Canas, P.M.; Valadas, J.S.; Shmidt, T.; Batalha, V.L.; Ferreira, D.G.; Ribeiro, J.A.; Bader, M.; Cunha, R.A.; et al. Overexpression of adenosine A2A receptors in rats: Effects on depression, locomotion, and anxiety. Front. Psychiatry 2014, 5, 67. [Google Scholar] [CrossRef]
- Chen, J.-F. Chapter twelve – Adenosine receptor control of cognition in normal and disease. Int. Rev. Neurobiol. 2014, 119, 257–307. [Google Scholar] [CrossRef]
- Kao, Y.-H.; Lin, M.-S.; Chen, C.-M.; Wu, Y.-R.; Chen, H.-M.; Lai, H.-L.; Chern, Y.; Lin, C.-J. Targeting ENT1 and adenosine tone for the treatment of Huntington’s disease. Hum. Mol. Genet. 2017, 26, 467–478. [Google Scholar] [CrossRef]
- Lee, C.-C.; Chang, C.-P.; Lin, C.-J.; Lai, H.-L.; Kao, Y.-H.; Cheng, S.-J.; Chen, H.-M.; Liao, Y.-P.; Faivre, E.; Buée, L.; et al. Adenosine augmentation evoked by an ENT1 inhibitor improves memory impairment and neuronal plasticity in the APP/PS1 mouse model of Alzheimer’s disease. Mol. Neurobiol. 2018, 55, 8936–8952. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-F.; Cunha, R.A. The belated US FDA approval of the adenosine A2A receptor antagonist istradefylline for treatment of Parkinson’s disease. Purinergic Signal. 2020, 16, 167–174. [Google Scholar] [CrossRef]
- Jenner, P.; Mori, A.; Aradi, S.D.; Hauser, R.A. Istradefylline—A first generation adenosine A2A antagonist for the treatment of Parkinson’s disease. Expert Rev. Neurother. 2021, 21, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-P.; Wu, K.-C.; Lin, C.-Y.; Chern, Y. Emerging roles of dysregulated adenosine homeostasis in brain disorders with a specific focus on neurodegenerative diseases. J. Biomed. Sci. 2021, 28, 70. [Google Scholar] [CrossRef]
- Heffner, T.G.; Wiley, J.N.; Williams, A.E.; Bruns, R.F.; Coughenour, L.L.; Downs, D.A. Comparison of the behavioral effects of adenosine agonists and dopamine antagonists in mice. Psychopharmacology 1989, 98, 31–37. [Google Scholar] [CrossRef]
- Dar, M.S. Central adenosinergic system involvement in ethanol-induced motor incoordination in mice. J. Pharmacol. Exp. Ther. 1990, 255, 1202–1209. [Google Scholar]
- Dar, M. Cerebellar CB1 receptor mediation of Δ9-THC-induced motor incoordination and its potentiation by ethanol and modulation by the cerebellar adenosinergic A1 receptor in the mouse. Brain Res. 2000, 864, 186–194. [Google Scholar] [CrossRef] [PubMed]
- DeSanty, K.P.; Dar, M. Involvement of the cerebellar adenosine A1 receptor in cannabinoid-induced motor incoordination in the acute and tolerant state in mice. Brain Res. 2001, 905, 178–187. [Google Scholar] [CrossRef]
- Dar, M.; Mustafa, S. Acute ethanol/cannabinoid-induced ataxia and its antagonism by oral/systemic/intracerebellar A1 adenosine receptor antisense in mice. Brain Res. 2002, 957, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.E.; Yu, A.E.; Wen, Y.; Yang, S.-H.; Simpkins, J.W. Estrogen prevents oxidative damage to the mitochondria in Friedreich’s ataxia skin fibroblasts. PLoS ONE 2012, 7, e34600. [Google Scholar] [CrossRef] [PubMed]
- Jauslin, M.L.; Wirth, T.; Meier, T.; Schoumacher, F. A cellular model for Friedreich Ataxia reveals small-molecule glutathione peroxidase mimetics as novel treatment strategy. Hum. Mol. Genet. 2002, 11, 3055–3063. [Google Scholar] [CrossRef] [PubMed]
- Jauslin, M.L.; Vertuani, S.; Durini, E.; Buzzoni, L.; Ciliberti, N.; Verdecchia, S.; Palozza, P.; Meier, T.; Manfredini, S. Protective effects of Fe-Aox29, a novel antioxidant derived from a molecular combination of idebenone and vitamin E, in immortalized fibroblasts and fibroblasts from patients with Friedreich Ataxia. Mol. Cell Biochem. 2007, 302, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Lew, S.-Y.; Yow, Y.-Y.; Lim, L.-W.; Wong, K.-H. Antioxidant-mediated protective role of Hericium erinaceus (Bull.: Fr.) Pers. against oxidative damage in fibroblasts from Friedreich’s ataxia patient. Food Sci. Technol. 2020, 40 (Suppl. 1), 264–272. [Google Scholar] [CrossRef]
- Igarashi, J.; Okamoto, R.; Yamashita, T.; Hashimoto, T.; Karita, S.; Nakai, K.; Kubota, Y.; Takata, M.; Yamaguchi, F.; Tokuda, M.; et al. A key role of PGC-1α transcriptional coactivator in production of VEGF by a novel angiogenic agent COA-Cl in cultured human fibroblasts. Physiol. Rep. 2016, 4, e12742. [Google Scholar] [CrossRef]
- Jasoliya, M.J.; McMackin, M.Z.; Henderson, C.K.; Perlman, S.L.; Cortopassi, G.A. Frataxin deficiency impairs mitochondrial biogenesis in cells, mice and humans. Hum. Mol. Genet. 2017, 26, 2627–2633. [Google Scholar] [CrossRef]
- Cheng, X.; Qian, W.; Chen, F.; Jin, Y.; Wang, F.; Lu, X.; Lee, S.R.; Su, D.; Chen, B. ATRA protects skin fibroblasts against UV-induced oxidative damage through inhibition of E3 ligase Hrd1. Mol. Med. Rep. 2019, 20, 2294–2302. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Lee, M.-S.; Jung, S.; Kim, S.; Park, H.; Park, S.; Kim, S.-Y.; Kim, C.-T.; Jo, Y.-H.; Kim, I.-H.; et al. Ginger extract increases muscle mitochondrial biogenesis and serum HDL-cholesterol level in high-fat diet-fed rats. J. Funct. Foods 2017, 29, 193–200. [Google Scholar] [CrossRef]
- Kwong, S.C.; Jamil, A.H.A.; Rhodes, A.; Taib, N.A.; Chung, I. Metabolic role of fatty acid binding protein 7 in mediating triple-negative breast cancer cell death via PPAR-α signaling. J. Lipid Res. 2019, 60, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Cook, A.; Giunti, P. Friedreich’s ataxia: Clinical features, pathogenesis and management. Br. Med. Bull. 2017, 124, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.S.; Poon, C.H.; Roy, J.; Tsui, K.C.; Lew, S.Y.; Phang, M.W.L.; Tan, R.J.Y.; Cheng, P.G.; Fung, M.-L.; Wong, K.H.; et al. Neurogenesis-dependent antidepressant-like activity of Hericium erinaceus in an animal model of depression. Chin. Med. 2021, 16, 132. [Google Scholar] [CrossRef]
- Samberkar, S.; Gandhi, S.; Naidu, M.; Wong, K.-H.; Raman, J.; Sabaratnam, V. Lion’s mane, Hericium erinaceus and Tiger milk, Lignosus rhinocerotis (higher basidiomycetes) medicinal mushrooms stimulate neurite outgrowth in dissociated cells of brain, spinal cord, and retina: An in vitro study. Int. J. Med. Mushrooms 2015, 17, 1047–1054. [Google Scholar] [CrossRef]
- Wong, K.-H.; Ng, C.-C.; Kanagasabapathy, G.; Yow, Y.-Y.; Sabaratnam, V. An Overview of culinary and medicinal mushrooms in neurodegeneration and neurotrauma research. Int. J. Med. Mushrooms 2017, 19, 191–202. [Google Scholar] [CrossRef]
- Lew, S.Y.; Lim, S.H.; Lim, L.W.; Wong, K.H. Neuroprotective effects of Hericium erinaceus (Bull.: Fr.) Pers. against high-dose corticosterone-induced oxidative stress in PC-12 cells. BMC Complement. Med. Ther. 2020, 20, 340. [Google Scholar] [CrossRef]
- Lin, T.S.; Woon, C.K.; Hui, W.K.; Abas, R.; Haron, M.H.; Das, S. Natural product-based nanomedicine: Recent advances and issues for the treatment of Alzheimer’s disease. Curr. Neuropharmacol. 2022, 20, 1498–1518. [Google Scholar] [CrossRef]
- Yanshree; Yu, W.S.; Fung, M.L.; Lee, C.W.; Lim, L.W.; Wong, K.H. The monkey head mushroom and memory enhancement in Alzheimer’s disease. Cells 2020, 11, 2284. [Google Scholar] [CrossRef]
- Lew, S.Y.; Phang, M.W.L.; Chong, P.S.; Roy, J.; Poon, C.H.; Yu, W.S.; Lim, L.W.; Wong, K.H. Discovery of therapeutics targeting oxidative stress in autosomal recessive cerebellar ataxia: A systematic review. Pharmaceuticals 2022, 15, 764. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.S.; Khairuddin, S.; Tse, A.C.K.; Hiew, L.F.; Lau, C.L.; Tipoe, G.L.; Fung, M.-L.; Wong, K.H.; Lim, L.W. Hericium erinaceus potentially rescues behavioural motor deficits through ERK-CREB-PSD95 neuroprotective mechanisms in rat model of 3-acetylpyridine-induced cerebellar ataxia. Sci. Rep. 2020, 10, 14945. [Google Scholar] [CrossRef] [PubMed]
- Phang, M.W.L.; Lew, S.Y.; Chung, I.; Lim, W.K.-S.; Lim, L.W.; Wong, K.H. Therapeutic roles of natural remedies in combating hereditary ataxia: A systematic review. Chin. Med. 2021, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Valdes, F.; Brown, N.; Morales-Bayuelo, A.; Prent-Peñaloza, L.; Gutierrez, M. Adenosine derivates as antioxidant agents: Synthesis, characterization, in vitro activity, and theoretical insights. Antioxidants 2019, 8, 468. [Google Scholar] [CrossRef] [PubMed]
- Gholinejad, M.; Anarkooli, I.J.; Taromchi, A.; Abdanipour, A. Adenosine decreases oxidative stress and protects H2O2-treated neural stem cells against apoptosis through decreasing Mst1 expression. Biomed. Rep. 2018, 8, 439–446. [Google Scholar] [CrossRef]
- Gueven, N.; Woolley, K.; Smith, J. Border between natural product and drug: Comparison of the related benzoquinones idebenone and coenzyme Q10. Redox Biol. 2015, 4, 289–295. [Google Scholar] [CrossRef]
- Jaber, S.; Polster, B.M. Idebenone and neuroprotection: Antioxidant, pro-oxidant, or electron carrier? J. Bioenerg. Biomembr. 2015, 47, 111–118. [Google Scholar] [CrossRef]
- Mercuri, E.; Muntoni, F. Efficacy of idebenone in Duchenne muscular dystrophy. Lancet 2015, 385, 1704–1706. [Google Scholar] [CrossRef]
- Nohara, Y.; Suzuki, J.; Yamazaki, Y.; Kubo, H. Determination of idebenone in plasma by HPLC with post-column fluorescence derivatization using 2-cyanoacetamide. Chem. Pharm. Bull. 2012, 60, 598–602. [Google Scholar] [CrossRef][Green Version]
- Giorgio, V.; Schiavone, M.; Galber, C.; Carini, M.; Da Ros, T.; Petronilli, V.; Argenton, F.; Carelli, V.; Lopez, M.J.A.; Salviati, L.; et al. The idebenone metabolite QS10 restores electron transfer in complex I and coenzyme Q defects. Biochim. Biophys. Acta (BBA) Bioenerg. 2018, 1859, 901–908. [Google Scholar] [CrossRef]
- Rustin, P.; von Kleist-Retzow, J.-C.; Chantrel-Groussard, K.; Sidi, D.; Munnich, A.; Rötig, A. Effect of idebenone on cardiomyopathy in Friedreich’s ataxia: A preliminary study. Lancet 1999, 354, 477–479. [Google Scholar] [CrossRef]
- Seznec, H.; Simon, D.; Monassier, L.; Criqui-Filipe, P.; Gansmuller, A.; Rustin, P.; Koenig, M.; Puccio, H. Idebenone delays the onset of cardiac functional alteration without correction of Fe-S enzymes deficit in a mouse model for Friedreich ataxia. Hum. Mol. Genet. 2004, 13, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Soriano, S.; Llorens, J.V.; Blanco-Sobero, L.; Gutiérrez, L.; Calap-Quintana, P.; Morales, M.P.; Moltó, M.D.; Martínez-Sebastián, M.J. Deferiprone and idebenone rescue frataxin depletion phenotypes in a Drosophila model of Friedreich’s ataxia. Gene 2013, 521, 274–281. [Google Scholar] [CrossRef]
- Calap-Quintana, P.; Soriano, S.; Llorens, J.V.; Al-Ramahi, I.; Botas, J.; Moltó, M.D.; Martínez-Sebastián, M.J. TORC1 inhibition by rapamycin promotes antioxidant defences in a Drosophila model of Friedreich’s ataxia. PLoS ONE 2015, 10, e0132376. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, S.; D’Amico, J.; La Rosa, P.; Bertini, E.S.; Piemonte, F. Targeting NRF2 for the treatment of Friedreich’s ataxia: A comparison among drugs. Int. J. Mol. Sci. 2019, 20, 5211. [Google Scholar] [CrossRef]
- Di Prospero, N.A.; Baker, A.; Jeffries, N.; Fischbeck, K.H. Neurological effects of high-dose idebenone in patients with Friedreich’s ataxia: A randomised, placebo-controlled trial. Lancet Neurol. 2007, 6, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.R.; Perlman, S.L.; Meier, T. A phase 3, double-blind, placebo-controlled trial of idebenone in Friedreich ataxia. Arch. Neurol. 2010, 67, 941–947. [Google Scholar] [CrossRef]
- Lagedrost, S.J.; Sutton, M.S.J.; Cohen, M.S.; Satou, G.M.; Kaufman, B.D.; Perlman, S.L.; Rummey, C.; Meier, T.; Lynch, D.R. Idebenone in Friedreich ataxia cardiomyopathy—Results from a 6-month phase III study (IONIA). Am. Heart J. 2011, 161, 639–645.e1. [Google Scholar] [CrossRef]
- Meier, T.; Perlman, S.L.; Rummey, C.; Coppard, N.J.; Lynch, D.R. Assessment of neurological efficacy of idebenone in pediatric patients with Friedreich’s ataxia: Data from a 6-month controlled study followed by a 12-month open-label extension study. J. Neurol. 2012, 259, 284–291. [Google Scholar] [CrossRef]
- Paredes-Fuentes, A.J.; Cesar, S.; Montero, R.; Latre, C.; Genovès, J.; Martorell, L.; Cuadras, D.; Colom, H.; Pineda, M.; O’Callaghan, M.D.M.; et al. Plasma idebenone monitoring in Friedreich’s ataxia patients during a long-term follow-up. Biomed. Pharmacother. 2021, 143, 112143. [Google Scholar] [CrossRef]
- Barth, A.; Newell, D.W.; Nguyen, L.B.; Winn, H.; Wender, R.; Meno, J.R.; Janigro, D. Neurotoxicity in organotypic hippocampal slices mediated by adenosine analogues and nitric oxide. Brain Res. 1997, 762, 79–88. [Google Scholar] [CrossRef]
- Jennings, J.A. Locally delivered adenosine and glutathione improve fibroblast proliferation and collagen production. Biomed. Eng. Rev. 2015, 2, 1–22. Available online: https://esmed.org/MRA/bme/article/view/167 (accessed on 6 December 2022).
- Ouyang, X.; Ghani, A.; Malik, A.; Wilder, T.; Colegio, O.R.; Flavell, R.A.; Cronstein, B.N.; Mehal, W.Z. Adenosine is required for sustained inflammasome activation via the A2A receptor and the HIF-1α pathway. Nat. Commun. 2013, 4, 2909. [Google Scholar] [CrossRef] [PubMed]
- Bansal, Y.; Kuhad, A. Mitochondrial dysfunction in depression. Curr. Neuropharmacol. 2016, 14, 610–618. [Google Scholar] [CrossRef]
- Konishi, H.; Kanou, S.-E.; Yukimatsu, R.; Inui, M.; Sato, M.; Yamamoto, N.; Nakano, M.; Koshiba, M. Adenosine inhibits TNFα-induced MMP-3 production in MH7A rheumatoid arthritis synoviocytes via A2A receptor signaling. Sci. Rep. 2022, 12, 6033. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Gimeno, A.; Gonzalez-Cabo, P.; Dasí, F.; Bolinches-Amorós, A.; Mollá, B.; Palau, F.; Pallardó, F.V. Differential expression of PGC-1α and metabolic sensors suggest age-dependent induction of mitochondrial biogenesis in Friedreich ataxia fibroblasts. PLoS ONE 2011, 6, e20666. [Google Scholar] [CrossRef]
- Xu, Z.; Park, S.-S.; Mueller, R.A.; Bagnell, R.C.; Patterson, C.; Boysen, P.G. Adenosine produces nitric oxide and prevents mitochondrial oxidant damage in rat cardiomyocytes. Cardiovasc. Res. 2005, 65, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Olatunji, O.J.; Feng, Y.; Olatunji, O.O.; Tang, J.; Ouyang, Z.; Su, Z.; Wang, D.; Yu, X. Neuroprotective effects of adenosine isolated from Cordyceps cicadae against oxidative and ER stress damages induced by glutamate in PC12 cells. Environ. Toxicol. Pharmacol. 2016, 44, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kalogeris, T.J.; Baines, C.; Korthuis, R.J. Adenosine prevents TNFα-induced decrease in endothelial mitochondrial mass via activation of eNOS-PGC-1α regulatory axis. PLoS ONE 2014, 9, e98459. [Google Scholar] [CrossRef]
- Janier, M.; Vanoverschelde, J.L.; Bergmann, S.R. Adenosine protects ischemic and reperfused myocardium by receptor-mediated mechanisms. Am. J. Physiol. Circ. Physiol. 1993, 264, H163–H170. [Google Scholar] [CrossRef]
- Fujii, W.; Funahashi, H. Exogenous adenosine reduces the mitochondrial membrane potential of murine oocytes during the latter half of in vitro maturation and pronuclear formation following chemical activation. J. Reprod. Dev. 2009, 55, 187–193. [Google Scholar] [CrossRef][Green Version]
- Delatycki, M.B.; Williamson, R.; Forrest, S.M. Friedreich ataxia: An overview. J. Med. Genet. 2000, 37, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.-Y.; Stringer, J.L. Energy failure in astrocytes increases the vulnerability of neurons to spreading depression. Eur. J. Neurosci. 2004, 19, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Augustyniak, J.; Lenart, J.; Zychowicz, M.; Stepien, P.P.; Buzanska, L. Mitochondrial biogenesis and neural differentiation of human iPSC is modulated by idebenone in a developmental stage-dependent manner. Biogerontology 2017, 18, 665–677. [Google Scholar] [CrossRef]
- Khdour, O.M.; Bandyopadhyay, I.; Visavadiya, N.P.; Chowdhury, S.R.; Hecht, S.M. Phenothiazine antioxidants increase mitochondrial biogenesis and frataxin levels in Friedreich’s ataxia cells. MedChemComm 2018, 9, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Aquilano, K.; Baldelli, S.; Cardaci, S.; Rotilio, G.; Ciriolo, M.R. Nitric oxide is the primary mediator of cytotoxicity induced by GSH depletion in neuronal cells. J. Cell Sci. 2011, 124, 1043–1054. [Google Scholar] [CrossRef]
- Rocha, M.C.; Grady, J.P.; Grünewald, A.; Vincent, A.; Dobson, P.F.; Taylor, R.W.; Turnbull, D.M.; Rygiel, K.A. A novel immunofluorescent assay to investigate oxidative phosphorylation deficiency in mitochondrial myopathy: Understanding mechanisms and improving diagnosis. Sci. Rep. 2015, 5, 15037. [Google Scholar] [CrossRef]
- Vincent, A.E.; Rosa, H.S.; Pabis, K.; Lawless, C.; Chen, C.; Grünewald, A.; Rygiel, K.A.; Rocha, M.C.; Reeve, A.K.; Falkous, G.; et al. Subcellular origin of mitochondrial DNA deletions in human skeletal muscle. Ann. Neurol. 2018, 84, 289–301. [Google Scholar] [CrossRef]
- Vincent, A.E.; Grady, J.P.; Rocha, M.C.; Alston, C.L.; Rygiel, K.A.; Barresi, R.; Taylor, R.W.; Turnbull, D.M. Mitochondrial dysfunction in myofibrillar myopathy. Neuromuscul. Disord. 2016, 26, 691–701. [Google Scholar] [CrossRef]
- Gordon, J.W.; Rungi, A.A.; Inagaki, H.; Hood, D.A. Selected Contribution: Effects of contractile activity on mitochondrial transcription factor A expression in skeletal muscle. J. Appl. Physiol. 2001, 90, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Li, P.A.; Hou, X.; Hao, S. Mitochondrial biogenesis in neurodegeneration. J. Neurosci. Res. 2017, 95, 2025–2029. [Google Scholar] [CrossRef] [PubMed]
- Virbasius, J.V.; Scarpulla, R.C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA 1994, 91, 1309–1313. [Google Scholar] [CrossRef] [PubMed]
- Marmolino, D.; Manto, M.; Acquaviva, F.; Vergara, P.; Ravella, A.; Monticelli, A.; Pandolfo, M. PGC-1alpha down-regulation affects the antioxidant response in Friedreich’s ataxia. PLoS ONE 2010, 5, e10025. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-R.; Cho, J.-M.; Shin, D.-H.; Yong, C.S.; Choi, H.-G.; Wakabayashi, N.; Kwak, M.-K. Adaptive response to GSH depletion and resistance to L-buthionine-(S,R)-sulfoximine: Involvement of Nrf2 activation. Mol. Cell Biochem. 2008, 318, 23–31. [Google Scholar] [CrossRef]
- Aquilano, K.; Baldelli, S.; Pagliei, B.; Cannata, S.M.; Rotilio, G.; Ciriolo, M.R. p53 orchestrates the PGC-1α-mediated antioxidant response upon mild redox and metabolic imbalance. Antioxid. Redox Signal. 2013, 18, 386–399. [Google Scholar] [CrossRef]
- Walsh, M.A.; Zhang, Q.; Musci, R.V.; Hamilton, K.L. The combination of NRF1 and Nrf2 activators in myoblasts stimulate mechanisms of proteostasis without changes in mitochondrial respiration. Redox Muscle Physiol. Exerc. Sport 2022, 1, 100001. [Google Scholar] [CrossRef]
- Min, K.-J.; Kim, J.-H.; Jou, I.; Joe, E.-H. Adenosine induces hemeoxygenase-1 expression in microglia through the activation of phosphatidylinositol 3-kinase and nuclear factor E2-related factor 2. Glia 2008, 56, 1028–1037. [Google Scholar] [CrossRef]
- Sheth, S.; Brito, R.; Mukherjea, D.; Rybak, L.P.; Ramkumar, V. Adenosine receptors: Expression, function and regulation. Int. J. Mol. Sci. 2014, 15, 2024–2052. [Google Scholar] [CrossRef]
- Castro, C.M.; Corciulo, C.; Solesio, M.E.; Liang, F.; Pavlov, E.V.; Cronstein, B.N. Adenosine A2A receptor (A2AR) stimulation enhances mitochondrial metabolism and mitigates reactive oxygen species-mediated mitochondrial injury. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 5027–5045. [Google Scholar] [CrossRef]
- Navia, A.M.; Ben, D.D.; Lambertucci, C.; Spinaci, A.; Volpini, R.; Coelho, J.E.; Lopes, L.V.; Marques-Morgado, I.; Marucci, G.; Buccioni, M. Adenosine receptors as neuroinflammation modulators: Role of A1 agonists and A2A antagonists. Cells 2020, 9, 1739. [Google Scholar] [CrossRef] [PubMed]



| Gene | Origin | 5′–3′ Primer Sequence |
|---|---|---|
| NFE2L2 | Human | Forward: ACA CGG TCC ACA GCT CAT C |
| Reverse: TGT CAA TCA AAT CCA TGT CCT G | ||
| NRF1 | Human | Forward: AGG AAC ACG GAG TGA CCC AA |
| Reverse: TAT GCT CGG TGT AAG TAG CCA | ||
| PPARGC1A | Human | Forward: TTG ACT GGC GTC ATT CAG GA |
| Reverse: GGG CAA TCC GTC TTC ATC CA | ||
| TFAM | Human | Forward: GTG ATT CAC CGC AGG AAA AGC |
| Reverse: GTG CGA CGT AGA AGA TCC TTT C | ||
| ACTB | Human | Forward: GCC AAC ACA GTG CTG TCT GG |
| Reverse: CTG CTT GCT GAT CCA CAT CTG C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lew, S.Y.; Mohd Hisam, N.S.; Phang, M.W.L.; Syed Abdul Rahman, S.N.; Poh, R.Y.Y.; Lim, S.H.; Kamaruzzaman, M.A.; Chau, S.C.; Tsui, K.C.; Lim, L.W.; et al. Adenosine Improves Mitochondrial Function and Biogenesis in Friedreich's Ataxia Fibroblasts Following L-Buthionine Sulfoximine-Induced Oxidative Stress. Biology 2023, 12, 559. https://doi.org/10.3390/biology12040559
Lew SY, Mohd Hisam NS, Phang MWL, Syed Abdul Rahman SN, Poh RYY, Lim SH, Kamaruzzaman MA, Chau SC, Tsui KC, Lim LW, et al. Adenosine Improves Mitochondrial Function and Biogenesis in Friedreich's Ataxia Fibroblasts Following L-Buthionine Sulfoximine-Induced Oxidative Stress. Biology. 2023; 12(4):559. https://doi.org/10.3390/biology12040559
Chicago/Turabian StyleLew, Sze Yuen, Nur Shahirah Mohd Hisam, Michael Weng Lok Phang, Syarifah Nur Syed Abdul Rahman, Rozaida Yuen Ying Poh, Siew Huah Lim, Mohd Amir Kamaruzzaman, Sze Chun Chau, Ka Chun Tsui, Lee Wei Lim, and et al. 2023. "Adenosine Improves Mitochondrial Function and Biogenesis in Friedreich's Ataxia Fibroblasts Following L-Buthionine Sulfoximine-Induced Oxidative Stress" Biology 12, no. 4: 559. https://doi.org/10.3390/biology12040559
APA StyleLew, S. Y., Mohd Hisam, N. S., Phang, M. W. L., Syed Abdul Rahman, S. N., Poh, R. Y. Y., Lim, S. H., Kamaruzzaman, M. A., Chau, S. C., Tsui, K. C., Lim, L. W., & Wong, K. H. (2023). Adenosine Improves Mitochondrial Function and Biogenesis in Friedreich's Ataxia Fibroblasts Following L-Buthionine Sulfoximine-Induced Oxidative Stress. Biology, 12(4), 559. https://doi.org/10.3390/biology12040559










