Taxonomic Novelties of Woody Litter Fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Morphological Observation, and Fungal Isolation
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analyses
| Taxon | Strain Number | GenBank Accession Numbers | Reference | |||
|---|---|---|---|---|---|---|
| SSU | LSU | ITS | tef1-α | |||
| Alloconiothyrium aptrootii | CBS 980.95 T | JX496121 | JX496234 | NA | NA | [28] |
| A. aptrootii | CBS 981.95 T | JX496122 | JX496235 | NA | NA | [28] |
| A. camelliae | NTUCC 17-032-1 T | MT112294 | MT071270 | MT071221 | MT232967 | [63] |
| A. camelliae | NTUCC 17-032-2 | MT112295 | MT071271 | MT071222 | MT232965 | [63] |
| A. camelliae | NTUCC 17-032-3 | MT112296 | MT071272 | MT071223 | MT232966 | [63] |
| A. encephalarti | CPC: 35980 | MN562102 | MN567610 | NA | NA | [63] |
| Austropleospora archidendri | MFLUCC 17-2429 | MK347757 | MK347974 | MK347863 | MK360044 | [33] |
| A. archidendri | CBS 168.77 | NA | JX496162 | JX496049 | NA | [28] |
| A. archidendri | KUMCC 21-0680 | OP059006 | OP059055 | OP058964 | OP135941 | This study |
| A. keteleeriae | MFLUCC 18-1551 T | MK347802 | MK348021 | MK347910 | MK360045 | [33] |
| A. ochracea | KUMCC 20-0020 T | MT799859 | MT799860 | MT808321 | MT872714 | [30] |
| A. osteospermi | BRIP51628 | FJ481946 | NA | NA | NA | [64] |
| Bambusistroma didymosporum | MFLU 15-0057 T | KP761733 | KP761730 | KP761737 | KP761727 | [65] |
| B. didymosporum | MFLU 15-0058 | KP761734 | KP761731 | KP761738 | KP761728 | [65] |
| Bimuria novae-zelandiae | CBS 107.79 T | MH861181 | AY016356 | AY016338 | DQ471087 | [66] |
| Chromolaenicola chiangraiensis | MFLUCC 17-1493 T | MN325017 | MN325005 | MN325011 | MN335650 | [39] |
| C. clematidis | MFLUCC 17-2075 T | MT310601 | MT214554 | MT226671 | NA | [67] |
| C. lampangensis | MFLUCC 17-1462 T | MN325016 | MN325004 | MN325010 | MN335649 | [39] |
| C. thailandensis | MFLUCC 17-1510 T | MN325018 | MN325006 | MN325012 | MN335651 | [39] |
| C. thailandensis | MFLUCC 17-1475 | MN325019 | MN325007 | MN325013 | MN335652 | [39] |
| C. nanensis | MFLUCC 17-1473 T | MN325015 | MN325003 | MN325009 | MN335648 | [39] |
| C. nanensis | MFLUCC 17-1477 | MN325014 | MN325002 | MN325008 | MN335647 | [39] |
| C. siamensis | MFLUCC 17-2527 T | NR_163337 | NG_066311 | NA | NA | [33] |
| C. sapindi | KUMCC 21-0564 T | OP059009 | OP059058 | OP058967 | OP135943 | This study |
| C. sapindi | KUMCC 21-0594 | OP059010 | OP059059 | OP058968 | OP135944 | This study |
| Cylindroaseptospora leucaenae | MFLUCC 17-2424 T | NR_163333 | NG_066310 | MK347856 | MK360047 | [33] |
| C. siamensis | MFLUCC 17-2527 T | MK347760 | MK347976 | MK347866 | MK360048 | [33] |
| Deniquelata barringtoniae | MFLUCC 11-0422 T | NR_111779 | NG_042696 | JX254656 | NA | [68] |
| D. vittalii | NFCCI4249 T | MF406218 | MF182395 | MF622059 | MF182398 | [69] |
| Dictyoarthrinium hydei | SQUCC 13296 T | MW077145 | NA | MW077161 | MW075771 | [26] |
| D. musae | MFLUCC 20-0105 T | MT482323 | MT482320 | MT482326 | MT495602 | [52] |
| D. musae | MFLUCC 20-0106 T | MT482324 | MT482321 | MT482327 | MT495603 | [52] |
| D. sacchari | MFLUCC 20-0107 | MT482325 | MT482322 | MT482328 | NA | [52] |
| D. sacchari | CBS 529.73 | NA | MH872479 | NA | NA | [70] |
| D. thailandicum | KUMCC 21-0664 T | OP059007 | OP059056 | OP058965 | NA | This study |
| D. thailandicum | KUMCC 21-0665 | OP059008 | OP059057 | OP058966 | OP135942 | This study |
| Didymocrea sadasivanii | CBS 438.65 T | MH858658 | DQ384103 | NA | NA | [70] |
| Didymosphaeria rubi-ulmifolii | MFLUCC 14-0023 T | NA | KJ436586 | NG_063557 | NA | [40] |
| D. rubi-ulmifolii | MFLUCC 14-0024 | NA | KJ436585 | KJ436587 | NA | [40] |
| Kalmusia italica | MFLUCC 14-0560 T | KP325440 | KP325441 | KP325442 | NA | [71] |
| K. variispora | CBS 121517 T | MH863113 | MH874668 | NA | NA | [28] |
| K. ebuli | CBS 123120 T | KF796674 | JN644073 | JN851818 | NA | [72] |
| Kalmusibambusa triseptata | MFLUCC 13-0232 T | KY682697 | KY682695 | KY682696 | NA | [41] |
| Karstenula lancangensis | KUMCC 21-0670 T | OP059011 | OP059060 | OP058969 | NA | This study |
| K. lancangensis | KUMCC 21-0677 | OP059012 | OP059061 | OP058970 | NA | This study |
| K. rhodostoma | CBS 690.94 | NA | GU301821 | GU296154 | GU349067 | [73] |
| K. rhodostoma | CBS 691.94 | LC014559 | AB807531 | AB797241 | AB808506 | [73] |
| Laburnicola hawksworthii | MFLUCC 13-0602 T | KU743194 | KU743195 | KU743196 | NA | [42] |
| L. muriformis | MFLUCC 14-0921 T | KU743200 | KU743201 | KU743202 | NA | [42] |
| Letendraea cordylinicola | MFLUCC 11-0150 | KM213996 | KM213999 | KM214002 | NA | [40] |
| L. cordylinicola | MFLUCC 11-0148 T | NR_154118 | NG_059530 | KM214001 | NA | [40] |
| Montagnula aloes | CPC 19671 T | JX069863 | JX069847 | NA | NA | [74] |
| M. aloes | CBS 132531 T | NR_111757 | NG_042676 | NA | NA | [74] |
| M. appendiculata | CBS 109027 T | DQ435529 | AY772016 | NA | NA | [75] |
| M. bellevaliae | MFLUCC 14-0924 T | KT443906 | KT443902 | KT443904 | NA | [76] |
| M. camporesii | MFLUCC 16-1369 T | MN401746 | NG_070946 | NG_068418 | MN397908 | [77] |
| M. chiangraiensis | MFLUCC 17-1420 T | NR_168864 | NG_068707 | NG_070155 | NA | [39] |
| M. chromolaenae | MFLUCC 17-1435 T | NR_168865 | NG_068708 | NG_070156 | NA | [39] |
| M. chromolaenicola | MFLUCC 17-1469 T | NR_168866 | NG_070948 | NG_070157 | MT235773 | [39] |
| M. cirsii | MFLUCC 13-0680 | KX274242 | KX274249 | KX274255 | KX284707 | [78] |
| M. cylindrospora | UTHSC: DI16-208 T | LT796834 | LN907351 | NA | LT797074 | [74] |
| M. donacina | HFG07004 | MF967419 | MF183940 | NA | NA | [79] |
| M. donacina | HVVV01 | KJ628375 | KJ628377 | KJ628376 | NA | [80] |
| M. donacina | KUMCC 21-0653 | OP059003 | OP059052 | OP058961 | OP135938 | This study |
| M. donacina | KUMCC 21-0579 | OP059005 | OP059054 | OP058963 | OP135940 | This study |
| M. donacina | KUMCC 21-0631 | OP059004 | OP059053 | OP058962 | OP135939 | This study |
| M. graminicola | MFLUCC 13-0352 T | KM658314 | KM658315 | KM658316 | NA | [81] |
| M. jonesii | MFLUCC 16-1448 T | KY313619 | KY273276 | KY313618 | KY313620 | [49] |
| M. krabiensis | MFLUCC 16-0250 T | NR168179 | NG068826 | NG068385 | MH412776 | [82] |
| M. puerensis | KUMCC 20-0225 T | MW567739 | MW575866 | MW575864 | MW575859 | [83] |
| M. puerensis | KUMCC 20-0331 | MW567740 | MW575867 | MW575865 | MW575860 | [83] |
| M. saikhuensis | MFLUCC 16-0315 T | KU743209 | KU743210 | KU743211 | NA | [42] |
| M. scabiosae | MFLUCC 14-0954 T | KT443907 | KT443903 | KT443905 | NA | [76] |
| M. thailandica | MFLUCC 17-1508 T | MT214352 | NG070949 | NG070158 | MT235774 | [39] |
| Neokalmusia brevispora | KT 1466 T | LC014573 | AB524600 | AB524459 | AB539112 | [73] |
| N. scabrispora | KT 1023 | LC014575 | AB524593 | AB524452 | AB539106 | [73] |
| Neptunomyces aureus | CMG12 T | MK912121 | NA | NA | MK948000 | [84] |
| N. aureus | CMG13 | MK912122 | NA | NA | MK948001 | [84] |
| Paraconiothyrium cyclothyrioides | CBS 972.95 T | JX496119 | JX496232 | AY642524 | NA | [28] |
| P. cyclothyrioides | CBS 432.75 | MH860933 | MH872689 | NA | NA | [28] |
| P. estuarinum | CBS 109850 | MH862842 | MH874432 | NA | NA | [28] |
| Paracamarosporium fagi | CPC 24890 | KR611886 | KR611904 | NA | NA | [35] |
| P. fagi | CPC 24892 T | KR611887 | KR611905 | NA | NA | [35] |
| Paramassariosphaeria anthostomoides | CBS 615.86 | MH862005 | GU205223 | GU205246 | NA | [28] |
| P. anthostomoides | MFLU 16-0172 T | KU743206 | KU743207 | KU743208 | NA | [42] |
| Paraphaeosphaeria rosae | MFLUCC 17-2547 | MG828935 | MG829044 | MG829150 | MG829222 | [85] |
| P. rosae | MFLUCC 17-2549 T | MG828937 | MG829046 | MG829152 | MG829223 | [85] |
| P. rosicola | MFLUCC 15-0042 T | NR_157528 | MG829047 | MG829153 | NA | [85] |
| Phaeodothis winteri | CBS 182.58 | NA | GU301857 | GU296183 | NA | [86] |
| Pseudocamarosporium propinquum | MFLUCC 13-0544 | KJ747049 | KJ813280 | KJ819949 | NA | [36] |
| P. pteleae | MFLUCC 17-0724 T | NR_157536 | MG829061 | MG829166 | MG829233 | [85] |
| Pseudopithomyces entadae | MFLUCC 17-0917 T | NA | NG_066305 | MK347835 | MK360083 | [33] |
| P. rosae | MFLUCC 15-0035 T | MG828953 | MG829064 | MG829168 | NA | [85] |
| Septofusispora thailandica | KUMCC 21-0647 T | OP059013 | OP059062 | OP058971 | OP135945 | This study |
| S. thailandica | KUMCC 21-0652 | OP059014 | OP059063 | OP058972 | NA | This study |
| Spegazzinia bromeliacearum | URM 8084 T | MK804501 | MK809513 | NA | NA | [87] |
| S. deightonii | MFLUCC 20-0002 T | MN956768 | MN956772 | MN956770 | MN927133 | [73] |
| S. intermedia | CBS 249.89 T | MH862171 | MH873861 | NA | NA | [70] |
| S. jinghaensis | KUMCC 21-0495 T | OP059015 | OP059064 | OP058973 | OP135946 | This study |
| S. jinghaensis | KUMCC 21-0496 | OP059016 | OP059065 | OP058974 | OP135947 | This study |
| S. lobulata | CBS 361.58 T | MH857812 | MH869344 | NA | NA | [70] |
| S. musae | MFLUCC 20-0001 T | MN930512 | MN930514 | MN930513 | MN927132 | [52] |
| S. neosundara | MFLUCC 15-0456 T | KX965728 | KX954397 | KX986341 | NA | [41] |
| S. radermacherae | MFLUCC 17-2285 T | MK347740 | MK347957 | MK347848 | MK360088 | [33] |
| S. tessarthra | SH 287 | JQ673429 | AB807584 | AB797294 | AB808560 | [73] |
| Tremateia arundicola | MFLU 16-1275 T | KX274241 | KX274248 | KX274254 | KX284706 | [49] |
| T. guiyangensis | GZAAS01 T | KX274240 | KX274247 | KX274253 | KX284705 | [49] |
| T. murispora | GZCC 18-2787 T | NR_165916 | MK972751 | MK972750 | MK986482 | [88] |
| Verrucoconiothyrium nitidae | CBS: 119209 | EU552112 | EU552112 | NA | NA | [89] |
| Xenocamarosporium acaciae | CBS: 139895 T | NR_137982 | NG_058163 | NA | NA | [35] |
| X. acaciae | MFLUCC 17-2432 | MK347766 | MK347983 | MK347873 | MK360093 | [33] |
3. Results
3.1. Phylogeny
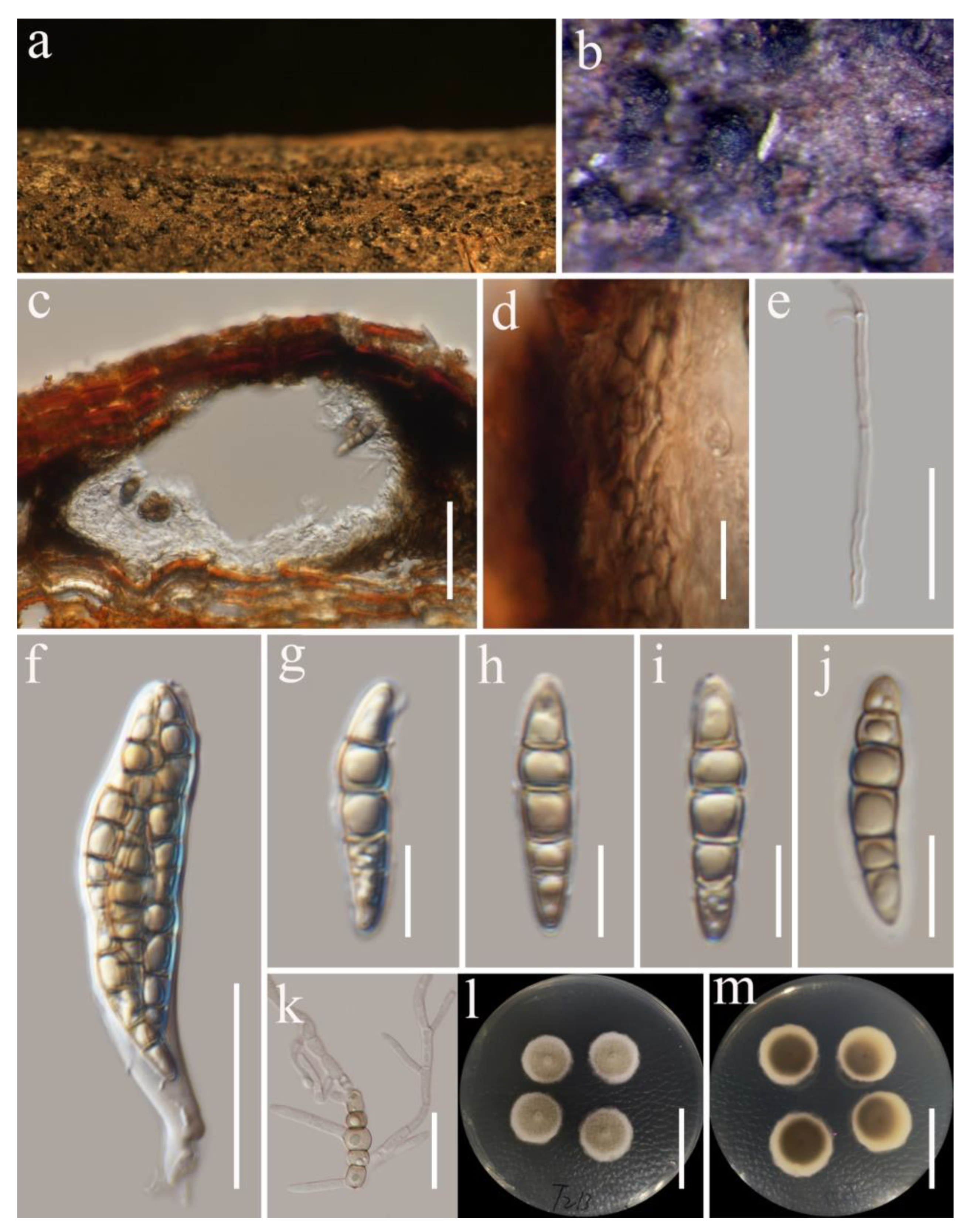
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hapuarachchi, K.K.; Karunarathna, S.C.; Phengsintham, P.; Yang, H.D.; Kakumyan, P.; Hyde, K.D.; Wen, T.C. Ganodermataceae (Polyporales): Diversity in Greater Mekong Subregion countries (China Laos Myanmar Thailand and Vietnam). Mycosphere 2019, 10, 221–309. [Google Scholar] [CrossRef]
- Costenbader, J.; Varns, T.; Vidal, A.; Stanley, L.; Broadhead, J. Drivers of Deforestation in the Greater Mekong Subregion Regional Report; USAID Lowering Emissions in Asia’s Forests (USAID LEAF): Bangkok, Thailand, 2015; pp. 1–38. [Google Scholar] [CrossRef]
- Li, H.; Guo, J.; Karunarathna, S.C.; Ye, L.; Xu, J.; Hyde, K.D.; Mortimer, P.E. Native forests have a higher diversity of macrofungi than comparable plantation forests in the Greater Mekong Subregion. Forests 2018, 9, 402. [Google Scholar] [CrossRef]
- Feng, B.; Yang, Z. Studies on diversity of higher fungi in Yunnan southwestern China: A review. Plant Divers. 2018, 40, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Hyde, K.D.; Bhat, D.J.; Jeewon, R.; Maharachchikumbura, S.S.N.; Bao, D.F.; Li, W.L.; Su, X.J.; Yang, X.Y.; Su, H.Y. Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan China. Mycol. Prog. 2018, 17, 511–530. [Google Scholar] [CrossRef]
- Ye, L.; Li, H.; Mortimer, P.E.; Xu, J.; Gui, H.; Karunarathna, S.C.; Kumar, A.; Hyde, K.D.; Shi, L. Substrate preference determines macrofungal biogeography in the Greater Mekong sub-region. Forests 2019, 10, 824. [Google Scholar] [CrossRef]
- Dong, W.; Wang, B.; Hyde, K.D.; McKenzie, E.H.C.; Raja, H.A.; Tanaka, K.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Doilom, M.; Phookamsak, R.; et al. Freshwater Dothideomycetes. Fungal Divers. 2020, 105, 319–575. [Google Scholar] [CrossRef]
- Chaiwan, N.; Tibpromma, S.; Jayawardena, R.S.; Mapook, A.; Wanasinghe, D.N.; Mortimer, P.E.; Lumyong, S.; Hyde, K.D. Colletotrichum dracaenigenum, a new species on Dracaena fragrans. Phytotaxa 2021, 491, 143–157. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Chen, J.; Dissanayake, A.J.; Doilom, M.; Hongsanan, S.; Jayawardena, R.S.; Jeewon, R.; Perera, R.H.; Thongbai, B.; et al. Thailand’s amazing diversity—Up to 96 % of fungi in northern Thailand are novel. Fungal Divers. 2018, 93, 215–239. [Google Scholar] [CrossRef]
- Kodsueb, R.; McKenzie, E.H.C.; Lumyong, S.; Hyde, K.D. Diversity of saprobic fungi on Magnoliaceae. Fungal Divers. 2008, 30, 37–53. [Google Scholar]
- Kodsueb, R.; McKenzie, E.H.C.; Lumyong, S.; Hyde, K.D. Fungal succession on woody litter of Magnolia liliifera (Magnoliaceae). Fungal Divers. 2008, 30, 55–72. [Google Scholar]
- Seephueak, P.; Phongpaichit, S.; Hyde, K.D.; Petcharat, V. Diversity of saprobic fungi on decaying branch litter of the rubber tree (Hevea brasiliensis). Mycosphere 2011, 2, 307–330. [Google Scholar]
- Monkai, J.; Boonmee, S.; Ren, G.C.; Wei, D.P.; Phookamsak, R.; Mortimer, P.E. Distoseptispora hydei sp. nov. (Distoseptisporaceae) a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 2020, 459, 093–107. [Google Scholar] [CrossRef]
- Ren, G.C.; Wanasinghe, D.N.; Wei, D.P.; Monkai, J.; Yasanthika, E.; Gui, H.; Mortimer, P.E.; Xu, J.C.; Hyde, K.D. Loculosulcatispora thailandica gen. et sp. nov. (Sulcatisporaceae) saprobic on woody litter in Thailand. Phytotaxa 2020, 475, 067–078. [Google Scholar] [CrossRef]
- Ren, G.C.; Wanasinghe, D.N.; Monkai, J.; Hyde, K.D.; Mortimer, P.E.; Xu, J.C.; Pang, A.; Gui, H. Introduction of Neolophiotrema xiaokongense gen. et sp. nov. to the poorly represented Anteagloniaceae (Pleosporales, Dothideomycetes). Phytotaxa 2021, 482, 25–35. [Google Scholar] [CrossRef]
- Ren, G.C.; Wanasinghe, D.N.; Monkai, J.; Mortimer, P.E.; Hyde, K.D.; Xu, J.C.; Pang, A.; Gui, H. Novel saprobic Hermatomyces species (Hermatomycetaceae Pleosporales) from China (Yunnan Province) and Thailand. MycoKeys 2021, 82, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.C.; Wanasinghe, D.N.; Jeewon, R.; Monkai, J.; Mortimer, P.E.; Hyde, K.D.; Xu, J.C.; Gui, H. Taxonomy and phylogeny of the novel rhytidhysteron-like collections in the Greater Mekong Subregion. MycoKeys 2021, 86, 65–85. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Wijayawardene, N.N.; Xu, J.C.; Cheewangkoon, R.; Mortimer, P.E. Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan China). PLoS ONE 2020, 15, e0235855. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Mortimer, P.E.; Xu, J. Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaea viscosa in Honghe (China). J. Fungi 2021, 7, 180. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Ren, G.C.; Xu, J.C.; Cheewangkoon, R.; Mortimer, P.E. Insight into the Taxonomic Resolution of the Pleosporalean Species Associated with Dead Woody Litter in Natural Forests from Yunnan, China. J. Fungi 2022, 8, 375. [Google Scholar] [CrossRef]
- Mortimer, P.E.; Jeewon, R.; Xu, J.C.; Lumyong, S.; Wanasinghe, D.N. Morpho-phylo taxonomy of novel dothideomycetous fungi associated with dead woody twigs in Yunnan Province China. Front. Microbiol. 2021, 12, 654683. [Google Scholar] [CrossRef]
- Calabon, M.S.; Jones, E.B.G.; Boonmee, S.; Doilom, M.; Lumyong, S.; Hyde, K.D. Five novel freshwater ascomycetes indicate high undiscovered diversity in lotic habitats in Thailand. J. Fungi 2021, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Calabon, M.S.; Hyde, K.D.; Jones, E.B.G.; Luo, Z.L.; Dong, W.; Hurdeal, V.G.; Gentekaki, E.; Rossi, W.; Leonardi, M.; Thiyagaraja, V.; et al. Freshwater fungal numbers. Fungal Divers. 2022, 114, 3–235. [Google Scholar] [CrossRef]
- Munk, A. The system of the pyrenomycetes. A contribution to a natural classification of the group Sphaeriales sensu Lindau. Dan. Bot. Ark. 1953, 15, 1–163. [Google Scholar]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.T.; Saxena, R.K.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of Fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Wanasinghe, D.N.; Cheewangkoon, R.; Al-Sadi, A.M. Uncovering the hidden taxonomic diversity of fungi in Oman. Fungal Divers. 2021, 106, 229–268. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y. Fungal Databases U.S. National Fungus Collections ARS USDA. Available online: http://nt.ars-grin.gov/fungaldatabases/ (accessed on 20 July 2022).
- Verkley, G.J.M.; Dukik, K.; Renfurm, R.; Göker, M.; Stielow, J.B. Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 2014, 32, 25–51. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Bhat, D.J.; Liu, N.G.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Dissanayake, L.S.; Wijayawardene, N.N.; Samarakoon, M.C.; Hyde, K.D.; Kang, J.C. The taxonomy and phylogeny of Austropleospora ochracea sp. nov. (Didymosphaeriaceae) from Guizhou China. Phytotaxa 2021, 491, 217–229. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Tanaka, K.; Thambugala, K.M.; Phookamsak, R.; Tian, Q.; Camporesi, E.; Hongsanan, S.; Monkai, J.; Wanasinghe, D.N.; Mapook, A.; et al. A molecular phylogenetic reappraisal of the Didymosphaeriaceae (= Montagnulaceae). Fungal Divers. 2014, 68, 69–104. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungilike taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Jeewon, R.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; Liu, J.K.; Lu, Y.Z.; et al. Diversity morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 2019, 10, 1–186. [Google Scholar] [CrossRef]
- Samarakoon, B.C.; Wanasinghe, D.N.; Samarakoon, M.C.; Phookamsak, R.; McKenzie, E.H.C.; Chomnunti, P.; Hyde, K.D.; Lumyong, S.; Karunarathna, S.C. Multi-gene phylogenetic evidence suggests Dictyoarthrinium belongs in Didymosphaeriaceae (Pleosporales, Dothideomycetes) and Dictyoarthrinium musae sp. nov. on Musa from Thailand. MycoKeys 2020, 71, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Schumacher, R.K.; Wingfeld, M.J.; Lombard, L.; Giraldo, A.; Christensen, M.; Gardiennet, A.; Nakashima, C.; Pereira, O.; Smith, A.J.; et al. Fungal systematics and evolution: FUSE 1. Sydowia 2015, 67, 81–118. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Bhat, D.J.; Camporesi, E.; Schumacher, R.K.; Chethana, K.W.T.; Wikee, S.; Bahkali, A.H.; Wang, Y. Camarosporium-like species are polyphyletic in Pleosporales; introducing Paracamarosporium and Pseudocamarosporiumgen. nov. in Montagnulaceae. Cryptogam. Mycol. 2014, 35, 177–198. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Thambugala, K.M.; Manamgoda, D.S.; Jayawardena, R.; Camporesi, E.; Boonmee, S.; Wanasinghe, D.N.; Phookamsak, R.; Hongsanan, S.; Singtripop, C.; et al. Towards a natural classification and backbone tree for Pleosporaceae. Fungal Divers. 2015, 71, 85–139. [Google Scholar] [CrossRef]
- Mena-Portales, J.; Cantillo-Pérez, T.; Minter, D.W. A new species of the conidial fungal genus Spegazzinia (Pleosporales Didymosphaeriaceae) collected on sugarcane in Cuba. Phytotaxa 2017, 331, 295–298. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, 101, 1–175. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Camporesi, E.; Thambugala, K.M.; Mapook, A.; Kang, J.C.; Alias, S.A.; Chukeatirote, E.; Thines, M.; McKenzie, E.H.C.; Hyde, K.D. Confusion surrounding Didymosphaeria–phylogenetic and morphological evidence suggest Didymosphaeriaceae is not a distinct family. Phytotaxa 2014, 176, 102–119. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Wanasinghe, D.N.; Phillips, A.J.L.; Camporesi, E.; Bulgakov, T.S.; Phukhamsakda, C.; Ariyawansa, H.A.; Goonasekara, I.D.; Phookamsak, R.; Dissanayake, A.; et al. Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 2017, 8, 697–796. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Jones, E.G.; Camporesi, E.; Dissanayake, A.J.; Kamolhan, S.; Mortimer, P.E.; Xu, J.C.; Hyde, K.D. Taxonomy and phylogeny of Laburnicola gen. nov. and Paramassariosphaeria gen. nov. (Didymosphaeriaceae, Massarineae Pleosporales). Fungal Biol. 2016, 120, 1354–1373. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection examination isolation sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/names/Names.asp (accessed on 1 August 2022).
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Hyde, K.D.; Wanasinghe, D.N.; Bahkali, A.H.; Camporesi, E.; Khan, S.; Phookamsak, R. Taxonomy and phylogenetic appraisal of Montagnula jonesii sp. nov. (Didymosphaeriaceae, Pleosporales). Mycosphere 2016, 7, 1346–1356. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Samarakoon, B.C.; Phookamsak, R.; Wanasinghe, D.N.; Chomnunti, P.; Hyde, K.D.; McKenzie, E.H.C.; Promputtha, I.; Xu, J.C.; Li, Y.J. Taxonomy and phylogenetic appraisal of Spegazzinia musae sp. nov. and S. deightonii (Didymosphaeriaceae Pleosporales) on Musaceae from Thailand. MycoKeys 2020, 70, 19–37. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 4. [Google Scholar] [CrossRef]
- Nylander, J.A.A.; Wilgenbusch, J.C.; Warren, D.L.; Swofford, D.L. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 2008, 24, 581–583. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree Version 1.4.0. 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 30 May 2022).
- Ariyawansa, H.A.; Tsai, I.; Thambugala, K.M.; Chuang, W.Y.; Lin, S.R.; Hozzein, W.N.; Cheewangkoon, R. Species diversity of Pleosporalean taxa associated with Camellia sinensis (L.) Kuntze in Taiwan. Sci. Rep. 2020, 10, 12762. [Google Scholar] [CrossRef]
- Morin, L.; Shivas, R.G.; Piper, M.C.; Tan, Y.P. Austropleospora osteospermi gen. et sp. nov. and its host specificity and distribution on Chrysanthemoides monilifera ssp. rotundata in Australia. Fungal Divers. 2010, 40, 65–74. [Google Scholar] [CrossRef]
- Adamčík, S.; Cai, L.; Chakraborty, D.; Chen, X.H.; Cotter, H.V.T.; Dai, D.Q.; Dai, Y.C.; Das, K.; Deng, C.Y.; Ghobad-Nejhad, M.; et al. Fungal biodiversity profiles 1–10. Cryptogamie Mycologie 2015, 36, 121–166. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Hindemith, R. Major lineages of Dothideomycetes (Ascomycota) inferred from SSU and LSU rDNA sequences. Mycol. Res. 2001, 105, 901–908. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.J.L.; Jones, E.B.G.; Bhat, D.J.; Marc, S.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Maharachchikumbura, S.S.N.; Karunarathne, S.C.; Chukeatirote, E.; Bahkali, A.H.; Kang, J.C.; Bhat, J.B.; Hyde, K.D. Deniquelata barringtoniae gen. et sp. nov. associated with leaf spots of Barringtonia asiatica. Phytotaxa 2013, 105, 11–20. [Google Scholar] [CrossRef]
- Devadatha, B.; Sarma, V.V.; Ariyawansa, H.A.; Gareth, J.E.B. Deniquelata vittalii sp.nov. a novel indian saprobic marine fungus on Suaeda monoica and two new records of marine fungi from Muthupet mangroves East coast of India. Mycosphere 2018, 9, 565–582. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Thambugala, K.M.; Hyde, K.D.; Tanaka, K.; Tian, Q.; Wanasinghe, D.N.; Ariyawansa, H.A.; Jayasiri, S.C.; Boonmee, S.; Camporesi, E.; Hashimoto, A.; et al. Towards a natural classifcation and backbone tree for Lophiostomataceae Floricolaceae and Amorosiaceae fam. nov. Fungal Divers. 2015, 74, 199–266. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Wang, Z.; Fournier, J.; Crous, P.W.; Zhang, X.; Li, W.; Hyde, K.D. Neotypification and phylogeny of Kalmusia. Phytotaxa 2014, 176, 164–173. [Google Scholar] [CrossRef][Green Version]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef]
- Aptroot, A. Two new ascomycetes with long gelatinous appendages collected from monocots in the tropics. Stud. Mycol. 2004, 50, 307–311. [Google Scholar]
- Hongsanan, S.; Hyde, K.D.; Bahkall, A.H.; Camporesi, B.E.; Chomnunti, P.; Ekanayaka, H.; Gomes, A.A.M.; Hofstetter, V.; Jones, E.B.G.; Pinho, D.B.; et al. Fungal biodiversity profiles 11–20. Cryptogam. Mycol. 2015, 36, 355–380. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.P.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Zhao, Z.Z.; Zhao, K.; Chen, H.P.; Bai, X.; Zhang, L.; Liu, J.K. Terpenoids from the mushroom-associated fungus Montagnula donacina. Phytochemistry 2017, 147, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Pitt, W.; Úrbez-Torres, J.R.; Trouillas, F.P. Munkovalsaria donacina from grapevines and Desert Ash in Australia. Mycosphere 2014, 5, 656–661. [Google Scholar] [CrossRef]
- Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Ariyawansa, H.A.; Bhat, D.J.; Boonmee, S.; Maharachchikumbura, S.; McKenzie, E.H.C.; Phookamsak, R.; Phukhamsakda, C.; et al. Fungal Diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015, 72, 1–197. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, J.D.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Du, T.; Hyde, K.D.; Mapook, A.; Mortimer, P.E.; Xu, J.C.; Karunarathna, S.C.; Tibpromma, S. Morphology and phylogenetic analyses reveal Montagnula puerensis sp. nov. (Didymosphaeriaceae, Pleosporales) from southwest China. Phytotaxa 2021, 514, 1–25. [Google Scholar] [CrossRef]
- Goncalves, M.F.M.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Neptunomyces aureus gen. et sp. nov. (Didymosphaeriaceae Pleosporales) isolated from algae in Ria de Aveiro Portugal. MycoKeys 2019, 60, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Jones, E.B.G.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.A.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef]
- Crous, P.W.; Carnegie, A.J.; Wingfield, M.J.; Sharma, R.; Mughini, G.; Noordeloos, M.E.; Santini, A.; Shouche, Y.S.; Bezerra, J.D.P.; Dima, B.; et al. Fungal planet description sheets: 868–950. Persoonia 2019, 42, 291–473. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, S.N.; Liu, Z.Y. Tremateia murispora sp. nov. (Didymosphaeriaceae Pleosporales) from Guizhou China. Phytotaxa 2019, 416, 79–87. [Google Scholar] [CrossRef]
- Marincowitz, S.; Crous, P.W.; Groenewald, J.Z.; Wingfield, M.J. Microfungi occurring on Proteaceae in the fynbos. In CBS Biodiversity Series; No. 7; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2008; p. 166. [Google Scholar]
- Niessl, G. Beiträge zur Kenntniss der Pilze. Beschreibung neuerund wenig bekannter Pilze. Verh Nat. Ver. Brünn 1872, 10, 153–217. [Google Scholar]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.W.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal diversity notes 111–252—Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Hughes, S.J. Fungi from the Gold Coast. I. Mycol. Pap. 1952, 48, 1–91. [Google Scholar]
- Ellis, M.B. Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1971; p. 608. [Google Scholar]
- Hyde, K.D.; Fröhlich, J.; Taylor, J.E. Fungi from palms XXXVI Reflections on unitunicate ascomycetes with apiospores. Sydowia 1998, 50, 21–80. [Google Scholar]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota—2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Barr, M.E. Melanommatales (Loculoascomycetes). In North American Flora Series II Part; NYBG Press: New York, NY, USA, 1990; Volume 13, pp. 1–129. [Google Scholar]
- Constantinescu, O. Teleomorph-anamorph connections in ascomycetes: Microdiplodia anamorph of Karstenula rhodostoma. Mycol. Res. 1993, 97, 377–380. [Google Scholar] [CrossRef]
- Berlese, A.N. Icones fungorum. Pyrenomycetes 1896, 2, 1–216. [Google Scholar]
- Aptroot, A. Redisposition of some species excluded from Didymosphaeria (Ascomycotina). Nova Hedwig. 1995, 60, 325–379. [Google Scholar]
- Saccardo, P.A. Conspectus generum fungorum Italiae inferorium. Michelia 1880, 2, 1–38. [Google Scholar]
- Tianyu, Z. Flora Fungorum Sincorum: 26 Genera of Dematiaceous Dictyosporous Hyphomycetes Excluding Alternaria; Science Press: Beijing, China, 2009; Volume 31, pp. 97–101. [Google Scholar]
- Leão-Ferreira, S.M.; Gusmão, L.F.P. Conidial fungi from the semi-arid Caatinga biome of Brazil. New species of Endophragmiella, Spegazzina and new records for Brazil South America and Neotropica. Mycotaxon 2010, 111, 1–10. [Google Scholar] [CrossRef]
- Whitton, S.R.; McKenzie, E.H.C.; Hyde, K.D. Fungi associated with Pandanaceae. Fungal Divers. Res. Ser. 2012, 21, 1–458. [Google Scholar]
- Tennakoon, D.S.; Kuo, C.H.; Maharachchikumbura, S.S.N.; Thambugala, K.M.; Gentekaki, E.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; de Silva, N.I.; Promputtha, I.; et al. Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Divers. 2021, 108, 1–215. [Google Scholar] [CrossRef]
- Manish, T.; Gupta, R.C.; Yogesh, J. Spegazzinia tessarthra isolated as a true endophyte from lichen Heterodermia flabellate. Indian Phytopathol. 2014, 67, 109–110. [Google Scholar]
- Borut, S.Y.; Johnson, T.W. Some biological observations on fungi in estuarine sediments. Mycologia 1962, 54, 181–193. [Google Scholar] [CrossRef]
- Ellis, M.B. More Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1976; p. 507. [Google Scholar]
- Chethana, K.W.T.; Niranjan, M.; Dong, W.; Samarakoon, M.C.; Bao, D.F.; Calabon, M.S.; Chaiwan, N.; Chuankid, B.; Dayarathne, M.C.; de Silva, N.I.; et al. AJOM new records and collections of fungi: 101–150. Asian J. Mycol. 2021, 4, 113–260. [Google Scholar] [CrossRef]
- Urríes, M.J. Hongos microscópicos de Canarias. In El Museo Canaria; Años XVII-XVIII: Las Palmas De Gran Canaria, Espana, 1957; pp. 43–44. Available online: http://www.elmuseocanario.com/images/documentospdf/revistaelmuseo/Revistas/1956-1957.pdf (accessed on 20 August 2022).
- Komarovii, V.L. Species Novae Ascomycetum Turkomania. In Novitates Systematicae Plantarum Non Vasculariul VII; Academia Scientiarum Urss Institutum Botanicum: Leningrad, Russia, 1970; pp. 189–197. Available online: https://www.binran.ru/files/journals/NSNR/1970_7/NSNR_1970_7_Frolov_2.pdf (accessed on 20 August 2022).
- Komarovii, V.L. Species Novae Ascomycetum Turkomania. In Novitates Systematicae Plantarum Non Vasculariul; Academia Scientiarum Urss Institutum Botanicum: Leningrad, Russia, 1967; pp. 233–234. Available online: https://www.binran.ru/files/journals/NSNR/1967_4/NSNR_1967_4_Frolov.pdf (accessed on 20 August 2022).
- Hyde, K.D.; Aptroot, A.; Frohlich, J.; Taylor, J.E. Fungi from palms. XLII. Didymosphaeria and similar ascomycetes from palms. Nova Hedwig. 1999, 69, 449–471. [Google Scholar] [CrossRef]
- Thaung, M.M. Pathologic and taxonomic analysis of leaf spot and tar spot diseases in a tropical dry to wet monsoon ecosystem of lowland Burma. Australas. Plant Pathol. 2008, 37, 180–197. [Google Scholar] [CrossRef]
- Francisco, C.S.; Ma, X.; Zwyssig, M.M.; Mcdonald, B.A.; Palma-Guerrero, J. Morphological changes in response to environmental stresses in the fungal plant pathogen Zymoseptoria tritici. Sci. Rep. 2019, 9, 9642. [Google Scholar] [CrossRef] [PubMed]
- Chethana, K.W.T.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E.; Raspé, O.; Promputtha, I.; Hyde, K.D. What are fungal species and how to delineate them? Fungal Divers. 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Pem, D.; Jeewon, R.; Chethana, K.W.T.; Hongsanan, S.; Doilom, M.; Suwannarach, N.; Hyde, K.D. Species concepts of Dothideomycetes: Classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Divers. 2021, 109, 283–319. [Google Scholar] [CrossRef]








| Species Name | Ascomata | Asci | Ascospores | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| Color | Shape | Size | Rows of Ascospores in Asci | Septation | Surface | ||||
| M. chromolaenicola | Uni-loculate, 310 × 275 μm diam. | 90 × 12 μm, bitunicate, elongate-clavate, 8-spored, long pedicellate | Brown to dark brown | Broadly fusiform to ellipsoid | 15.5 × 6 μm | Overlapping 1–2-seriate | 1 | Guttulate | [39] |
| M. donacina | Multi-loculate, 500 μm diam. | 90–100 × 12–13 μm, bitunicate, clavate, 8-spored, long pedicellate | Brown | Ellipsoid | 14.8–15.2 × 7.5–7.7 μm | Irregularly biseriate | 1 | Guttulate | [80,100] |
| M. donacina (HKAS 122778) | Uni-loculate, 490 × 410 µm diam. | 110 × 13 µm, bitunicate, elongate-clavate, slightly curved, 8-spored, long pedicellate | Pale brown to brown | Broadly fusiform | 15 × 5 µm | Overlapping 1–2-seriate | 1 | Guttulate | This study |
| M. donacina graminicola | Uni-loculate, 37–117.22 μm diam. | 81.3 × 10.1 μm, bitunicate, cylindrical to clavate, 8-spored, long pedicellate | Brown | Ellipsoid | 11.3 × 4.9 μm | Biseriate | 1 | Verruculose, mucilaginous sheath | [81] |
| M. puerensis | Uni-loculate, 300–600 × 230–380 μm diam. | 92 × 11 μm, bitunicate, elongate-clavate, 8-spored, long, furcate pedicellate | Brown to dark brown | Ellipsoid | 14 × 6 μm | Biseriate | 1 | Guttulate | [83] |
| M. saikhuensis | Uni-loculate, 411.7 × 460.5 μm diam. | 84.2 × 11.2 μm, bitunicate, elongate-clavate to short cylindrical, 8-spored, long pedicellate | Brown to blackish | Ellipsoid | 14.6 × 5.1 μm | Overlapping 1–2-seriate | 1 | Guttulate | [42] |
| M. thailandica | Uni-loculate, 380 × 340 μm diam. | 90 × 11 μm, bitunicate, elongate-clavate, slightly curved, 8-spored, long pedicellate | Brown to reddish-brown | Broadly fusiform to ellipsoid | 15 × 5.5 μm | Overlapping 1–2-seriate | 1 | Guttulate | [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, G.; Wanasinghe, D.N.; de Farias, A.R.G.; Hyde, K.D.; Yasanthika, E.; Xu, J.; Balasuriya, A.; Chethana, K.W.T.; Gui, H. Taxonomic Novelties of Woody Litter Fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion. Biology 2022, 11, 1660. https://doi.org/10.3390/biology11111660
Ren G, Wanasinghe DN, de Farias ARG, Hyde KD, Yasanthika E, Xu J, Balasuriya A, Chethana KWT, Gui H. Taxonomic Novelties of Woody Litter Fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion. Biology. 2022; 11(11):1660. https://doi.org/10.3390/biology11111660
Chicago/Turabian StyleRen, Guangcong, Dhanushka N. Wanasinghe, Antonio Roberto Gomes de Farias, Kevin D. Hyde, Erandi Yasanthika, Jianchu Xu, Abhaya Balasuriya, Kandawatte Wedaralalage Thilini Chethana, and Heng Gui. 2022. "Taxonomic Novelties of Woody Litter Fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion" Biology 11, no. 11: 1660. https://doi.org/10.3390/biology11111660
APA StyleRen, G., Wanasinghe, D. N., de Farias, A. R. G., Hyde, K. D., Yasanthika, E., Xu, J., Balasuriya, A., Chethana, K. W. T., & Gui, H. (2022). Taxonomic Novelties of Woody Litter Fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion. Biology, 11(11), 1660. https://doi.org/10.3390/biology11111660








