Simple Summary
Species of Sarcoscyphaceae are saprobic on branches, stumps, trunks, or twigs. The majority of members in this family are widespread in tropical areas, with only a fraction of the known species found in temperate areas. All species have typical disc- or cup-shaped fruiting bodies in a variety of colours ranging from white, grey, orange, red to brown. A high diversity of Sarcoscyphaceae has been reported in southwestern China and Thailand. In this study, we provide redescriptions of five known species and establish three new species in Sarcoscyphaceae from these regions based on morphology and phylogeny. We also propose an amendment for Phillipsia gelatinosa. Cookeina sinensis, a common species in China, is reported from Thailand for the first time.
Abstract
Sarcoscyphaceae (Pezizales) is distinguished by small to large, vividly-coloured sessile to stipitate apothecia, plurinucleate and pigmented paraphyses, operculate asci with thick walls, and plurinucleate, uniguttulate to multiguttulate ascospores with smooth walls or ornamentations. We collected more than 40 Sarcoscyphaceae specimens from dead twigs or wood. Based on morphology and phylogeny, these species belong to Cookeina, Nanoscypha, Phillipsia, Pithya, and Sarcoscypha. Among these, we introduce three new species–Nanoscypha aequispora, Pithya villosa, and Sarcoscypha longitudinalis. Phylogenetic analyses based on ITS, LSU, SSU, rpb2, and tef-1α gene regions indicate the relationships of these species within Sarcoscyphaceae. Meanwhile, we propose Ph. gelatinosa as a synonym of Ph. domingensis. One new record of C. sinensis is reported from Thailand.
1. Introduction
Sarcoscyphaceae comprises discomycetous fungi that occur abundantly in tropical areas but are also found in temperate regions [1,2,3]. Le Gal [4] improperly introduced Sarcoscyphaceae without supplying a Latin description. Eckblad [5] provided a legitimate description according to proper nomenclature standards. However, transfer of the type genus of Sarcosomataceae to Sarcoscyphaceae caused a long-term conceptual confusion between these two families [4,5,6,7]. It was not until clarification by Korf [8] that Sarcoscyphaceae had a clear concept. Sarcoscyphaceae has typical apothecia of Pezizomycetes (commonly referred to as cup fungi) and comprises one of the few families with no records of hypogeous taxa [2]. The ascal apical apparatus, one of the most distinctive characters of the whole group, was a point of confusion for decades. Chadefaud [9] and Le Gal [4] considered some ascal apical structures that were apparently hypothetical structures and/or the artifacts that occurred during the process of material fixation. They proposed that these structures represented a transition between an inoperculate apical ring and a true operculum. They called that kind of ascus ‘paraoperculate’ or ‘suboperculate’. They proposed that this ascus represented an intermediate stage towards the evolution of operculate forms. Eckblad [5,10], van Brummelen [11,12], Samuelson [13], and Samuelson et al. [14] showed that the apical apparatus type of Sarcoscypha (Fr.) Boud. is in fact operculate and by no means a transitional form between inoperculate ascus apical ring and true pezizalean operculum. This has subsequently been supported in numerous phylogenetic analyses. Sarcoscyphaceae is characterized by vividly-coloured, sessile to stipitate apothecia, pigmented paraphyses containing carotenoids, thick-walled asci equipped with narrow and thick lenticular operculum encircled by a subapical markedly thickened wall (suboperculum), and uniguttulate to multiguttulate ascospores with smooth walls or cyanophobic lateral striation/reticulation [2,6]. There are only a few reports of anamorphs in this family. Pfister [2] and Ekanayaka et al. [3] have provided the most recent summaries. There are 13 genera in the family, namely, Aurophora Rifai, Cookeina Kuntze, Geodina Denison, Kompsoscypha Pfister, Microstoma Bernstein, Nanoscypha Denison, Phillipsia Berk., Pithya Fuckel, Pseudopithyella Seaver, Rickiella Syd. & P. Syd. ex Rick, Sarcoscypha, Thindia Korf & Waraitch and Wynnea Berk. & M.A. Curtis, with a total of 83 estimated species in this family [15,16]. Within Sarcoscyphaceae, several species have been used as food and medicine. For example, Cookeina speciosa (Fr.) Dennis and C. tricholoma (Mont.) Kuntze are treated as edible fungi in Mexico, while there are also records of their use in treating ear infections in Cameroon [17,18,19]. The Scarlet elf cup, Sarcoscypha coccinea (Jacq.) Lambotte, has also been said to be edible [20].
The earliest phylogenetic study of Sarcoscyphaceae traces to Harrington et al. [21], who used the nuclear small subunit rRNA (SSU) gene region to reconstruct the phylogeny of Pezizales. Phylogenetic analysis of nine sequences involving nine genera revealed the monophyly of Sarcoscyphaceae and its placement in Pezizales [21]. Romero et al. [15] added molecular data for a known species of Rickiella and explored phylogenetic relationships within Sarcoscyphaceae based on nuclear large subunit rRNA (LSU) and SSU rDNA sequences. Angelini et al. [22] introduced a new species of Geodina based on morphology and phylogenetic analysis using LSU, but it was later shown to be a synonym of the type species by Pfister et al. [23]. Pfister et al. [23] proposed the new family Wynneaceae, which contained Geodina and Wynnea, thus separating these taxa from Sarcoscyphaceae based on phylogenetic analyses of four genetic markers and morphology. The two genera also exhibit morphological characteristics and habitat preferences that distinguish them from other genera of Sarcoscyphaceae. However, gene regions from different strains were combined to represent certain taxa, which were then used for phylogenetic inference. Hence, establishing a new family should be put on hold until genetic markers from the taxa of interest are available to avoid confusion [23]. Unfortunately, Aurophora and Thindia still lack molecular data, and therefore their phylogenetic placement currently remains unknown.
In this study, we collected 45 specimens related to Sarcoscyphaceae from southwestern China and Thailand. Through morphological examinations and phylogenetic inferences based on ITS, LSU, SSU, rpb2, and tef-1α, we introduce three new species within Nanoscypha, Pithya and Sarcoscypha. Cookeina collections separate into four distinct clades, which mainly belong to four species. Following re-examination of the type specimen of Ph. gelatinosa Ekanayaka, Q. Zhao & K.D. Hyde, we suggest that Ph. domingensis (Berk.) Berk. ex Denison takes precedence over Ph. gelatinosa.
2. Materials and Methods
2.1. Sample Collection, Morphological Examination, and Deposition
All specimens were collected from dead wood or twigs from southwestern China and southern Thailand. Fresh specimens were dried in a dehydrator at 25–30 °C shortly after collection to prevent decay. At the same time, a small amount of tissue material from each fresh sample was put into a labeled zipper sealed bag containing allochroic silica gel for moisture absorption to be used for molecular work. All materials were brought back to the laboratory for morphological and molecular studies. Four herbarium specimens labeled Phillipsia gelatinosa (MFLU 15-2360, MFLU 16-2956, MFLU 16-2992) and Phillipsia subpurpurea (MFLU 16-0612) were borrowed from the Herbarium of Mae Fah Luang University (MFLU) for further morphological investigation.
Documentations, descriptions, and measurements of macroscopic features, including colour, shape, and size of ascomata, were recorded before fresh specimens were processed. Morphological features indistinguishable to the naked eye were photographed using a Leica M125 C stereo microscope (Leica Microsystems GmbH, Wetzlar, Germany). Colour descriptions follow RAL Colour Chart [24]. Hand sections of ascomata were performed using a Motic SMZ-168 stereoscope (Speed Fair Co., Ltd., Hong Kong, China). Dried specimens were rehydrated in distilled water, or treated with 5% or 10% KOH solution, and stained with Cotton Blue (CB), Congo Red (CR), and Melzer’s reagent (MLZ) solutions. A Nikon Eclipse Ni compound microscope with a Nikon DS-Ri2 camera (Nikon Instruments Inc., Tokyo, Japan) were used for microscopic photography. The Tarosoft® Image Frame Work program v.0.9.7 (Tarosoft, Nontha Buri, Thailand) was used for measuring microscopical features. The measured number of ascospores (n), ascomata (m) and specimens (p) was denoted as [n/m/p]. Minimal (a–) and maximal (–b) values of length and width of ascospores, the 90% confidence interval (b–c) were provided as (a–)b–c(–d). Ascospore length/width ratio was referred to as Q, and Q values (average Q ± standard deviation) were provided to indicate the ascospore shape [25]. Photoplates were assembled using Adobe Photoshop CS6 (Adobe Systems, San Jose, CA, USA).
Specimens were deposited at the Herbarium of Mae Fah Luang University (MFLU) and Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (HKAS). Facesoffungi and Index Fungorum numbers were obtained as in Jayasiri et al. [26] and Index Fungorum [27]. The newly-generated data were added to the Greater Mekong Subregion webpage [28].
2.2. DNA Extraction, PCR Amplification and Sequencing
DNA was extracted from treated ascomata tissues (see Section 2.1) using the TreliefTM Plant Genomic DNA Extraction Kit (Tsingke Biotechnology Co., Ltd., Beijing, China). Polymerase chain reaction (PCR) was used to amplify the internal transcribed spacer (ITS), the large subunit rRNA (LSU), the small subunit rRNA (SSU), the second-largest subunit of RNA polymerase II (rpb2), and the translation elongation factor-1 alpha (tef-1α). Amplifications of ITS, LSU, SSU, rpb2 and tef-1α loci were performed using primer pairs ITS5/ITS4 [29], LR0R/LR5 [30], NS1/NS4 [29], fRPB2-5f/fRPB2-7cR [31], and 983F/2218R [32], respectively. The total volume of each PCR reaction mixture was 25 μL containing 9.5 μL sterile deionized water, 12.5 μL of 2× Power Taq PCR MasterMix, 1 μL of each primer (10 μM stock) and 1 μL DNA template. Amplifications were carried out using an Applied Biosystems 2720 thermocycler (Foster City, CA, USA). The cycling conditions of PCR amplification included initial denaturation at 94 °C for 5 min, followed by 35 cycles (ITS, LSU, SSU and tef-1α) or 40 cycles (rpb2) of: denaturation at 94 °C for 50 s, annealing at 56 °C for 50 s (ITS, LSU, SSU and tef-1α) or 55 °C for 2 min (rpb2), extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. The obtained PCR products were purified and sequenced by Tsingke Company, Beijing, P.R. China.
2.3. Phylogenetic Analysis
The raw sequences were assembled using DNASTAR Lasergene SeqMan Pro v.7.1.0 (44.1) (DNAStar Inc., Madison, WI, USA). Sequences spanning the spectrum of available diversity of Sarcoscyphaceae were downloaded from GenBank (Table 1). Individual sequence datasets of five gene regions were aligned using MAFFT v.7 available online [33]. All datasets were trimmed by TrimAl v.1.2 with the user-defined option (ITS: 0.9 value for gap threshold; LSU and SSU: 0.5 value for gap threshold) and gappyout option (rpb2 and tef-1α) [34]. Individual datasets were used to construct phylogenetic trees for each genetic marker to assess the topological congruence of the five datasets (data not shown). A dataset combining all five genetic markers was assembled into a matrix using Sequence Matrix v.1.8 [35]. AliView v.1.19-betalk was used to convert file format [36].
Maximum likelihood (ML) and Bayesian inference (BI) analyses were carried out on CIPRES Science Gateway v.3.3 platform [37] using RAxML-HPC2 v.8.2.12 [38] and MrBayes v.3.2.7a on XSEDE [39,40]. Maximum likelihood analysis was performed using the GTR + I + G substitution model with 1000 rapid bootstrap replicates. For BI analysis, GTR + I + G substitution was selected as best-fit model of evolution for each gene using MrModeltest v.2.3 [41] as performed by MrMTgui [42] based on the Akaike information criterion [43]. Markov Chain Monte Carlo Sampling (MCMC) was used to calculate posterior probabilities (PP) [39,44]. Two runs comprising of six simultaneous Markov Chains each were run for 635,000 generations for ITS tree and 9000 generations for combined gene tree, and trees were sampled every 100th generation [45]. The first 25% of the trees were discarded as burn-in and analysis was stopped when the standard deviation of split frequencies reached 0.01.
Phylogenetic trees were viewed in FigTree v.1.4.2 [46] and edited using Adobe Illustrator CS5 (Adobe Systems, San Jose, CA, USA).

Table 1.
Sequences used in this study.
Table 1.
Sequences used in this study.
| Species Name | Country | Voucher/Strain Number | ITS | LSU | SSU | rpb2 | tef-1α | References |
|---|---|---|---|---|---|---|---|---|
| Chorioactis geaster ♦ | USA | ZZ2 FH | AY307935 | AY307943 | – | DQ017608 | – | [47] |
| Cookeina colensoi | Mexico | CUP 62500 | AF394040 | – | – | – | – | [48] |
| Cookeina colensoi | Australia | DAR 63642 | AF394038 | – | – | – | – | [48] |
| Cookeina colensoi | India | FH 00432432 | AF394532 | – | – | – | – | [48] |
| Cookeina colensoi | New Zealand | PDD 55306 | AF394037 | – | – | – | – | [48] |
| Cookeina cremeirosea | American Samoa | UTC000275474 | KU306964 | – | – | – | – | [49] |
| Cookeina cremeirosea | American Samoa | UTC000275475 | KU306963 | – | – | – | – | [49] |
| Cookeina garethjonesii ♦ | China | HKAS90509 | KY094617 | MG871315 | – | MG980711 | MG980686 | [50] |
| Cookeina garethjonesii ♦ | China | HKAS90513 | KY094622 | MG871316 | – | MG980712 | MG980687 | [50] |
| Cookeina indica | China | C.ind119 | AF394029 | – | – | – | – | [48] |
| Cookeina indica ♦ | China | MFLU 16-0610 | KY094621 | MG871343 | – | MG980727 | – | [3,50] |
| Cookeina indica | Thailand | MFLU 20-0548 | MT941004 | – | – | – | – | [51] |
| Cookeina indica ♦ | China | HKAS 121171 | OK170053 | OK398387 | OK398409 | – | OK557973 | This study |
| Cookeina indica | China | HKAS 121172 | OK170054 | – | – | – | – | This study |
| Cookeina indica | China | HKAS 121173 | OK170055 | – | – | – | – | This study |
| Cookeina indica ♦ | China | HKAS 121174 | OK170058 | OK398386 | OK398408 | – | OK557972 | This study |
| Cookeina insititia | China | FH Wang sp 2 | AF394033 | – | – | – | – | [48] |
| Cookeina insititia | China | HMAS 70078 | AF394030 | – | – | – | – | [48] |
| Cookeina insititia | China | HMAS 71942 | AF394031 | – | – | – | – | [48] |
| Cookeina korfii | Philippines | CUP-SA-1797 | KT893782 | – | – | – | – | [52] |
| Cookeina korfii | Philippines | CUP-SA-2454 | KT893781 | – | – | – | – | [52] |
| Cookeina sinensis | China | HKAS 14679 | AF394028 | – | – | – | – | [52] |
| Cookeina sinensis | China | HMAS 70088 | AF394027 | – | – | – | – | [52] |
| Cookeina sinensis ♦ | China | HKAS 121175 | OK170056 | OK398385 | OK398407 | – | OK557971 | This study |
| Cookeina sinensis | China | HKAS 121176 | OK170057 | – | – | – | – | This study |
| Cookeina sinensis ♦ | China | HKAS 121177 | OK170059 | OK398384 | OK398406 | – | OK557970 | This study |
| Cookeina sinensis | China | HKAS 121178 | OK170060 | – | – | – | – | This study |
| Cookeina sinensis | China | HKAS 121179 | OK170067 | – | – | – | – | This study |
| Cookeina sinensis ♦ | Thailand | MFLU 21-0155 | OK413269 | OK398383 | OK398405 | – | OK557969 | This study |
| Cookeina speciosa | Malaysia | C TL 6035 | AF394018 | – | – | – | – | [48] |
| Cookeina speciosa | Venezuela | FH Iturriaga 1C-D4 | AF394011 | – | – | – | – | [48] |
| Cookeina speciosa | Venezuela | FH Iturriaga 1D-D6 | AF394016 | – | – | – | – | [48] |
| Cookeina speciosa | Venezuela | FH Iturriaga 1E-D5 | AF394003 | – | – | – | – | [48] |
| Cookeina speciosa | Venezuela | FH Iturriaga 2610 | AF394005 | – | – | – | – | [48] |
| Cookeina speciosa | Venezuela | FH Iturriaga 2D-D4 | AF394017 | – | – | – | – | [48] |
| Cookeina speciosa | Venezuela | FH Iturriaga 4A-D4 | AF394014 | – | – | – | – | [48] |
| Cookeina speciosa | Venezuela | FH Iturriaga 7A-D4 | AF394006 | – | – | – | – | [48] |
| Cookeina speciosa | Colombia | FH Muneton 296 | AF394013 | – | – | – | – | [48] |
| Cookeina speciosa | Thailand | FH Pfister 7131 | AF394009 | – | – | – | – | [48] |
| Cookeina speciosa | Thailand | FH Pfister 7143 | AF394010 | – | – | – | – | [48] |
| Cookeina speciosa ♦ | Thailand | MFLU 21-0156 | OK413270 | OK398390 | OK398412 | OK585150 | OK557976 | This study |
| Cookeina speciosa ♦ | Thailand | MFLU 21-0157 | OK413271 | OK398391 | OK398413 | OK585151 | OK557977 | This study |
| Cookeina speciosa ♦ | Thailand | MFLU 21-0158 | OK413272 | OK398392 | OK398414 | OK585152 | OK557978 | This study |
| Cookeina speciosa ♦ | Thailand | MFLU 21-0159 | OK413273 | OK398393 | OK398415 | OK585153 | OK557979 | This study |
| Cookeina speciosa | Thailand | MFLU 21-0160 | OK413274 | – | – | – | – | This study |
| Cookeina speciosa | Thailand | MFLU 21-0161 | OK413275 | – | – | – | – | This study |
| Cookeina speciosa | Thailand | MFLU 21-0162 | OK413276 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121180 | OK170044 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121181 | OK170045 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121182 | OK170047 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121183 | OK170048 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121184 | OK170049 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121185 | OK170050 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121186 | OK170064 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121187 | OK170065 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 121188 | OK170066 | – | – | – | – | This study |
| Cookeina speciosa | China | HKAS 124640 | OP364889 | – | – | – | – | This study |
| Cookeina sulcipes | Thailand | MFLU 15-2358 | KY094620 | – | – | – | – | [50] |
| Cookeina tricholoma | Thailand | FH Pfister 7170 | AF394020 | – | – | – | – | [48] |
| Cookeina tricholoma ♦ | China | HKAS87041 | KY094619 | MG871317 | – | – | MG980688 | [3,50] |
| Cookeina tricholoma ♦ | Thailand | MFLU 15-2359 | KY094618 | MG871318 | MG859240 | – | MG980689 | [3,50] |
| Cookeina tricholoma ♦ | Thailand | MFLU 21-0165 | OK413279 | OK398394 | OK398416 | – | – | This study |
| Cookeina tricholoma ♦ | Thailand | MFLU 21-0166 | OK413280 | OK398395 | OK398417 | – | OK557980 | This study |
| Cookeina tricholoma ♦ | Thailand | MFLU 21-0167 | OK413281 | OK398396 | OK398418 | – | OK557981 | This study |
| Cookeina tricholoma | Thailand | MFLU 21-0168 | OK413282 | OK398397 | OK398419 | – | – | This study |
| Cookeina tricholoma | Thailand | MFLU 21-0169 | OK413283 | OK398398 | OK398420 | – | – | This study |
| Cookeina tricholoma | Thailand | MFLU 21-0163 | OK413277 | – | – | – | – | This study |
| Cookeina tricholoma | Thailand | MFLU 21-0164 | OK413278 | – | – | – | – | This study |
| Cookeina tricholoma | China | HKAS 121189 | OK170043 | – | – | – | – | This study |
| Cookeina tricholoma | China | HKAS 121190 | OK170046 | – | – | – | – | This study |
| Cookeina tricholoma | China | HKAS 121191 | OK170061 | – | – | – | – | This study |
| Cookeina venezuelae | Puerto Rico | FH00432502 | AF394041 | – | – | – | – | [48] |
| Cookeina venezuelae | Venezuela | FH Iturriaga 6065 | AF394044 | – | – | – | – | [48] |
| Cookeina venezuelae | Venezuela | FH Iturriaga 6066 | AF394043 | – | – | – | – | [48] |
| Cookeina venezuelae | Guadeloupe | FH00432503 | AF394042 | – | – | – | – | [48] |
| Geodina guanacastensis ♦ | Bahamas | FH | MN096939 | MN096940 | MN096941 | MN103424 | MN090946 | [23] |
| Geodina guanacastensis | Costa Rica | CUP CA84 | MN096938 | – | – | – | – | [23] |
| Geodina guanacastensis | Dominican Republic | JBSD 127408 | MG597289 | – | – | – | – | [22,23] |
| Geodina guanacastensis | Dominican Republic | JBSD 127409 | MG597290 | – | – | – | – | [22,23] |
| Kompsoscypha chudei ♦ | China | HKAS 107663 | MT907443 | MT907444 | – | – | – | [51] |
| Kompsoscypha phyllogena ♦ | Puerto Rico | DHP 10-690 | – | JQ260810 | JQ260820 | MN103430 | – | [15] |
| Microstoma floccosum | Mexico | FH K. Griffith (Micro45) | AF394046 | – | – | – | – | [48] |
| Microstoma floccosum | Mexico | FH K. Griffith (Micro46) | AF394045 | – | – | – | – | [48] |
| Nanoscypha striatispora | China | HMAS 61133 | U66016 | – | – | – | – | [21] |
| Nanoscypha tetraspora ♦ | Puerto Rico | FH 00464570 | AF117352 | DQ220374 | AF006314 | – | – | [21,53,54] |
| Nanoscypha aequispora ♦ | Thailand | MFLU 21-0170 | OK413284 | OK398399 | OK398421 | OK585154 | – | This study |
| Nanoscypha aequispora ♦ | Thailand | MFLU 21-0171 | OK413285 | OK398400 | OK398422 | OK585155 | OK557982 | This study |
| Neournula pouchetii | USA | MO 205345 | KT968605 | – | – | – | – | [55] |
| Phillipsia carnicolor ♦ | Thailand | DHP-7126 (FH) | AF117353 | JQ260811 | JQ260821 | MN103426 | MN090948 | [53] |
| Phillipsia carnicolor | Thailand | MFLU 18-0713 | MH602282 | – | – | – | – | [56] |
| Phillipsia chinensis | China | HMAS 76094 | AY254710 | – | – | – | – | [57] |
| Phillipsia crispata | Ecuador | T. Læssøe AAU-44801 | AF117354 | – | – | – | – | [53] |
| Phillipsia crispata ♦ | Ecuador | T. Læssøe AAU-44895a | AF117355 | AY945845 | – | DQ017599 | – | [47] |
| Phillipsia domingensis | USA | CO-1864 (NO) | AF117363 | – | – | – | – | [53] |
| Phillipsia domingensis | Costa Rica | CO-2032 (NO) | AF117361 | – | – | – | – | [53] |
| Phillipsia domingensis ♦ | Thailand | DHP 7169 (FH) | AF117373 | JQ260817 | JQ260827 | – | – | [53] |
| Phillipsia domingensis | Dominican Republic | DR-321 (CFMR) | AF117370 | – | – | – | – | [53] |
| Phillipsia domingensis | Costa Rica | Franco-M 1270 (NY) | AF117358 | – | – | – | – | [53] |
| Phillipsia domingensis | Puerto Rico | PR-1583 (FH) | AF117365 | – | – | – | – | [47] |
| Phillipsia domingensis ♦ | China | HKAS 121192 | OK170062 | OK398388 | OK398410 | OK585148 | OK557974 | This study |
| Phillipsia domingensis ♦ | China | HKAS 121193 | OK170063 | OK398389 | OK398411 | OK585149 | OK557975 | This study |
| Phillipsia gelatinosa ♦ | Thailand | MFLU 15-2360 | KY498595 | KY498589 | – | MG980728 | – | [58] |
| Phillipsia gelatinosa | Thailand | MFLU 16-2956 | KY498593 | – | – | – | – | [58] |
| Phillipsia hydei | Thailand | MFLU 18-0714 | MH602283 | – | – | – | – | [56] |
| Phillipsia hydei | Thailand | MFLU 18-1329 | MH602284 | – | – | – | – | [56] |
| Phillipsia lutea ♦ | French Guiana | NY-4113 (NY) | AF117374 | JQ260816 | JQ260826 | [53] | ||
| Phillipsia olivacea | Costa Rica | Franco-M 1360 (NY) | AF117375 | – | – | – | – | [53] |
| Phillipsia olivacea ♦ | Venezuela | Halling-5456 (NY) | AF117376 | JQ260814 | JQ260824 | – | – | [53] |
| Phillipsia olivacea | Ecuador | T. Læssøe AAU-43162 (C) | AF117378 | – | – | – | – | [47] |
| Phillipsia subpurpurea | China | MFLU 16-0612 | KY498596 | – | – | – | – | [58] |
| Pithya cupressina ♦ | USA | mh 208 | U66009 | JQ260818 | AF006316 | – | – | [23,59] |
| Pithya sp. | China | DWS8m3 | KJ188703 | [60] | ||||
| Pithya sp. | USA | T5N32c | AY465469 | – | – | – | – | [61] |
| Pithya vulgaris | – | RK 90.01 | U66008 | – | – | – | – | [59] |
| Pithya villosa ♦ | China | HKAS 104653 | OK170069 | OK398401 | OK398423 | OK585156 | – | This study |
| Pithya villosa ♦ | China | HKAS 121194 | OK170068 | OK398402 | OK398424 | – | – | This study |
| Plectania nannfeldtii ♦ | USA | FH 00822732 | – | AY945853 | – | DQ017592 | KC109214 | [47,62] |
| Pseudopithyella minuscula ♦ | USA | FH 00465568 | – | AY945849 | AF006317 | DQ017600 | FJ238387 | [47] |
| Rickiella edulis ♦ | Argentina | BAFC 51697 | JQ260808 | JQ260809 | JQ260819 | MN103425 | MN090947 | [15] |
| Sarcoscypha austriaca | Norway | CUP 62771 | U66010 | – | – | – | – | [59] |
| Sarcoscypha austriaca | USA | CUP 63162 | U66011 | – | – | – | – | [59] |
| Sarcoscypha coccinea ♦ | – | AFTOL-ID 50 | DQ491486 | AY544647 | – | DQ497612 | – | [63] |
| Sarcoscypha coccinea ♦ | France | AFTOL-ID 930 | – | FJ176859 | FJ176805 | FJ713615 | – | [63] |
| Sarcoscypha coccinea | USA | CUP 62113 | U66013 | – | – | – | – | [59] |
| Sarcoscypha coccinea | USA | CUP 63160 | U66015 | – | – | – | – | [59] |
| Sarcoscypha dudleyi | USA | CUP 62775 | U66018 | – | – | – | – | [59] |
| Sarcoscypha dudleyi | China | HMJAU36044 | KU234218 | – | – | – | – | [64] |
| Sarcoscypha dudleyi | – | mh 192 | U66019 | – | – | – | – | [59] |
| Sarcoscypha emarginata | Luxembourg | CUP 62723 | U66020 | – | – | – | – | [59] |
| Sarcoscypha emarginata | – | HB2861 | U66021 | – | – | – | – | [59] |
| Sarcoscypha hosoyae | – | TRL 456 | U66031 | – | – | – | – | [59] |
| Sarcoscypha humberiana | China | TNM F28630 | KT716833 | – | – | – | – | [65] |
| Sarcoscypha humberiana | China | CUP 63489 | U66028 | – | – | – | – | [59] |
| Sarcoscypha javensis | China | HMAS 61198 | U66026 | – | – | – | – | [59] |
| Sarcoscypha knixoniana | – | TRL 1006 | U66030 | – | – | – | – | [59] |
| Sarcoscypha korfiana | – | mh 705 | AF026308 | – | – | – | – | [21] |
| Sarcoscypha longitudinalis ♦ | China | HKAS 121195 | OK170051 | OK398403 | OK398425 | OK585157 | – | This study |
| Sarcoscypha longitudinalis ♦ | China | HKAS 121196 | OK170052 | OK398404 | OK398426 | – | – | This study |
| Sarcoscypha macaronesica | Canary Islands | CUP-MM 2628 | U66022 | – | – | – | – | [59] |
| Sarcoscypha macaronesica | – | TFC-MIC 6460 | U66023 | – | – | – | – | [59] |
| Sarcpscypha mesocyatha | China | TNM F3688 | KT936558 | – | – | – | – | [65] |
| Sarcpscypha mesocyatha | China | TNM F5134 | KT936559 | – | – | – | – | [65] |
| Sarcoscypha mesocyatha | USA | CUP 62699 | U66029 | – | – | – | – | [59] |
| Sarcoscypha minuta | China | TNM F28831 | KT716834 | – | – | – | – | [65] |
| Sarcoscypha occidentalis | USA | CUP 62777 | U66024 | – | – | – | – | [59] |
| Sarcoscypha occidentalis | USA | CUP 63484 | U66025 | – | – | – | – | [59] |
| Sarcoscypha sp. | China | HMAS 61202 | U66027 | – | – | – | – | [59] |
| Sarcoscypha tatakensis | China | TNM F0754 | KT716835 | – | – | – | – | [65] |
| Sarcoscypha tatakensis | China | TNM F0993 | KT716836 | – | – | – | – | [65] |
| Sarcoscypha vassiljevae ♦ | Chian | HKAS 89817 | MG871302 | MG871337 | – | MG980724 | MG980700 | [3] |
| Sarcoscypha vassiljevae | China | HMAS 61210 | U66017 | – | – | – | – | [59] |
| Urnula craterium ♦ | USA | DHP 04-511 | – | AY945851 | – | DQ017595 | KC109216 | [47,62] |
| Wynnea americana ♦ | USA | FH 00445979 | MK599141 | AY945848 | MK592785 | MN103435 | MN103417 | [23,66] |
| Wynnea americana | USA | HKAS 75484 | MG871308 | – | – | – | – | [3] |
| Wynnea gigantea | China | HKAS 101385 | MG871307 | – | – | – | – | [3] |
| Wynnea macrospora ♦ | China | FH 00445975 | MK335784 | MK335803 | MK335793 | MN103432 | MN103419 | [23,66] |
| Wynnea macrospora ♦ | – | CUP 2684 | – | MK335804 | MK335795 | – | MN103420 | [23,66] |
| Wynnea sparassoides ♦ | USA | FH 00445986 | – | EU360917 | MK335796 | MN103431 | MN103418 | [23,47] |
1 Names in red indicate newly-described species in this study. Names in bold indicate type collections. Names in blue indicate newly-sequenced collections. 2 Species names marked by “♦” refer to taxa used in the combined tree. 3 Abbreviations: AAU: Herbarium of Aarhus University, Denmark; BAFC: Facultad de Ciencias Exactas y Naturales, Argentina; CFMR: Center for Forest Mycology Research, USDA Forest Service, USA; CUP: The Cornell Plant Pathology Herbarium, New York, USA; DAR: NSW Plant Pathology & Mycology Herbarium, Australia; FH: Farlow Herbarium, Harvard University Herbaria, Cambridge, Massachusetts, USA; HKAS: Herbarium of Cryptogams of Kunming Institute of Botany, Chinese Academy of Sciences, China; HMAS: Herbarium Mycologicum Academiae Sinicae, Beijing, China; HMJAU: Herbarium of Mycology of Jilin Agricultural University, Changchun, China; JBSD: Herbarium of the Santo Domingo National Botanical Garden, Dominican Republic; MFLU: Mae Fah Luang University Herbarium, Chiang Rai, Thailand; MO: Missouri Botanical Garden Herbarium, Missouri, USA; NY: New York Botanical Garden, USA; PDD: New Zealand Fungarium, New Zealand; TFC: La Laguna University Herbarium, Spain; TNM: Department of Botany, National Museum of Natural Science, Taiwan, China; UTC: Intermountain Herbarium, Utah, USA.
3. Results
3.1. Phylogenetic Analysis
The ITS phylogenetic tree was inferred (Figure 1) using 151 taxa and 476 sites including sequences from Chorioactis geaster (Peck) Kupfer (ZZ2 FH) and Neournula pouchetii (Berthet & Riousset) Paden (MO 205345) as outgroup. The best sorting RAxML tree had a final likelihood value of −9665.136236. Sequences of the ITS region are available for nearly all taxa of Sarcoscyphaceae for which molecular data exist. Genera are grouped in distinct monophyletic clades except for Phillipsia, Nanoscypha and Rickiella, which are paraphyletic. In the ITS tree, taxa are grouped in eight main clades, which mainly represent genera. Our new collections were placed in four clades, namely clade 1, clade 3, clade 4, and clade 8. Within clade 1 (Sarcoscypha clade), the newly-described Sarcoscypha species, S. longitudinalis (represented by two collections), formed an individual branch as sister to S. vassiljevae Raitv., but this relationship was not strongly supported (52BS/0.87PP). Clade 2 comprised a single species, K. chudei (Pat. ex Le Gal) Pfister, and was sister to clade 3 (100BS/1.00PP). Within clade 3 (Pithya clade), our new Pithya species (represented by two collections) branched sister to Pi. cupressina (Batsch) Fuckel (mh 208) and to one unknown Pithya species (DWS8m3) with strong statistical support (94BS/1.00PP). Clade 4 comprised Phillipsia, Rickiella, and Nanoscypha. The new species, N. aequispora, was sister to N. tetraspora (Seaver) Denison (DHP PR-61) with moderate support (65BS/0.97PP). Two new collections of Ph. domingensis (HKAS 121192 and HKAS 121193) were placed within the Ph. domingensis complex. Clades 5, 6, and 7 comprised Geodina (one species), Wynnea (four species) and Microstoma (one species). Clade 8 (Cookeina clade) contained the rest of the new collections. Seventeen new collections were distributed in two of the five subclades within the C. speciosa complex, ten grouped with C. tricholoma, six clustered with C. sinensis Zheng Wang, while four grouped with C. indica Pfister & R. Kaushal.
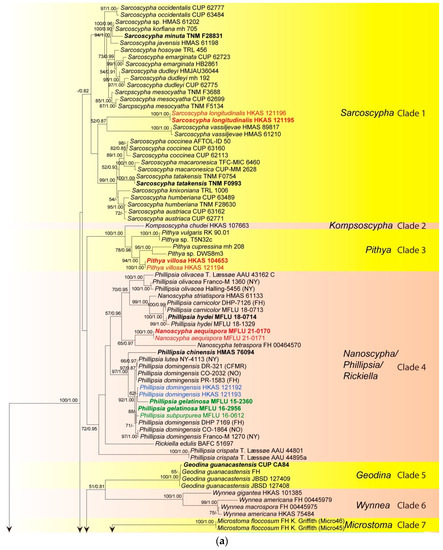
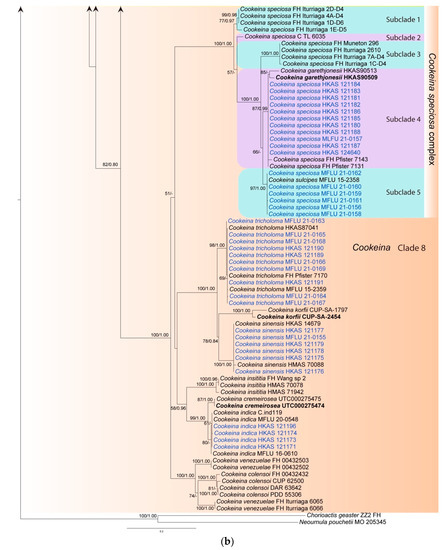
Figure 1.
Maximum likelihood tree of ITS sequence data inferred from 151 taxa and 476 sites under the GTR (general time reversible) + G + I model of nucleotide substitution. Bootstrap support values for maximum likelihood (BS) and Bayesian posterior probabilities (PP) greater than 50% and 0.80 are indicated above or below the nodes in this order. Names in red indicate newly-described species and names in blue stand for newly-sequenced collections. Names in green indicate correction to Phillipsia domingensis (a). Chorioactis geaster (ZZ2 FH) and Neournula pouchetii (MO 205345) are used as the outgroup taxa (b).
Initially, a phylogenetic tree was inferred using a combined dataset of ITS, LSU, SSU, rpb2, and tef-1α data containing all available strains in the preliminary analysis (data not shown). However, the large amount of missing data (for many strains only ITS was available, while for others only LSU) confounded the results as indicated by unstable placement of taxa and very low statistical support in deep and shallow nodes. Therefore, a smaller representative dataset was assembled containing 49 taxa, for which a minimum of three genes was available for each taxon (Figure 2). The alignment comprised 4366 total characters (ITS: 1–482 bp; LSU: 483–1386 bp; SSU: 1387–2432 bp; rpb2: 2433–3485 bp; tef-1α: 3486–4366 bp). The best-sorting RAxML tree had a final likelihood value of −30228.032462. In the combined data tree, taxa grouped in seven main clades. The Microstoma clade (clade 7 in the ITS tree) is missing due to lack of data. Kompsoscypha, Pithya, Wynnea, Geodina and Cookeina were monophyletic. Species of Phillipsia, Nanoscypha and Rickiella interspersed within a clade, while Pseudopithyella grouped within Sarcoscypha. The phylogenetic placements of our new species and collections were almost identical to that of the ITS tree, but statistical supports were much higher in the combined data tree (Figure 1 and Figure 2). Pseudopithyella did not separate from Sarcoscypha but grouped as sister to S. coccinea in the combined genes tree with nearly maximum statistical support (99BS/1.00PP). It is unknown if this relationship is recovered in the ITS tree as the sequence is not available. However, the relationship was recovered in the single gene trees that contain sufficient taxon sampling for Sarcoscypha, i.e., rpb2 and SSU.
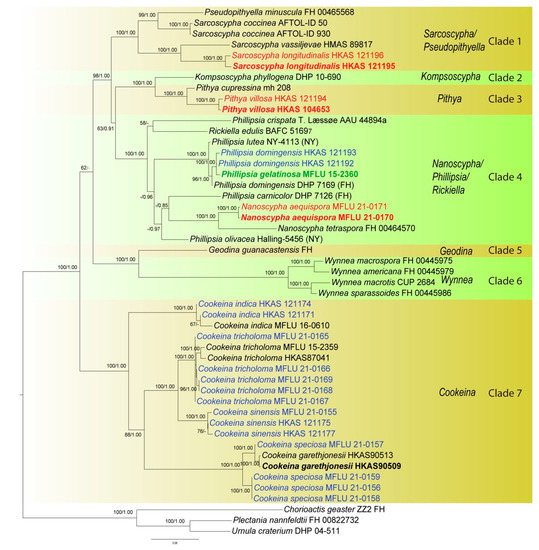
Figure 2.
Phylogenetic tree of combined ITS, LSU, SSU, rpb2, and tef-1α sequence data inferred from 49 taxa and 4366 sites under the GTR + G + I model of nucleotide substitution. Numerical values at the nodes indicate maximum likelihood bootstrap support (BS) and posterior probabilities (PP). Values of BS greater than 50% and PP over 0.80 are indicated above or below the nodes in this order. Names in red indicate newly-described species and names in blue stand for newly-sequenced collections. Names in green indicate correction to Phillipsia domingensis. Tree is artificially rooted to Chorioactis geaster (ZZ2 FH), Plectania nannfeldtii (FH 00822732) and Urnula criterium (DHP 04-511).
3.2. Taxonomy

Figure 3.
Cookeina indica. (A–I) Fresh apothecia [(A,B) HKAS 121171. (C) HKAS 121172. (D–H) HKAS 121173. (I) HKAS 121174.] (J) Margin (HKAS 121173). (K) Receptacle surface of an apothecium (HKAS 121173). (L) A concentric sulcus (HKAS 121173). Scale bars (A,B,F) = 5 cm; (C–E) = 10 cm; (G–I) = 3 cm; (J,K) = 1000 μm; (L) = 500 μm.
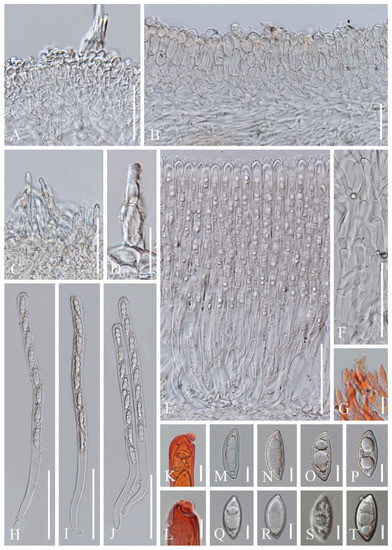
Figure 4.
Cookeina indica (HKAS 121173). (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C) Hyphoid hairs at the margin. (D) A moniliform hair-like processes. (E) Hymenium. (F) Paraphyses. (G) Apical part of the paraphyses in CR. (H–J) Asci and ascospores. (K,L) Apices of asci in CR. (M–T) Ascospores ornamented with longitudinal striate ridges. Scale bars (A–C,F) = 50 μm; (D) = 20 μm; (E,H–J) = 100 μm; (G,K–T) = 10 μm.
Index Fungorum number: IF 302844; Facesoffungi number: FoF 02671
Saprobic on dead wood. Teleomorph: Apothecia up to 7 cm high, 1–4 cm broad, solitary or scattered, deeply cupulate, rarely ear-shaped, fleshy, with short to long stipe. Stipe up to 4 cm long, up to 3 mm broad, central to eccentrical, terete, or sharply reduced to a basal, sulcate attachment, solid, usually white to yellowish when fresh, yellow when dry, nearly smooth. Receptacle cup-shaped, surface light ivory (RAL 1015), yellowish to orange when fresh, nearly smooth, with one concentric sulcus, margin broadly entire, or rarely deeply split on one side. Disc deeply concave, mostly concolorous with the receptacle surface, or somewhat darker in colour. Stipal ecto-excipulum 70–140 µm broad, composed of hyaline to yellowish textura globulosa-angularis, cells 14–19 × 11–14 μm, some outermost globose cells form irregularly loose aggregates to a pruinose-like surface, rarely with hyphoid hairs which are 3–6 µm wide, hyaline, septate, slightly tapering towards a rounded apex, usually fasciculate. Stipal medulla composed of hyaline to subhyaline textura intricata, hyphae 3–6.5 μm broad. Ectal excipulum 80–130 µm thick, mainly divided to two sub-layers delimited by outer and inner cells: outer layer composed of hyaline to yellowish textura globulosa to textura prismatica, terminal cells globose, 13–19 μm diameter, with a pruinose-like surface, prismatic cells 23–31 × 11–16 μm, hyphoid hairs forming abundant fascicles at margin, composed of 4–6 µm wide subhyaline, septate hyphae, sometimes with individual monilioid hairlike processes having 2–3 ellipsoid cells; inner layer composed of hyaline textura angularis to textura epidermoidea, angular cells 14–18 × 11–14 μm, and elongated cells 16–23 × 6–9 μm. Medullary excipulum 200–260 μm broad, composed of hyaline textura intricata, hyphae 4–7 μm broad. Hymenium 340–400 μm thick, subhyaline, paraphyses exceeding the asci slightly when dehydrated. Paraphyses 4–6 μm broad in the middle part, cylindric, septate, constricted at septa, normally anastomosing to form a network, branched in the apical part, apical cell tapered. Asci 330–360 × 15–19 μm, 8-spored, eccentrically operculate, cylindrical, with obtuse apices and attenuated basal part. Ascospores [20/1/1, in H2O] (26.3–)29.8–36(–38) × (10.8–)11.5–13.4(–14.2) μm (Q = 2.08–3.13, Q = 2.64 ± 0.28), fusiform to subreniform, inequilateral, subpapillate at the poles, uniseriate, often biguttulate to 3-guttulate, some multiguttulate, ornamented with parallel ridges arranged longitudinally. Anamorph: not seen.
Material examined: CHINA, YUNNAN, XISHUANGBANNA: Menghai, on unidentified dead branch and trunk under broadleaved forest, 19 August 2019, Ming Zeng, ZM255 (HKAS 121171), ZM257 (HKAS 121172); Mengyang, on unidentified dead branch under broadleaved forest, 21 August 2019, Ming Zeng, ZM280 (HKAS 121173); Jinghong, on unidentified dead twigs under broadleaved forest, 23 August 2019, Ming Zeng, ZM306 (HKAS 121174).
Notes: This species is distinguished by nearly smooth apothecia with a concentric sulcus close to margin, paraphyses with tapering ends, fusiform and inequilateral ascospores with subpapillate ends, and longitudinal striae on surface of ascospores [67]. Cookeina indica was first discovered in India and has since then been reported in southwestern China [6,50,67,68,69]. It was also recently discovered in Thailand [51]. This species has a nearly smooth surface when observed with the naked eye, compared to most other species that have easily visible hairs [67], while the furfuraceous receptacle surface can be seen with a magnifying hand lens. Cookeina cremeirosea Kropp has smooth apothecia, and it showed a close phylogenetic relationship with C. indica (Figure 1) [49]. Cookeina cremeirosea is distinct in having pinkish apothecia and smooth-walled ascospores [49]. Compared with the type description in the protologue [68], our collections have large apothecia (up to 7 cm high vs up to 3.5 cm high) and a long stipe (up to 4 cm long vs up to 2.2 cm long). In addition, ascospores (11.5–13.4 μm vs 10–11.5 μm) also vary significantly in width.
- 2.
 Figure 5. Cookeina sinensis. (A–G) Fresh apothecia [(A) HKAS 121177. (B) HKAS 121176. (C) HKAS 121178. (D,E) HKAS 121179. (F) HKAS 121175. (G) MFLU 21-0155.] (H) Dry apothecium (HKAS 121175). (I) Stipe (HKAS 121175). (J) Compound hairs (HKAS 121177). (K) Margin (HKAS 121178). (L,M) Receptacle surface of an apothecium (HKAS 121175). Scale bars (A,E,F) = 5 cm; (B–D,G) = 3 cm; (H,J) = 2000 μm; (I,K) = 1000 μm; (L,M) = 500 μm.
Figure 5. Cookeina sinensis. (A–G) Fresh apothecia [(A) HKAS 121177. (B) HKAS 121176. (C) HKAS 121178. (D,E) HKAS 121179. (F) HKAS 121175. (G) MFLU 21-0155.] (H) Dry apothecium (HKAS 121175). (I) Stipe (HKAS 121175). (J) Compound hairs (HKAS 121177). (K) Margin (HKAS 121178). (L,M) Receptacle surface of an apothecium (HKAS 121175). Scale bars (A,E,F) = 5 cm; (B–D,G) = 3 cm; (H,J) = 2000 μm; (I,K) = 1000 μm; (L,M) = 500 μm.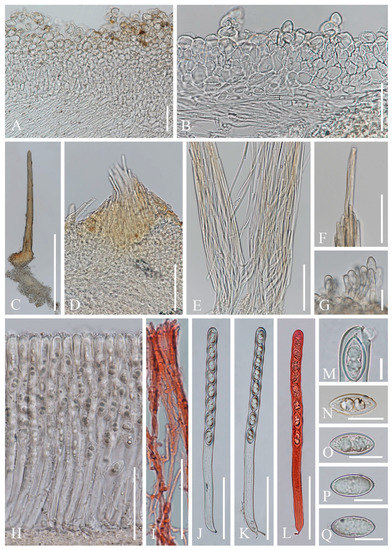 Figure 6. Cookeina sinensis (HKAS 121179). (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C) Compound hair. (D) Base of the compound hair arising from medullary excipulum. (E) Loose compound hairs. (F) Tips of compound hairs. (G) Hyphoid hairs. (H) Hymenium. (I) Paraphyses in CR. (J–L) Asci and ascospores (L) Ascus and ascospores in CR. (M) Apex of an ascus. (N–Q) Ascospores. Scale bars (A,B,F,I) = 50 μm; (C) = 500 μm; (D,E,H,J–L) = 100 μm; (G,N–Q) = 20 μm; (M) = 10 μm.
Figure 6. Cookeina sinensis (HKAS 121179). (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C) Compound hair. (D) Base of the compound hair arising from medullary excipulum. (E) Loose compound hairs. (F) Tips of compound hairs. (G) Hyphoid hairs. (H) Hymenium. (I) Paraphyses in CR. (J–L) Asci and ascospores (L) Ascus and ascospores in CR. (M) Apex of an ascus. (N–Q) Ascospores. Scale bars (A,B,F,I) = 50 μm; (C) = 500 μm; (D,E,H,J–L) = 100 μm; (G,N–Q) = 20 μm; (M) = 10 μm.
Index Fungorum number: IF 437499; Facesoffungi number: FoF 02674
Saprobic on dead wood. Teleomorph: Apothecia up to 4 cm high, up to 5 cm broad, scattered, cupulate, fleshy, with conspicuous stipe. Stipe up to 2 cm long, up to 8 mm broad, central, terete, solid, white to yellowish when fresh, yellow when dry, furfuraceous or tomentose, with long compound hairs as on the receptacle. Receptacle cup-shaped, surface yellowish to orange, or rarely oyster white (RAL 1013) when fresh, with long compound hairs, tomentose, margin entire, leveled or inrolled. Disc deeply concave, yellow to orange, or pinkish when fresh, mostly concolorous with the receptacle surface. Stipal ecto-excipulum 90–260 µm broad, composed of hyaline to yellowish textura globulosa-angularis, cells 16–19 × 12–15 μm, some outer globose cells irregularly loosely aggregated forming a pruinose-like surface. Stipal medulla composed of hyaline to subhyaline textura intricata, hyphae 4–6.5 μm broad. Ectal excipulum 50–110 µm thick, composed of hyaline textura globulosa-angularis, cells 15–20 × 12–15 μm, with hyphoid hairs forming abundant fascicles, composed of 4–7 µm wide, subhyaline, septate, broad hyphae, sometimes with monilioid processes having 1–2 globose cells forming a pruinose-like surface. Medullary excipulum 140–220 μm thick, composed of hyaline textura intricata, hyphae interwoven, 3–6 μm wide. Compound hairs up to 5 mm long, up to 150 µm diameter at base of fascicle, fasciculate, yellow to brown, arising from the medullary excipulum, composed of parallel, yellowish, septate, thick-walled individial hairs, 4–5 µm diameter, with a rounded end. Hymenium 280–310 μm thick, subhyaline. Paraphyses 3–4 μm broad at the middle, filiform, septate, anastomosing to form a network in middle parts, branched, with a rounded end. Asci 290–300 × 15–18 μm, 8-spored, eccentrically operculate, cylindrical, with obtuse apices and slightly attenuated basal part. Ascospores [20/1/1, in H2O] (25–)26.3–29.6(–30.4) × (12.2–)12.4–13.7(–14.2) μm (Q = 1.91–2.38, Q = 2.15 ± 0.14), fusiform, pointed at ends, uniseriate, equilateral, biguttulate, rarely apiculi-like solidifications present at one pole, smooth. Anamorph: not seen.
Material examined: CHINA, YUNNAN, XISHUANGBANNA: Dadugang, on unidentified dead branch under broadleaved forest, 22 August 2019, Ming Zeng, ZM287 (HKAS 121175); Jinghong, on unidentified dead branch under broadleaved forest, 23 August 2019, Ming Zeng, ZM297 (HKAS 121176); Manshan, on unidentified dead branch under broadleaved forest, 25 August 2019, Ming Zeng, ZM320 (HKAS 121177), ZM322 (HKAS 121178); Mengla, on unidentified dead branch under broadleaved forest, 27 August 2019, Ming Zeng, ZM351 (HKAS 121179); THAILAND: Chiangmai, Mushroom Research Center (MRC), on unidentified dead branch under broadleaved forest, 16 August 2020, Deping Wei, ZM370 (MFLU 21-0155).
Notes: This species has long compound hairs covering receptacle, arising from medullary excipulum. The ascospores are fusiform, smooth, with apiculi-like structure [6,67]. Cookeina sinensis is similar to C. tricholoma and C. korfii Iturr., F. Xu & Pfister, as all three share the conspicuous long compound hairs. Compared with C. korfii and C. sinensis, C. korfii has smaller ascospores (22–25 × 9–11.5 μm) [52]. The major difference between C. sinensis and C. tricholoma is that the ascospores of C. tricholoma have longitudinal striae, whereas C. sinensis has smooth ascospores [67]. Cookeina sinensis had only been reported from China [6,67,70], until Patil et al. [71] described new collections from India. Comparing Indian specimens with ours, the Indian specimens have larger ascospores (30–40 × 15.6 μm) [71]. The ascospores of our specimens have similar size with type specimens (25–28 × 12–12.5 μm) described by Wang [70], which have smaller apothecia (2.5 cm high) than ours. In this study, we report on a new record collected from Thailand (MFLU 21-0155).
- 3.
- Cookeina speciosa (Fr.) Dennis, Mycotaxon 51: 239 (1994); Figure 7, Figure 8, Figure 9 and Figure 10
 Figure 7. Cookeina speciosa. (A–H) Fresh apothecia [(A,B). HKAS 121188. (C,D) HKAS 121186. (E,F) HKAS 121187. (G,H) MFLU 21-0157.] (I,K) Dry apothecia [(I) HKAS 121186. (K) HKAS 121188.] (J) Margin (HKAS 121186). (L,M) Concentric ridges [(L) MFLU 21-0157. (M) HKAS 121182.] (N) Compound hairs (MFLU 21-0157). (P) Triangular-shaped hairs (MFLU 21-0157). (O) Stipe (MFLU 21-0157). (Q) Receptacle surface of an apothecium (MFLU 21-0157). Scale bars (A,B,E,F) = 5 cm; (C,D,G,H) = 3 cm; (I–L) = 2000 μm; (M–O) = 1000 μm; (P,Q) = 500 μm.
Figure 7. Cookeina speciosa. (A–H) Fresh apothecia [(A,B). HKAS 121188. (C,D) HKAS 121186. (E,F) HKAS 121187. (G,H) MFLU 21-0157.] (I,K) Dry apothecia [(I) HKAS 121186. (K) HKAS 121188.] (J) Margin (HKAS 121186). (L,M) Concentric ridges [(L) MFLU 21-0157. (M) HKAS 121182.] (N) Compound hairs (MFLU 21-0157). (P) Triangular-shaped hairs (MFLU 21-0157). (O) Stipe (MFLU 21-0157). (Q) Receptacle surface of an apothecium (MFLU 21-0157). Scale bars (A,B,E,F) = 5 cm; (C,D,G,H) = 3 cm; (I–L) = 2000 μm; (M–O) = 1000 μm; (P,Q) = 500 μm.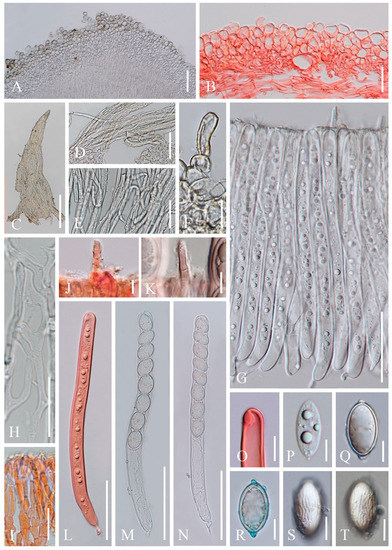 Figure 8. Cookeina speciosa (MFLU 21-0157). (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C–E) Triangular-shaped compound hairs. (F) Monilioid process. (G) Hymenium including setae from an immature apothecium. (H) Paraphyses. (I) Apices of paraphyses in CR. (J,K) Hymenial setae in CR. (L) Immature ascus and ascospores in CR from an immature apothecium. (M,N) Asci and ascospores from a mature apothecium. (O) Apex of ascus in CR. (P) Immature ascospore. (Q–T) Mature ascospores. [(R) Ascospore in CB.] Scale bars (A,D,G,N–L) = 100 μm; (B,F) = 50 μm. (C) = 200 μm; (E,H,I,O–T) = 20 μm.
Figure 8. Cookeina speciosa (MFLU 21-0157). (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C–E) Triangular-shaped compound hairs. (F) Monilioid process. (G) Hymenium including setae from an immature apothecium. (H) Paraphyses. (I) Apices of paraphyses in CR. (J,K) Hymenial setae in CR. (L) Immature ascus and ascospores in CR from an immature apothecium. (M,N) Asci and ascospores from a mature apothecium. (O) Apex of ascus in CR. (P) Immature ascospore. (Q–T) Mature ascospores. [(R) Ascospore in CB.] Scale bars (A,D,G,N–L) = 100 μm; (B,F) = 50 μm. (C) = 200 μm; (E,H,I,O–T) = 20 μm. Figure 9. Cookeina speciosa. (A–J) Fresh apothecia [(A–C) MFLU 21-0158. (D,E) MFLU 21-0162. (F,G) MFLU 21-0156. (H) MFLU 21-0159. (I) MFLU 21-0161. (J) MFLU 21-0160. (K) Dry apothecium (MFLU 21-0160). (M) Receptacle surface of an apothecium (MFLU 21-0160). (L) Hairs are arranged in concentric ridges (MFLU 21-0158). (N) Stipe (MFLU 21-0158). Scale bars (A) = 5 cm; (B,D–G,I,J) = 2 cm; (C,H) = 3 cm; (K,L,N) = 2000 μm; (M) = 300 μm.
Figure 9. Cookeina speciosa. (A–J) Fresh apothecia [(A–C) MFLU 21-0158. (D,E) MFLU 21-0162. (F,G) MFLU 21-0156. (H) MFLU 21-0159. (I) MFLU 21-0161. (J) MFLU 21-0160. (K) Dry apothecium (MFLU 21-0160). (M) Receptacle surface of an apothecium (MFLU 21-0160). (L) Hairs are arranged in concentric ridges (MFLU 21-0158). (N) Stipe (MFLU 21-0158). Scale bars (A) = 5 cm; (B,D–G,I,J) = 2 cm; (C,H) = 3 cm; (K,L,N) = 2000 μm; (M) = 300 μm.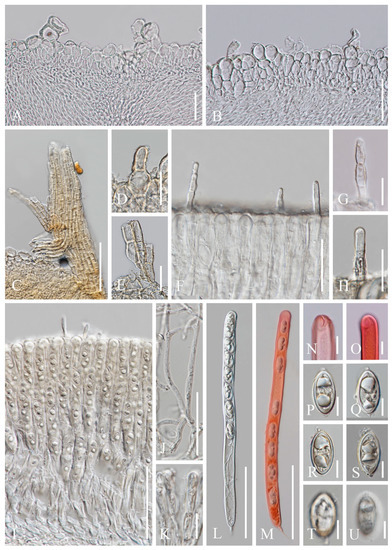 Figure 10. Cookeina speciosa (MFLU 21-0162). (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C) Triangular-shaped compound hair. (D,E) Hyphoid hairs. (F–H) Hymenial setae. (I) Hymenium. (J) Paraphyses. (K) Apices of the paraphyses. (L,M) Asci and ascospores [(M). Ascus and ascospores in CR.] (N,O) Apical part of asci in CR. (P–U) Ascospores. Scale bars (A,B,F) = 50 μm; (C) = 100 μm; (D,E,J) = 30 μm; (G,H) = 20 μm; (K,N–U) = 10 μm.
Figure 10. Cookeina speciosa (MFLU 21-0162). (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C) Triangular-shaped compound hair. (D,E) Hyphoid hairs. (F–H) Hymenial setae. (I) Hymenium. (J) Paraphyses. (K) Apices of the paraphyses. (L,M) Asci and ascospores [(M). Ascus and ascospores in CR.] (N,O) Apical part of asci in CR. (P–U) Ascospores. Scale bars (A,B,F) = 50 μm; (C) = 100 μm; (D,E,J) = 30 μm; (G,H) = 20 μm; (K,N–U) = 10 μm.
Index Fungorum number: IF 362244; Facesoffungi number: FoF 02675
Saprobic on dead wood. Teleomorph: Apothecia up to 4 cm high, up to 3 cm broad, rarely 6 cm broad, solitary or scattered, cupulate, funnel-shaped, usually with a long stipe. Stipe up to 2.5 cm long, up to 3 mm broad, central, terete, solid, white to yellowish when fresh, yellow when dry, furfuraceous or tomentose. Receptacle cup-shaped, surface yellowish to orange, pale rosy, light ivory (RAL 1015), or pinkish to deep coral, rarely white when fresh, with up to 5 distinct concentric ridges composed of compound hairs, margin entire, or slightly inrolled, with long hairs. Disc deeply concave, yellowish to orange or pale rosy to pink when fresh, mostly concolorous with the receptacle surface, or darker in colour. Stipal ecto-excipulum 50–140 µm broad, composed of hyaline to yellowish textura globulosa-angularis, cells 18–27 × 13–24 μm, some outer globose cells irregularly loosely aggregated forming a pruinose-like surface. Stipal medulla composed of hyaline to subhyaline textura intricata, hyphae 3–7 μm broad. Ectal excipulum 60–160 µm thick, composed of hyaline textura globulosa-angularis, cells 17–28 × 12–22 μm, monilioid processes usually composed of 1–2 rounded cells, sometimes with a sub-clavate terminal cell, forming a pruinose-like surface, compound hair bundles up to 800 µm long and up to 260 µm broad at the base, yellow, composed of 5–8 µm wide yellowish septate hyphae, which are fused to triangular-shaped fascicles. Medullary excipulum 60–250 μm thick, composed of hyaline textura intricata, hyphae interwoven, 2–6 μm wide. Hymenium 280–340 μm thick, subhyaline, with hymenial setae, 4–8 μm wide, exceeding the hymenium by 45 μm at most, 1–2 septate, with a rounded apex. Paraphyses 2–4 μm wide in the middle part, filiform, septate, mostly constricted at septa, anastomosing to form a network, branched, with a rounded apex. Asci 290–320 × 17–21 μm (subclade 4) or 266–300 × 14–17 μm (subclde 5), 8-spored, eccentrically operculate, cylindrical, with obtuse apices and narrow hyphoid base. Ascospores [20/1/1, in H2O] (24.8–)25.3–27.5(–28.1) × (13.1–)13.7–15(–15.5) μm (Q = 1.69–1.96, Q = 1.84 ± 0.07) (subclade 4) or (19.6–)22.2–24.7(–25) × (9.2–)10.3–11.9(–12.3) μm (Q = 1.93–2.38, Q = 2.12 ± 0.11) (subclade 5), ellipsoid, rounded at ends, uniseriate, equilateral, biguttulate when mature, immature multiguttulate, projecting apiculi present at one or both poles, perispore ornamented with anastomosing cyanophobic longitudinal striae. Anamorph: not seen.
Material examined: CHINA, YUNNAN, XISHUANGBANNA: on unidentified dead branch under broadleaved forest, 5 June 2018, Ming Zeng, Zeng003 (HKAS 121180), Zeng004 (HKAS 121181); ibid., 6 June 2018, Ming Zeng, Zeng006 (HKAS 121182), Zeng007 (HKAS 121183), Zeng008 (HKAS 121184); ibid., 12 June 2018, Ming Zeng, Zeng023 (HKAS 121185); ibid., 27 August 2019, Ming Zeng, ZM356 (HKAS 124640); Mengla, on unidentified dead branch under broadleaved forest, 27 August 2019, Ming Zeng, ZM332 (HKAS 121186), ZM336 (HKAS 121187); ibid., on unidentified trunk under broadleaved forest, ZM339 (HKAS 121188); THAILAND: Phangnga, Kapong, on unidentified dead branch under broadleaved forest, 29 August 2017, Chuangen Lin, 4-1 (MFLU 21-0157); ibid., Thap Phut, on unidentified dead branch under broadleaved forest, 30 August 2017, Chuangen Lin, 5-1 (MFLU 21-0158), 5-3 (MFLU 21-0159), 5-5 (MFLU 21-0160); ibid., 1 September 2017, Chuangen Lin, 11-1 (MFLU 21-0161); Prachuap Khiri Khan, Bang Saphan, on unidentified dead branch under broadleaved forest, 28 August 2017, Chuangen Lin, 1-1 (MFLU 21-0156); Ranong, on unidentified dead branch under broadleaved forest, 7 October 2017, Ming Zeng, ST09 (MFLU 21-0162).
Notes: This species is widely distributed in tropical areas [67]. The main feature of this species is the variable colour of the apothecia with up to five distinct concentric ridges close to margin, and compound hairs arranged on these ridges. In addition, hymenium has hymenial setae, broadly ellipsoid ascospores with obviously projecting apiculi, and longitudinal striae on surface of the ascospores, anastomosed in some parts [48,67]. This species was introduced by Dennis [72] based on Peziza speciosa Fr.; meanwhile, C. sulcipes was synonymized as a later epithet [72]. Although phylogenetic studies based on ITS show some genetic variation associated with colour differences within the C. speciosa clade [48], Iturriaga and Pfister [67] still consider this as a complex. In this study, Cookeina garethjonesii represented by two strains formed an independent branch, which is nested in the C. speciosa clade in subclade 4 (Figure 1). The phylogeny herein contains a comprehensive sampling of C. speciosa sequences. Cookeina garethjonesii was established as a separate species from C. speciosa due to the lack of hymenial setae and smooth-walled ascospores [50]. In the illustration of the holotype of C. garethjonesii provided in the original study, distinct hymenial setae are clearly visible [50] (Figure 2 in the original paper). In their phylogeny, the species was phylogenetically distinct; however, only a limited number of C. speciosa sequences were used in the dataset. Based on these contradictions, we suggest that the type specimen should be re-examined in the future. In our trees, six collections were grouped with a sequence designated as C. sulcipes (MFLU 15-2358), forming a distinct clade within C. speciosa complex (subclade 5, Figure 1). This well-defined clade has members that produce almost uniformly pink to coral apothecia (Figure 9). However, the coral-coloured apothecia are not unique to subclade 5; rather, collections with coral apothecia are spread across C. speciosa complex (e.g., FH Iturriaga 1E-D5, FH Iturriaga 4A-D4 and FH Iturriaga 7A-D4 from the subclades 1 and 3, Figure 1) [48] (this study). In our described collections, C. speciosa (MFLU 21-0157) (Figure 8) from subclade 4 has broader asci (17–21 μm vs. 14–17 μm) and larger ascospores (25.3–27.5 × 13.7–15.0 μm vs. 22.2–24.7 × 10.3–11.9 μm) when compared to C. speciosa (MFLU 21-0162) (Figure 10) from subclade 5. Nevertheless, C. sulcipes (MFLU 15-2358) described by Ekanayaka et al. [50] has an indistinguishable size of asci (280–380 × 15–22 μm) and ascospore (21–30 × 11–18 μm) compared to both our described collections. Hence, it seems that the sizes of asci and ascospores cannot distinguish these specimens at the species level. Within the C. speciosa complex, our collections placed in two subclades, both of which have high statistical support (subclade 4: 87BS/0.99PP, subclade 5: 97BS/1.00PP). More sampling and type studies are needed to resolve C. speciosa complex in the future.
- 4.
- Cookeina tricholoma (Mont.) Kuntze, Revis. gen. pl. (Leipzig) 2: 849 (1891); Figure 11 and Figure 12
 Figure 11. Cookeina tricholoma. (A–I) Fresh apothecia [(A) HKAS 121191. (B,C) MFLU 21-0165. (D) MFLU 21-0163. (E,F) MFLU 21-0168. (G–H) MFLU 21-0167. (I) MFLU 21-0166.] (J,K,M) Dry apothecia [(J) MFLU 21-0168. (K) MFLU 21-0165. M HKAS 121191.] (L) Margin (HKAS 121191). (N) Compound hairs (MFLU 21-0164). (O) Receptacle surface of an apothecium (MFLU 21-0163). Scale bars (A,E,G) = 3 cm; (B,C,H,I) = 1 cm; (D,F) = 2 cm; (J,K,M) = 5000 μm; (L) = 1000 μm; (N) = 500 μm; (O) = 200 μm.
Figure 11. Cookeina tricholoma. (A–I) Fresh apothecia [(A) HKAS 121191. (B,C) MFLU 21-0165. (D) MFLU 21-0163. (E,F) MFLU 21-0168. (G–H) MFLU 21-0167. (I) MFLU 21-0166.] (J,K,M) Dry apothecia [(J) MFLU 21-0168. (K) MFLU 21-0165. M HKAS 121191.] (L) Margin (HKAS 121191). (N) Compound hairs (MFLU 21-0164). (O) Receptacle surface of an apothecium (MFLU 21-0163). Scale bars (A,E,G) = 3 cm; (B,C,H,I) = 1 cm; (D,F) = 2 cm; (J,K,M) = 5000 μm; (L) = 1000 μm; (N) = 500 μm; (O) = 200 μm.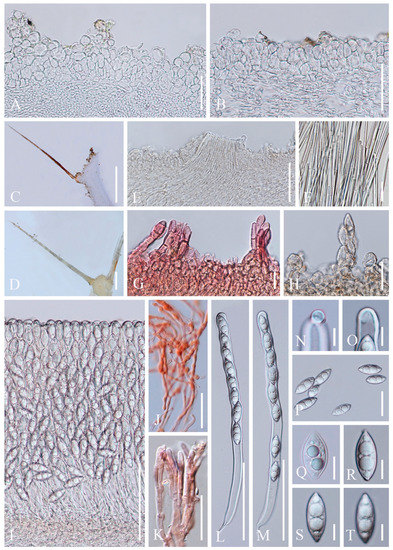 Figure 12. Cookeina tricholoma. (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C,D) Compound hairs. (E) Broken compound hair from medullary excipulum. (F) Loose compound hair. (G) Hyphoid hairs in CR. (H) Monilioid processes. (I) Hymenium. (J) Paraphyses in CR. (K) Apices of paraphyses in CR. (L,M) Asci and ascospores. (N,O) Apices of the asci. (P) Ascospores. (Q) Ascospore when young. (R–T) Ascospores ornamented by fine longitudinal striate ridges when mature. Scale bars (A,B,F) = 50 μm; (C,D) = 500 μm; (E,I,L,M) = 100 μm; (G,H,J,K) = 20 μm; (N–T) = 10 μm.
Figure 12. Cookeina tricholoma. (A) Vertical section of stipe ectal excipulum. (B) Vertical section of receptacle ectal excipulum. (C,D) Compound hairs. (E) Broken compound hair from medullary excipulum. (F) Loose compound hair. (G) Hyphoid hairs in CR. (H) Monilioid processes. (I) Hymenium. (J) Paraphyses in CR. (K) Apices of paraphyses in CR. (L,M) Asci and ascospores. (N,O) Apices of the asci. (P) Ascospores. (Q) Ascospore when young. (R–T) Ascospores ornamented by fine longitudinal striate ridges when mature. Scale bars (A,B,F) = 50 μm; (C,D) = 500 μm; (E,I,L,M) = 100 μm; (G,H,J,K) = 20 μm; (N–T) = 10 μm.
Index Fungorum number: IF 121551; Facesoffungi number: FoF 02677
Saprobic on dead wood. Teleomorph: Apothecia up to 3 cm high, up to 2 cm broad, solitary or scattered, cupulate, fleshy, with short to long stipe. Stipe up to 1.5 cm long, up to 2 mm broad, central, rarely eccentrical, terete, solid, yellowish or pinkish when fresh, yellow when dry, furfuraceous or tomentose, with long compound hairs as the receptacle. Receptacle cup-shaped, surface yellowish to orange, or pink when fresh, with long compound hairs, tomentose, margin entire, rarely deeply split on one side, inrolled when dry. Disc deeply concave, yellow to orange, or pinkish when fresh, mostly concolorous with the receptacle surface. Stipal ecto-excipulum 85–175 µm broad, composed of hyaline to yellowish textura globulosa-angularis, cells 12–17 × 11–15 μm, some outer globose cells irregularly loosely aggregated forming a pruinose-like surface. Stipal medulla composed of hyaline textura intricata, hyphae 5–7.5 μm broad. Ectal excipulum 50–80 µm thick, composed of hyaline textura globulosa-angularis, cells 13–19 × 11–15 μm, with two types of hairs mixed throughout the ectal excipular surface: fasciculate hyphoid hairs, composed of 5–9 µm wide subhyaline to yellowish, septate, broad hyphae; additionally with monilioid processes composed of 2 or more globose cells forming a pruinose-like surface. Medullary excipulum 100–160 μm broad, composed of hyaline textura intricata, hyphae interwoven, 3–5 μm wide. Compound hairs up to 5 mm long, up to 200 µm diameter at base of fascicle, fasciculate, brown, arising from the medullary excipulum, composed of 3–6 µm diameter, parallel, yellowish, septate, thick-walled hyphae, with a rounded end. Hymenium 280–350 μm thick, subhyaline. Paraphyses 1.5–3 μm broad in the middle part, filiform, septate, anastomosing to form a network, branched, with a rounded end. Asci 308–342 × 13–21 μm, 8-spored, eccentrically operculate, cylindrical, with obtuse apices and narrow bases. Ascospores [20/1/1, in H2O] (25.8–)28.0–32.9(–33.9) × (11–)11.6–12.8(–13.8) μm (Q = 2.33–2.68, Q = 2.50 ± 0.17), fusiform, pointed at ends, uniseriate, equilateral, biguttulate to multiguttulate, projecting apiculi sometime present at one pole, ornamentation with fine longitudinal striate ridges. Anamorph: not seen.
Material examined: CHINA, YUNNAN, XISHUANGBANNA: on unidentified dead branch under broadleaved forest, 5 June 2018, Ming Zeng, Zeng002 (HKAS 121189), Zeng005 (HKAS 121190); ibid., 26 August 2019, Ming Zeng, ZM328 (HKAS 121191). THAILAND: Chiang Rai, Song Khwae, on unidentified dead branch under broadleaved forest, 13 August 2017, Ming Zeng, N003 (MFLU 21-0163); Ranong, on unidentified dead branch under broadleaved forest, 7 October 2017, Ming Zeng, ST10 (MFLU 21-0164); Phangnga, Thap Phut, on unidentified dead branch under broadleaved forest, 30 August 2017, Chuangen Lin, 6-1 (MFLU 21-0165); ibid., 1 September 2017, Chuangen Lin, 11-2 (MFLU 21-0168), 11-3 (MFLU 21-0169); ibid., on unidentified dead trunk under broadleaved forest, Chuangen Lin, 6-2 (MFLU 21-0166), 6-3 (MFLU 21-0167).
Notes: This species is distinguished by yellow to orange, or coral apothecia with long compound hairs extending from medullary excipulum, fusiform ascospores with longitudinal striae [67]. It is similar to C. korfii and C. sinensis, both of which have smooth-walled ascospores [52]. This species is widely distributed in tropical areas and is also a common species in southwest China and Thailand [50,67].
- 5.
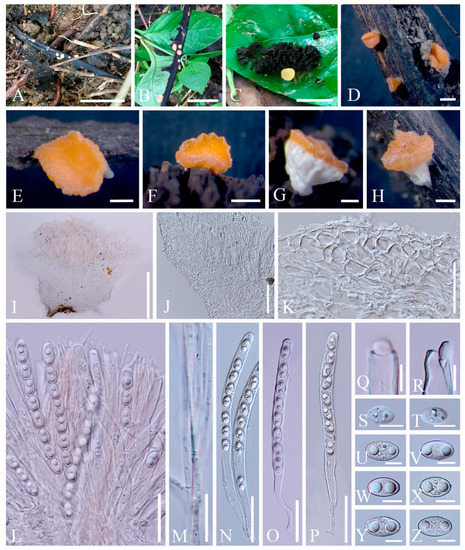 Figure 13. Nanoscypha aequispora (MFLU 21-0170, holotype). (A–H) Apothecia. (I) Vertical median section of apothecia. (J) Vertical median section of flank. (K) Vertical section of receptacle ectal excipulum. (L) Hymenium. (M) Paraphyses. (N–P) Asci and ascospores. (Q–R) Asci apices. (S,T) Immature ascospores. (U–Z) Ascospores when mature. Scale bars (A) = 2 cm; (B) = 1 cm; (C) = 5 mm; (D,F) = 1000 μm; (E,G–I) = 500 μm; (J) = 100 μm; (K,L,N–P) = 50 μm; (M) =20 μm; (Q–Z) =10 μm.
Figure 13. Nanoscypha aequispora (MFLU 21-0170, holotype). (A–H) Apothecia. (I) Vertical median section of apothecia. (J) Vertical median section of flank. (K) Vertical section of receptacle ectal excipulum. (L) Hymenium. (M) Paraphyses. (N–P) Asci and ascospores. (Q–R) Asci apices. (S,T) Immature ascospores. (U–Z) Ascospores when mature. Scale bars (A) = 2 cm; (B) = 1 cm; (C) = 5 mm; (D,F) = 1000 μm; (E,G–I) = 500 μm; (J) = 100 μm; (K,L,N–P) = 50 μm; (M) =20 μm; (Q–Z) =10 μm.
Index Fungorum number: IF 559928; Facesoffungi number: FoF 10410
Etymology: The specific epithet refers to equilateral ascospores.
Holotype: MFLU 21-0170
Diagnosis: This species is diagnosed by turbinate to shallowly cupulate apothecia with broadly whitish stipe, yellowish to orange disc, undulate margin, filiform paraphyses with yellowish granules, ellipsoid and equilateral ascospores with biguttulate.
Saprobic on dead wood and plant fruit. Teleomorph: Apothecia 0.5–1 mm high, 1–2 mm broad, scattered, shallowly cupulate when fresh, turbinate when dry, broadly stipitate, glabrous. Stipe 400–1500 µm long, 500–2000 µm broad, central, funnel-shaped, wrinkled on surface, solid, whitish to cream, rarely yellowish. Receptacle shallowly cupulate, surface yellowish to orange, margin undulate. Disc shallowly concave to discoid, concolorous with the receptacle surface. Stipal ecto-excipulum 62–166 µm, composed of hyaline textura angularis, cells 14–20 × 8–12 μm, mixed with textura prismatica, cells 18–25 × 9–12 µm, with some porrectoid cells arranged on surface, cells 4–6 µm wide. Stipal medulla composed of hyaline textura intricata, hyphae 4–6 μm broad. Ectal excipulum 56–94 µm thick, composed of hyaline to yellowish textura globulosa-angularis, cells 14–22 × 9–13 μm, mixed with textura prismatica, cells 19–25 × 9–13 µm, with some porrectoid cells arranged on surface, cells 4–6 µm wide. Medullary excipulum 76–192 µm thick, composed of hyaline textura intricata, hyphae 4–6 µm wide. Hymenium 280–310 µm thick, yellowish, paraphyses slightly exceeding the asci when dehydrated. Paraphyses 2–3 µm wide in the middle part, filiform, branched, septate, filled with yellowish granules. Asci 235–284 × 10–13 µm, 8-spored, subterminally operculate, apices obtuse, cylindrical, becoming narrow towards the base. Ascospores [20/1/1, in H2O] (14.8–)16.2–18.6(–19.2) × (12.4–)10.3–11.6(–9.9) µm (Q = 1.41–1.78, Q = 1.59 ± 0.14), ellipsoid, with round or slightly truncated ends, equilateral, rarely slightly inequilateral with one side flat, uniseriate, multiguttulate when immature, biguttulate when mature, smooth-walled. Anamorph: not seen.
Material examined: THAILAND: Ranong, on unidentified twigs and plant fruit, 7 October 2017, Ming Zeng, ST07 (MFLU 21-0170, holotype); ibid., 8 October 2017, Ming Zeng, ST11 (MFLU 21-0171, paratype).
Notes: Species in the genus Nanoscypha are small cup-fungi, normally less than 1 cm in diameter. Apothecia vary from discoid to cupulate, to turbinate or funnel-shaped, sessile to stipitate, with yellow, orange to red in colour. Cylindrical asci 3-, 4-, 6-, or 8-spored, and having tapered bases. Ascospores are ellipsoid to reniform, mostly inequilateral, rarely equilateral, mostly with two oil drops [6,73]. There are currently eight species assigned in this genus according to Species Fungorum [27], while the placement of N. striatispora (W.Y. Zhuang) F.A. Harr. [6,74] is still under debate (see discussion). Although our new species and N. tetraspora clustered together, it is difficult to confirm the correct position of new species in the phylogenetic tree due to the lack of available data for other species. Meanwhile, Nanoscypha strains are nested inside a clade together with Phillipsia and Rickiella in the ITS and multigene analyses. Through morphological comparison (Table 2), most species share inequilateral ascospores, except for N. macrospora Denison [6,74,75] and two vaguely-described species, N. bella (Berk. & M.A. Curtis) Pfister and N. euspora (Rick) S.E. Carp. Of these three, N. bella has larger-sized apothecia and ascospores [76], while N. euspora differs in its uniguttulate ascospores [77]. Nanoscypha macrospora is having equilateral ascospores, rarely inequilateral, same as our new species. The main difference of the N. macrospora is that the asci contain only 3 or 4 ascospores. In addition, the ascospores are elongated ellipsoid in shape [73]. Moreover, N. orissaensis C.M. Das & D.C. Pant is a rarely-recorded species, which lacks type material [75]. Thus, we proposed the new species Nanoscypha aequispora here based on morphology.

Table 2.
Morphological characteristics of Nanoscypha species.
- 6.
- Phillipsia domingensis (Berk.) Berk. ex Denison, Mycologia 61(2): 293 (1969); Figure 14
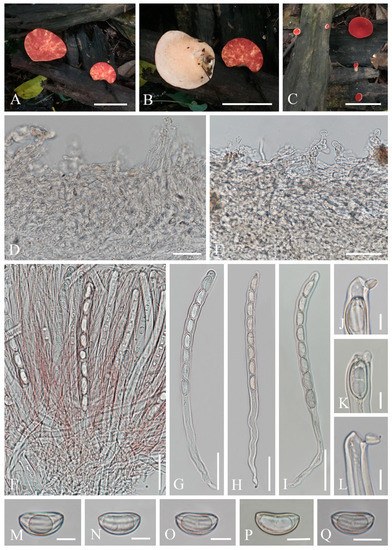 Figure 14. Phillipsia domingensis. (A–C) Fresh apothecia [(A,B) HKAS 121193. (C) HKAS 121192.] (D) Vertical section of stipal ecto-excipulum. (E) Vertical section of ectal excipulum. (F) Hymenium. (G–I) Asci and ascospores. (J–L) Asci apices. (M–Q) Ascospores. Scale bars (A–C) = 3 cm; (D,E) = 30 μm; (F–I) = 50 μm; (J–Q) = 10 μm.
Figure 14. Phillipsia domingensis. (A–C) Fresh apothecia [(A,B) HKAS 121193. (C) HKAS 121192.] (D) Vertical section of stipal ecto-excipulum. (E) Vertical section of ectal excipulum. (F) Hymenium. (G–I) Asci and ascospores. (J–L) Asci apices. (M–Q) Ascospores. Scale bars (A–C) = 3 cm; (D,E) = 30 μm; (F–I) = 50 μm; (J–Q) = 10 μm.
=Phillipsia gelatinosa Ekanayaka, Q. Zhao & K.D. Hyde, Phytotaxa 316(2): 142 (2017)
Index Fungorum number: IF 122362; Facesoffungi number: FoF 02868
Saprobic on dead wood. Teleomorph: Apothecia 7–11 mm high, up to 4 cm broad, scattered, leathery, shallowly cupulate to discoid, substipitate to shortly stipitate. Stipe up to 3 mm long, 5 mm broad, central to eccentrical, obconical, solid, bright beige red (RAL 3012), or reddish, or creamy yellowish, pubescent. Receptacle shallowly cupulate, surface concolorous with the stipe, pubescent, margin entire. Disc shallowly cup-shaped to discoid, pearl pink (RAL 3033) to orient red (RAL 3031), or with yellow patches. Stipal ecto-excipulum 60–90 µm thick, composed of yellowish to subhyaline textura porrecta to textura epidermoidea, hyphae 4–7 µm wide, with some outermost hyphae irregularly loosely aggregated to form pubescent surface. Stipal medulla composed of hyaline textura intricata, hyphae 3.5–5 μm broad. Ectal excipulum 60–100 µm thick, composed of yellowish to subhyaline textura porrecta to textura epidermoidea, hyphae 4–7 µm wide, with some loose hyphae in the outermost part. Medullary excipulum 280–650 µm thick, composed of hyaline textura intricata, hyphae 3–4 µm wide. Hymenium 300–350 µm thick, pink to red, paraphyses slightly exceeding the asci when dehydrated. Paraphyses 1–2 µm broad in the middle part, filiform, with reddish contents, septate, branched. Asci 285–346 × 11–14 µm, 8-spored, eccentrically operculate, cylindrical, apices obtuse, becoming narrow towards the base. Ascospores [20/1/1, in H2O] (19.8–)20.5–24.2(–27.7) × (10.4–)10.6–12(–13.2) µm (Q = 1.82–2.17, Q = 1.98 ± 0.08), subreniform or reniform with pointed or subpapillate ends, uniseriate, inequilateral, uniguttulate or biguttulate, ornamented with several longitudinal striations. Anamorph: not seen.
Material examined: CHINA, YUNNAN, XISHUANGBANNA: on unidentified dead branch under broadleaved forest, 26 August 2019, Ming Zeng, ZM329 (HKAS 121192), ZM330 (HKAS 121193). THAILAND, CHIANGMAI: on rotten wood, 18 June 2016, H. Maoqiang, LE2016112 (MFLU 16-2992); Mushroom Research Center (MRC), on rotten wood, 12 December 2015, S.C. Karunarathna, HD044 (MFLU15-2360, holotype? of Ph. gelatinosa); ibid., 12 June 2016, A.H. Ekanayaka, HD057 (MFLU 16-2956, holotype? of Ph. gelatinosa).
Notes: This is a common species in the subtropical and tropical areas. This species has larger-sized apothecia, red to purple-red hymenium, subreniform or reniform ascospores with several conspicuous longitudinal striations [57,80]. Hansen et al. [53] suggested the Ph. domingensis complex based on ITS genetic marker, owing to the species Ph. lutea Denison and some Ph. domingensis collections featuring yellow apothecia, which nested in the typically red Ph. domingensis. Ekanayaka et al. [58] introduced Ph. gelatinosa based on three collections and provided a description of Ph. subpurpurea Berk. & Broome along with molecular data, which didn’t exist before. Even these two species are placed in the Ph. domingensis complex, Ekanayaka et al. [58] identified morphological differences to distinguish them from Ph. domingensis. Phillipsia gelatinosa is distinguished by its orange contents of paraphyses, larger-sized asci and ascospores (Table 3), and presence of a gelatinous sheath surrounding ascospores [58]. Phillipsia subpurpurea (MFLU16-0612) differs in that has smooth ascospores or with faint striations, a thick gelatinous sheath surrounding ascospores. We re-examined all specimens named Ph. gelatinosa and Ph. subpurpurea (MFLU16-0612) (Figure 15, Figure 16 and Figure 17). Three of them show morphological features almost consistent with Ph. domingensis, which have distinct reddish contents in paraphyses, subreniform or reniform ascospores with conspicuous longitudinal striations. Although conspicuous reddish contents and striate ascospores are difficult to observe in Ph. gelatinosa (MFLU 15-2360) due to the quality of the specimen, we still can find sporadic reddish contents in paraphyses and faint striations on surface of ascospores. For all specimens, sheath-like structure was only present when rehydrating in 10% KOH, not in water. By comparing the sizes of asci and ascospores of the re-examined specimens (Table 3), there are no significant differences among these specimens, while these sizes are largely different from Ekanayaka et al.’s descriptions. Most importantly, the “two holotypes” (MFLU 16-2956 and MFLU 15-2360) were assigned in the protologue of Ph. gelatinosa [58]. According to the International Code of Nomenclature for algae, fungi, and plants, specifically the Art. 8.1. in Shenzhen Code [81], Ph. gelatinosa is an invalid name. Based on these, Ph. gelatinosa should be a nomen invalidum and a synonym of Ph. domingensis. Meanwhile, the specimen (MFLU16-0612) was incorrectly assigned to Ph. subpurpurea which should be corrected to Ph. domingensis. Phillipsia domingensis is a complex lacking type sequences with almost solely ITS region known for most collections. Thus, for efficient differentiation at the species level, sequencing of additional DNA regions and more data on phenotypic characters of as many collections possible are needed.

Table 3.
Sizes comparison for asci and ascospores of Phillipsia gelatinosa and Phillipsia subpurpurea (MFLU16-0612).

Figure 15.
Herbarium materials of Phillipsia gelatinosa. (A–C) MFLU 16-2956 (? holotype). (D–F) MFLU 15-2360 (? holotype). (G–I) MFLU 16-2992. Scale bars (B,C,E,F,H,I) = 2 cm.
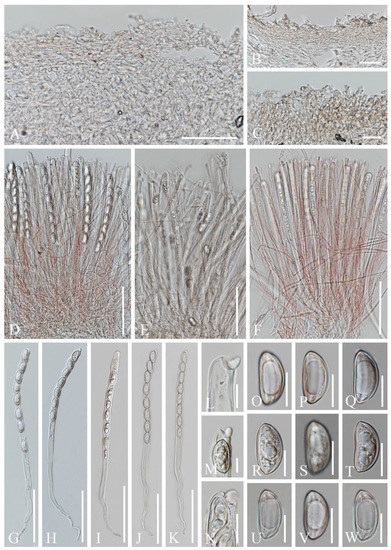
Figure 16.
Sections of herbarium materials of Phillipsia gelatinosa. (A,F,J,K,N,U–W) MFLU 16-2992. (B,E,I,M,R–T) MFLU 15-2360. (C,D,G,H,L,O–Q) MFLU 16-2956. (A–C) Vertical section of receptacle ectal excipulum. (D–F) Hymenium. (G–K) Asci and ascospores. (L–N) Apices of asci. (O–W) Ascospores. Scale bars (A) = 50 μm; (B,C) = 30 μm; (D–K) = 100 μm; (L–N) = 10 μm; (O–W) = 15 μm.
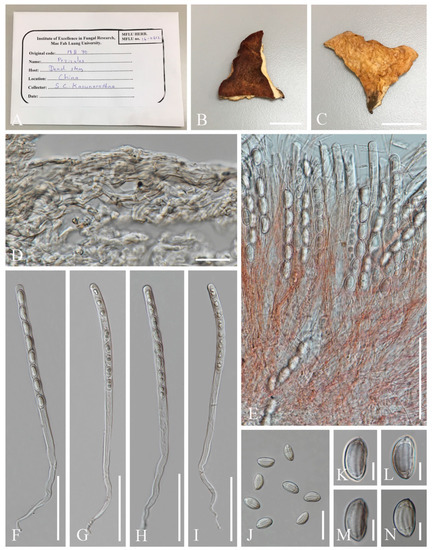
Figure 17.
Phillipsia subpurpurea (MFLU16-0612). (A–C) Herbarium materials. (D) Vertical section of receptacle ectal excipulum. (E) Hymenium. (F–I) Asci and ascospores. (J–N) Ascospores. Scale bars (B,C) = 1 cm; (D) = 20 μm; (E–I) = 100 μm; (J) = 40 μm; (K–N) = 10 μm.
- 7.
- Pithya villosa M. Zeng, Q. Zhao & K.D. Hyde, sp. nov.; Figure 18
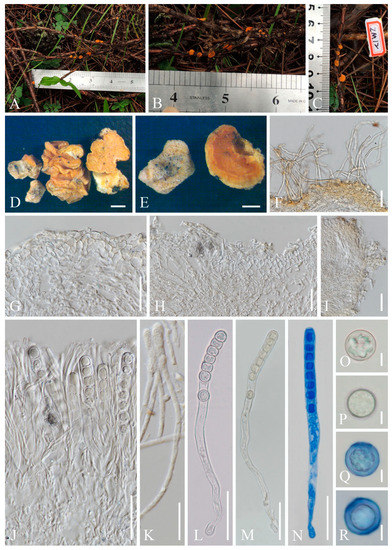 Figure 18. Pithyavillosa (HKAS 104653, holotype). (A–C) Fresh specimens. (D,E) Dry specimens. (F) Hyphoid hairs on the surface of receptacle ectal excipulum. (G,H,I) Vertical section of receptacle ectal excipulum on the upper flank. (J) Hymenium. (K) Paraphyses. (L–N) Asci and ascospores [(N) Ascus and ascospores in CB.] (O–R) Ascospores (Q,R) Ascospore in CB. Scale bars (D) = 1000 μm; (E) = 500 μm; (F–J,L–N) = 50 μm; (K) = 20 μm; (O–R) = 5 μm.
Figure 18. Pithyavillosa (HKAS 104653, holotype). (A–C) Fresh specimens. (D,E) Dry specimens. (F) Hyphoid hairs on the surface of receptacle ectal excipulum. (G,H,I) Vertical section of receptacle ectal excipulum on the upper flank. (J) Hymenium. (K) Paraphyses. (L–N) Asci and ascospores [(N) Ascus and ascospores in CB.] (O–R) Ascospores (Q,R) Ascospore in CB. Scale bars (D) = 1000 μm; (E) = 500 μm; (F–J,L–N) = 50 μm; (K) = 20 μm; (O–R) = 5 μm.
Index Fungorum number: IF 559929; Facesoffungi number: FoF 10411
Etymology: The specific epithet refers to villose receptacle surface.
Holotype: HKAS 104653
Diagnosis: This species is diagnosed by shallowly cupulate to discoid, or convex apothecia growing on Juniperus sp., yellowish excipular surface covered with hyphoid hairs, entire or lobate margin, subhyaline to yellowish paraphyses, spherical ascospores with granular contents.
Saprobic on twigs of Juniperus sp. Teleomorph: Apothecia 2–3 mm high, 3–6 mm broad, scattered to gregarious, fleshy, shallowly cupulate to discoid, or convex, sessile to substipitate. Receptacle shallowly cupulate, margin entire to lobate when fresh, or curled when dry, subglabrous to finely pubescent, whitish, flanks pubescent to villose towards the base, whitish on yellowish ground. Disc discoid to slightly convex, yellow to orange. Ectal excipulum 60–100 µm broad, hyaline on a wider marginal area, composed of textura porrecta, subhyaline to yellowish towards the base, and composed of textura epidermoidea with cells 5–8 µm broad, to textura angularis, with cells 13–17 × 9–12 µm. Hairs mostly arise from the excipular flank surface and apothecial base, subhyaline, flexuous, hyphoid, septate, 5–7 µm wide. Medullary excipulum 60–210 µm broad, of textura intricata, hyaline, composed of 4–6 µm broad hyphae. Hymenium 180–290 µm thick, yellow, paraphyses slightly exceeding the asci when dehydrated. Paraphyses 2–3 µm broad in the middle part, filiform, apex enlarged, 4–6 µm broad, branched, septate, subhyaline to yellowish. Asci 227–275 × 11–14 µm, 8-spored, terminally operculate, subcylindrical, apex obtuse, becoming narrow towards the base. Ascospores [20/1/1, in H2O] (11.4–)11.7–13.7(–14.4) × (11.3–)11.7–13.8(–14.0) µm (Q = 0.93–1.14, Q = 1.00 ± 0.05), spherical, uniseriate, subhyaline, with refractive granular contents, smooth-walled. Anamorph: not seen.
Material examined: CHINA: Yunnan, Shangri-La, on twigs of Juniperus sp., elev. 3413 m a.s.l., 14 August 2018, Ming Zeng, ZM12 (HKAS 104653, holotype); ibid. elev. 3390 m a.s.l., 15 August 2018, Ming Zeng, ZM23 (HKAS 121194, paratype).
Notes: Pithya is unique in Sarcoscyphaceae with its spherical ascospores [73]. Most species have similar features, namely yellow to orange apothecia, shallowly cupulate to discoid, sessile to substipitate, filiform paraphyses with enlarged apices, and smooth-walled spherical ascospores [73,82,83]. Kirk et al. [84] and Wijayawardene et al. [16,85] respectively accounted five and two species in this genus without listing species names, while there are 11 species records in Species Fungorum [27], excluding Pi. thujina (Peck) Sacc. which is synonymised to Pi. cupressina [82]. To our knowledge, the species richness does not appear to be clarified in this genus. Our introduction of the new species is based on a comparison of ten Pithya species according to Species Fungorum records. Pithya cupressina and Pi. vulgaris Fuckel are the oldest two species introduced in this genus, and they are also the most commonly recorded [73,82,83,86,87,88,89,90,91,92,93]. Molecular data are also most abundant in these two species [15,21,59]. Pithya vulgaris has larger apothecia and ascospores than Pi. cupressina, and whereas Pi. vulgaris mainly grows on the dead branches of Abies or Pinus, Pi. cupressina commonly grows on Cupressus or Juniperus [82,90,91]. Although our species is also found from Juniperus, it differs from Pi. cupressina in its apothecial vesture. While our new species has villose surface where hyphoid hairs are covering the whole excipular surface with margin, and to some extent also wider marginal area, Pi. cupressina has smooth excipular surface and white hyphal hairs are only present at the base [6,73,82,90]. The new species is phylogenetically distinct and is sister to Pi. cupressina. For other species that are rarely described, most of them are collected from Pinaceae with limited number of records, such as Pi. arctica L.I. Vassiljeva [94], Pi. epichrysea (Beck) Boud. [95], Pi. lacunosa (Ellis & Everh.) Seaver [82], Pi. malochi Velen. and Pi. microspora Velen. [96]. Pi. arethusa Velen. was collected from Ligustrum vulgare [96] and Pi. madothecae Buchloh was collected from Porella platyphylla [97]. Pithya fascicularis (Berk. & Broome) Sacc. was only described from bark, but its subglobose and small-sized ascosposes (7–8 µm diameter) is enough to distinguish it from our new species [95].
- 8.
- Sarcoscypha longitudinalis M. Zeng, Q. Zhao & K.D. Hyde, sp. nov.; Figure 19
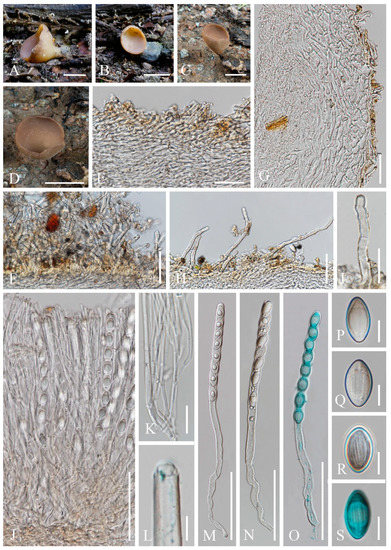 Figure 19. Sarcoscyphalongitudinalis. (A–D) Fresh specimens [(A,B) HKAS 121196. (C,D) HKAS 121195.] (E) Vertical section of stipe ectal excipulum. (F) Hyphoid hairs on the base. (G) Vertical section of receptacle ectal excipulum. (H,I) Hyphoid hairs from receptacle ectal excipulum. (J) Hymenium. (K) Paraphyses. (L) Apex of ascus. (M–O) Asci and ascospores [(O) Ascus and ascospores in CB.] (P–S) Ascospores [(S) Ascospore in CB.] Scale bars (A) = 1 cm; (B–D) = 2 cm; (E,G) = 30 μm; (F,H) = 50 μm; (I,K) = 20 μm; (J,M–O) = 100 μm; (L,P–S) = 10 μm.
Figure 19. Sarcoscyphalongitudinalis. (A–D) Fresh specimens [(A,B) HKAS 121196. (C,D) HKAS 121195.] (E) Vertical section of stipe ectal excipulum. (F) Hyphoid hairs on the base. (G) Vertical section of receptacle ectal excipulum. (H,I) Hyphoid hairs from receptacle ectal excipulum. (J) Hymenium. (K) Paraphyses. (L) Apex of ascus. (M–O) Asci and ascospores [(O) Ascus and ascospores in CB.] (P–S) Ascospores [(S) Ascospore in CB.] Scale bars (A) = 1 cm; (B–D) = 2 cm; (E,G) = 30 μm; (F,H) = 50 μm; (I,K) = 20 μm; (J,M–O) = 100 μm; (L,P–S) = 10 μm.
Index Fungorum number: IF 559930; Facesoffungi number: FoF 10412
Etymology: The specific epithet refers to ascospores with longitudinal striae.
Holotype: HKAS 121195
Diagnosis: This species is diagnosed by brown stipitate apothecia with villose receptacle, margin slightly wavy, or deeply split on one side, broadly fusiform ascospores with longitudinal striates.
Saprobic on dead wood. Teleomorph: Apothecia up to 4 cm high, 2.5 cm broad, solitary, cupulate, stipitate. Stipe up to 1 cm long, 5 mm broad, central, terete, solid, brown, with a thin inconspicuous hyphal pad close to base. Receptacle cupulate, villose, surface brown, margin slightly wavy, or deeply split on one side. Disc deeply concave, concolorous with the receptacle surface. Stipal ecto-excipulum 40–80 µm broad, composed of subhyaline to yellowish ochre, textura porrecta, hyphae 5–7 µm broad, hyphoid hairs abundant at the base, 4–6 µm broad, yellowish ochre. Stipal medulla composed of hyaline to subhyaline textura intricata, hyphae 3.5–6 μm broad. Ectal excipulum 60–100 µm broad, composed of hyaline textura porrecta to textura prismatica, brownish at the outermost part, hyphae 4–7 µm broad, mixed with 11–16 × 6–8 µm cells, with hyphoid hairs on the surface, 5–7 µm broad, septate, hyaline to subhyaline, with a rounded end. Medullary excipulum 235–310 µm broad, composed of hyaline to subhyaline textura intricata, hyphae 3–4 µm broad. Hymenium 280–345 µm thick, subhyaline, paraphyses slightly exceeding asci when dehydrated. Paraphyses 2–4 µm broad in the middle part, filiform, branched, septate, with a rounded end. Asci 297–359 × 12–14 µm, 8-spored, terminally operculate, subcylindrical, apex obtuse, becoming narrow towards the base. Ascospores [20/1/1, in H2O] (18.3–)19.3–21.4(–22.4) × (9.5–)10.7–12.1(–12.8) µm (Q = 1.57–1.99, Q = 1.79 ± 0.12), broadly fusiform, equilateral, uniseriate, multiguttulate when immature, uniguttulate when mature, ornamentation with several longitudinal striae. Anamorph: not seen.
Material examined: CHINA: Xishuangbanna, Bulangshan, on unidentified dead branch under broadleaved forest, 18 August 2019, Ming Zeng, ZM234 (HKAS 121195, holotype); ibid., 20 August 2019, Song Wang, ZM256 (HKAS 121196, paratype).
Notes: Sarcoscypha is distinguished by grey-white, yellow, orange to red apothecia which are substipitate to stipitate, glabrous to tomentose receptacle surface, ellipsoid to subcylindrical ascospores, normally with blunt ends or shallow depressions at both poles, smooth or ornamented wall, uniguttulate to multiguttulate [1,6,65,73]. Within Sarcoscypha, this new species is easily characterized by brown stipitate apothecia with villose receptacle, broadly fusiform, uniguttulate ascospores with longitudinal striates. According to our phylogenetic analyses, our new species is a sister group of S. vassiljevae. These two species share similar morphology in ascospores having a big oil drop, while S. vassiljevae differs from our species in grey-white hymenium and ellipsoid smooth-walled ascospores [6].
4. Discussion
Ekanayaka et al. [50,58] proposed the presence of ascospore sheath as a new taxonomically important character for some new and known species of Sarcoscyphaceae. While observing our recent collections and herbaria specimens, this particular feature appeared only in the ascospores that were treated with 5% or 10% KOH, but not in those that were mounted in water. This situation has been previously described by Pfister et al. [23] as ascospore walls loosening upon treatment with KOH, and it seems to be a universal characteristic within Sarcoscyphaceae. Thus, the gelatinous sheath is an invalid feature for species descriptions, much less an appropriate diagnostic feature for species identification as it is an artifact of the chemical treatment with KOH.
In this study, we introduce three new species, N. aequispora, Pi. villosa and S. longitudinalis, represented by two collections each based on morphology and phylogeny. In the phylogenetic analysis herein, Phillipsia, Rickiella, and Nanoscypha are not reciprocally monophyletic, but instead form a clade in both the ITS and combined data trees. This is a perennial unresolved problem that has also been noted in other studies [15,22]. In previous phylogenetic analyses lacking S. vassiljevae, Sarcoscypha and Pseudopithyella are sister taxa [15,22,23]. In studies where S. vassiljevae is included, Sarcoscypha is not monophyletic [3]. In our study, the new Sarcoscypha species is sister to S. vassiljevae; however, Sarcoscypha is not monophyletic in the combined gene tree. Instead, Pseudopithyella clustered in Sarcoscypha as sister to S. coccinea. Due to the absence of ITS data, the position of Pseudopithyella in the ITS tree cannot be inferred. Additional data from more Sarcoscypha and Pseudopithyella species would greatly clarify placement of these taxa.
Harrington [74] provided an ITS locus analysis and morphology of Sarcoscypha. In her study, S. striatispora clustered with two other Nanoscypha species, forming a distinct Nanoscypha clade separately from Sarcoscypha. Thus, Harrington [74] established the combination N. striatispora for S. striatispora W.Y. Zhuang. Furthermore, the author thought this species is more closely related to Nanoscypha because of its eccentrically operculate asci, and slightly equilateral ascospore with striae, which are more representative of Nanoscypha rather than Sarcoscypha [74]. The establishment of S. striatispora within Sarcoscypha was based on its distinct textura porrecta in the ectal excipulum [79], which differs from that of Nanoscypha, the latter having textura angularis to textura epidermoidea ectal excipulum [73]. Thus, Zhuang et al. [6] did not agree with this combination and proposed that the name S. striatispora should be retained. In the phylogeny (Figure 1), the only available N. striatispora strain (HMAS 61133) clusters independently from the Sarcoscypha species clade. Instead, N. striatispora groups within Phillipsia as sister to the clade formed by Ph. carnicolor Le Gal and Ph. hydei M. Zeng & Q. Zhao. Although there is only one strain to show the phylogenetic position of N. striatispora, the close relationship between Phillipsia and Nanoscypha has been demonstrated in other studies [15,22]. The relationship is also shown in the combined data tree herein, whereby Nanoscypha nests within Phillipsia. Phillipsia has textura porrecta ectal excipulum, which is similar to N. striatispora. Notably, the other Nanoscypha species represented by our new species and N. tetraspora also grouped within Phillipsia, but separately from N. striatispora. In terms of morphology, there are enough morphological features to distinguish Nanoscypha and Philipisia. Nanoscypha are often discoid to turbinate, normally less than 10 mm, centrally attached. In contrast, Phillipsia are often discoid, ear-shaped, and cup-shaped, normally more than 10 mm, centrally attached or eccentric [6,73,80]. The noted discrepancies warrant further exploration of the relationship between N. striatispora and Phillipsia from a phylogenetic and morphological point of view. At the moment, all Nanoscypha strains along with Phillipsia and Rickiella form a distinct clade, but their generic relationships remain unresolved. Additional taxa and genes will help resolve these complexities in the future.
Cookeina as a commonly-encountered genus of Sarcoscyphaceae in tropical and subtropical regions is adapted to growth in humid and hot environments [48,67]. Among the specimens we collected, a large number of species from tropical regions belong to the genus Cookeina, indicating a high abundance and diversity of the genus in China and Thailand. The phylogenetic position of the strains is identical in both trees. The new collections are accommodated in C. indica, C. sinensis, C. speciosa and C. tricholoma. Within Cookeina, the C. speciosa complex has high genetic variation and is divided into distinct subclades. Based on ITS and LSU phylogenies, Weinstein et al. [48] studied the correlation between colour differences of C. speciosa and different groups within the species complex. The colour of C. speciosa ranges from mauve, coral, orange, yellow to white, while there are no consistent anatomical differences among the colour variants [48]. In their study, two clades were segregated. One clade was associated with dark-coloured apothecia (mauve to deep coral), while members of the other had light-coloured apothecia (light coral, orange, yellow to white). It was then considered that the complex contains at least two taxa [48]. Taking into account our ITS inference, which contains multiple strains, their dark-coloured apothecia clade corresponds to subclades 1 and 2, while the light-coloured apothecia (light coral, orange, yellow to white) clade corresponds to subclades 3 and 4. Hence, it seems that this colour-based classification does not correspond to the phylogenetic inference herein. Six collections of C. speciosa cluster in a separate subclade expanding the existing diversity of the species complex. Within the C. speciosa complex, the placement of the sequences designated as C. sulcipes and C. garethjonesii is problematic. In our inferred phylogeny using a significantly expanded taxon sampling, neither appear as separate species from a phylogenetic point of view. Examining the type specimens and obtaining additional molecular data is necessary to disentangle this complex issue.
5. Conclusions
Southwestern China and Thailand are regions with high contributions to the species richness of Sarcoscyphaceae. Species of Cookeina, Phillipsia, and Sarcoscypha are very common in these areas, while Nanoscypha and Pithya have limited records. In the present study, we have redescribed five known species and established three new species in these genera. Meanwhile, Ph. gelatinosa is here proposed as a later epithet of Ph. domingensis. Our morphological and phylogenetic studies add a meaningful contribution to advancing this family toward natural classification. However, the lack of some type species and molecular data, and the presence of some species complexes, pose a challenge to future research.
Author Contributions
Conceptualization, M.Z.; methodology, M.Z. and E.G.; software, M.Z. and E.G.; validation, E.G., K.D.H. and Q.Z.; formal analysis, M.Z. and E.G.; investigation, M.Z., K.D.H. and Q.Z.; resources, M.Z., K.D.H. and Q.Z.; data curation, M.Z. and E.G.; writing—original draft preparation, M.Z.; writing—review and editing, M.Z., E.G., K.D.H., Q.Z, N.M., and I.K.; visualization, E.G., K.D.H., Q.Z., N.M., and I.K.; supervision, E.G., K.D.H. and Q.Z.; project administration, K.D.H. and Q.Z.; funding acquisition, K.D.H. and Q.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (Grant No. 2019QZKK0503), the open research project of “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences (Grant No. 292019312511043), Science and Technology Service Network Initiative, Chinese Academy of Sciences (KFJ-STS-QYZD-171), Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (Grant No. RDG6130001), the National Research Council of Thailand (NRCT) grant “Total fungal diversity in a given forest area with implications towards species numbers, chemical diversity and biotechnology” (Grant No. N42A650547). Ming Zeng is supported by MFLU grant number 5971105508 and the dissertation writing grant for Pezizomycetes studies.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Chuangen Lin, Deping Wei, and Song Wang for collecting samples. The authors would like to thank the herbarium of Mae Fah Luang University (MFLU) for allowing us to examine herbarium specimens. Shaun Pennycook (Manaaki Whenua-Landcare Research, New Zealand) is thanked for his help in checking the Latin names of new taxa.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baral, H.O. Taxonomische und ökologische Studien über Sarcoscypha coccinea agg., Zinnoberrote Kelchbecherlinge. Z. Mykol. 1984, 50, 117–145. [Google Scholar]
- Pfister, D.H. Chapter 2. Pezizomycotina: Pezizomycetes, Orbiliomycetes. In Systematics and Evolution. The Mycota (a Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research; McLaughlin, D., Spatafora, J., Eds.; Springer: Berlin, Germany, 2015; Volume 7B, pp. 35–56. [Google Scholar]
- Ekanayaka, A.H.; Hyde, K.D.; Jones, E.B.G.; Zhao, Q. Taxonomy and phylogeny of operculate discomycetes: Pezizomycetes. Fungal Divers. 2018, 90, 161–243. [Google Scholar] [CrossRef]
- Le Gal, M. Les Discomycètes suboperculés. Bull. Trimest. Soc. Mycol. Fr. 1946, 62, 218–240. [Google Scholar]
- Eckblad, F.E. The genera of the operculate discomycetes. A re-evaluation of their taxonomy, phylogeny and nomenclature. Nytt Mag. Bot. 1968, 15, 1–191. [Google Scholar]
- Zhuang, W.Y. Hyaloscyphaceae, Sarcoscyphaceae et Sarcosomataceae. In Flora Fungorum Sinicorum; Science Press: Beijing, China, 2004; Volume 21, pp. 1–192. [Google Scholar]
- Zeng, M.; Gentekaki, E.; Hyde, K.D.; Zhao, Q. Donadinia echinacea and Plectania sichuanensis, two novel species of Sarcosomataceae from southwestern China. Phytotaxa 2021, 508, 1–21. [Google Scholar] [CrossRef]
- Korf, R.P. Nomenclatural notes. VII. Family and tribe names in the Sarcoscyphineae (Discomycetes) and a new taxonomic disposition of the genera. Taxon 1970, 19, 782–786. [Google Scholar] [CrossRef]
- Chadefaud, M. Les asques para-operculés et la position systématique de la Pézize Sarcoscypha coccinea Fries ex Jacquin. C R Hebd. Seances Acad. Sci. 1946, 222, 753–755. [Google Scholar]
- Eckblad, F.E. The suboperculate ascus, a review. Persoonia 1972, 6, 439–443. [Google Scholar]
- van Brummelen, J. Light and electron microscopic studies of the ascus top in Sarcoscypha coccinea. Persoonia 1975, 8, 259–271. [Google Scholar]
- van Brummelen, J. The operculate ascus and allied forms. Persoonia 1978, 10, 113–128. [Google Scholar]
- Samuelson, D.A. The apical apparatus of the suboperculate ascus. Can. J. Bot. 1975, 53, 2660–2679. [Google Scholar] [CrossRef]
- Samuelson, D.A.; Benny, L.; Kimbrough, J.W. Asci of the Pezizales. VII. The apical apparatus of Galiella rufa and Sarcosoma globosum: Reevaluation of the suboperculate ascus. Can. J. Bot. 1980, 58, 1235–1243. [Google Scholar] [CrossRef]
- Romero, A.I.; Robledo, G.; LoBuglio, K.F.; Pfister, D.H. Rickiella edulis and its phylogenetic relationships within Sarcoscyphaceae. Kurtziana 2012, 37, 79–89. [Google Scholar]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Villarreal, L.; Perez-Moreno, J. Los hongos comestibles silvestres de Mexico, un enfoque integral. Micol. Neotrop. Apl. 1989, 2, 77–114. [Google Scholar]
- Sánchez, J.E.; Martin, A.M.; Sánchez, A.D. Evaluation of Cookeina sulcipes as an edible mushroom: Determination of its biomass composition. In Developments in Food Science; Elsevier: Amsterdam, The Netherlands, 1995; Volume 37, pp. 1165–1172. [Google Scholar]
- van Dijk, H.; Onguene, N.A.; Kuyper, T.W. Knowledge and utilization of edible mushrooms by local populations of the rain forest of south Cameroon. Ambio. 2003, 32, 19–23. [Google Scholar] [CrossRef]
- Arora, D. Mushrooms Demystified: A Comprehensive Guide to the Fleshy Fungi; Ten Speed Press: Berkeley, CA, USA, 1986; pp. 1–836. [Google Scholar]
- Harrington, F.A.; Pfister, D.H.; Potter, D.; Donoghue, M.J. Phylogenetic studies within the Pezizales. I. 18S rRNA sequence data and classification. Mycologia 1999, 91, 41–45. [Google Scholar] [CrossRef]
- Angelini, C.; Medardi, G.; Alvarado, P. Contribution to the study of neotropical discomycetes: A new species of the genus Geodina (Geodina salmonicolor sp. nov.) from the Dominican Republic. Mycosphere 2018, 9, 169–177. [Google Scholar] [CrossRef]
- Pfister, D.H.; Quijada, L.; LoBuglio, K.F. Geodina (Pezizomycetes: Wynneaceae) has a single widespread species in tropical America. Fungal Syst. Evol. 2020, 5, 131–138. [Google Scholar] [CrossRef]
- RAL Color Chart. Available online: https://www.ralcolor.com/ (accessed on 8 September 2022).
- Kušan, I.; Matočec, N.; Antonić, O.; Hairaud, M. Biogeographical variability and re-description of an imperfectly known species Hamatocanthoscypha rotundispora (Helotiales, Hyaloscyphaceae). Phytotaxa 2014, 170, 1–12. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/names/names.asp (accessed on 8 September 2022).
- Chaiwan, N.; Gomdola, D.; Wang, S.; Monkai, J.; Tibpromma, S.; Doilom, M.; Wanasinghe, D.N.; Mortimer, P.E.; Lumyong, S.; Hyde, K.D. https://gmsmicrofungi.org: An online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere 2021, 12, 1513–1526. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among Ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nylander, J.A.; Ronquist, J.P.; Huelsenbeck, F.; Nieves-Aldrey, J.L. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004, 53, 47–67. [Google Scholar] [CrossRef]
- Nuin, P.A.S. (University of Toronto, Toronto, ON, Canada). MTgui–A Simple Interface to ModelTest. Personal communication, 2005. [Google Scholar]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Cai, L.; Jeewon, R.; Hyde, K.D. Phylogenetic evaluation and taxonomic revision of Schizothecium based on ribosomal DNA and protein coding genes. Fungal Divers. 2005, 19, 1–21. [Google Scholar]
- FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 8 September 2022).
- Pfister, D.H.; Slater, C.; Hansen, K. Chorioactidaceae: A new family in the Pezizales (Ascomycota) with four genera. Mycol. Res. 2008, 112, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.N.; Pfister, D.H.; Iturriaga, T. A phylogenetic study of the genus Cookeina. Mycologia 2002, 94, 673–682. [Google Scholar] [CrossRef]
- Kropp, B.R. Cookeina cremeirosea, a new species of cup fungus from the South Pacific. Mycoscience 2016, 58, 40–44. [Google Scholar] [CrossRef]
- Ekanayaka, A.H.; Hyde, K.D.; Zhao, Q. The genus Cookeina. Mycosphere 2016, 7, 1399–1413. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Niranjan, M.; Dong, W.; Samarakoon, M.C.; Bao, D.; Calabon, M.S.; Chaiwan, N.; Chuankid, B.; Dayarathne, M.C.; de Silva, N.I.; et al. AJOM new records and collections of fungi: 101–150. Asian J. Mycol. 2021, 4, 113–260. [Google Scholar]
- Iturriaga, T.; Xu, F.; Pfister, D.H. Cookeina korfii, a new species hidden in Cookeina tricholoma. Ascomyceteorg. 2015, 7, 331–335. [Google Scholar]
- Hansen, K.; Pfister, D.H.; Hibbett, D.S. Phylogenetic relationships among species of Phillipsia inferred from molecular and morphological data. Mycologia 1999, 91, 299–314. [Google Scholar] [CrossRef]
- Perry, B.A.; Hansen, K.; Pfister, D.H. A phylogenetic overview of the family Pyronemataceae (Ascomycota, Pezizales). Mycol Res. 2007, 11, 549–571. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KT968605 (accessed on 8 September 2022).
- Zeng, M.; Ekanayaka, A.H.; Zhao, Q. Phylogeny and morphology of Phillipsia hydei sp. nov. (Sarcoscyphaceae) from Thailand. Phytotaxa 2019, 395, 277–286. [Google Scholar] [CrossRef]
- Zhuang, W.Y. Re-dispositions of Phillipsia (Pezizales) collections from China. Mycotaxon 2003, 86, 291–301. [Google Scholar]
- Ekanayaka, A.H.; Bhat, D.J.; Hyde, K.D.; Jones, E.B.G.; Zhao, Q. The genus Phillipsia from China and Thailand. Phytotaxa 2017, 316, 138–148. [Google Scholar] [CrossRef]
- Harrington, F.A.; Potter, D. Phylogenetic relationships within Sarcoscypha based upon nucleotide sequences of the internal transcribed spacer of nuclear ribosomal DNA. Mycologia 1997, 89, 258–267. [Google Scholar] [CrossRef]
- Luo, J.; Walsh, E.; Naik, A.; Zhuang, W.; Zhang, K.; Cai, L.; Zhang, N. Temperate pine barrens and tropical rain forests are both rich in undescribed fungi. PLoS ONE 2014, 9, e103753. [Google Scholar] [CrossRef]
- Ganley, R.J.; Newcombe, G. Fungal endophytes in seeds and needles of Pinus monticola. Mycol. Res. 2006, 110, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.; Perry, B.A.; Dranginis, A.W.; Pfister, D.H. A phylogeny of the highly diverse cup-fungus family Pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traits. Mol. Phylogenet. Evol. 2013, 67, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Celio, G.J.; Padamsee, M.; Dentinger, B.T.M.; Bauer, R.; McLaughlin, D.J. Assembling the Fungal Tree of Life: Constructing the Structural and Biochemical Database. Mycologia 2006, 98, 850–859. [Google Scholar] [CrossRef]
- Shi, C.H.; Bau, T.; Li, Y. Newly recorded genus and species of Pezizales in China. Mycosystem 2016, 35, 1348–1356. [Google Scholar]
- Wang, Y.Z.; Huang, C.L.; Wei, J.L. Two new species of Sarcoscypha (Sarcosyphaceae, Pezizales) from Taiwan. Phytotaxa 2016, 245, 169–177. [Google Scholar] [CrossRef]
- Xu, F.; LoBuglio, K.F.; Pfister, D.H. On the co-occurrence of species of Wynnea (Ascomycota, Pezizales, Sarcoscyphaceae) and Armillaria (Basidiomycota, Agaricales, Physalacriaceae). Fungal Syst. Evol. 2019, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Iturriaga, T.; Pfister, D.H. A monograph of the genus Cookeina (Ascomycota, Pezizales, Sarcoscyphaceae). Mycotaxon 2006, 95, 137–180. [Google Scholar]
- Pfister, D.H.; Kaushal, R. Cookeina indica, a new species from India with a key to the species of Cookeina. Mycotaxon 1984, 20, 117–121. [Google Scholar]
- Yang, Z.L. Several noteworthy higher fungi from southern Yunnan, China. Mycotaxon 1990, 38, 407–416. [Google Scholar]
- Wang, Z. Taxonomy of Cookeina in China. Mycotaxon 1997, 62, 289–298. [Google Scholar]
- Patil, A.; Patil, M.S.; Dangat, B.T. Cookeina Sinensis from India. Mycosphere 2012, 3, 603–605. [Google Scholar] [CrossRef]
- Dennis, R.W.G. Plumier’s Discomycetes. Mycotaxon 1994, 51, 237–239. [Google Scholar]
- Denison, W.C. Central American Pezizales. IV. The genera Sarcoscypha, Pithya, and Nanoscypha. Mycologia 1972, 64, 609–623. [Google Scholar] [CrossRef]
- Harrington, F.A. Relationships among Sarcoscypha species: Evidence from molecular and morphological characters. Mycologia 1998, 90, 235–243. [Google Scholar] [CrossRef]
- Pant, D.C.; Prasad, V. Indian Sarcoscyphaceous Fungi. In Technology & Engineering; Scientific Publishers: Jodhpur, India, 2008; pp. 1–124. [Google Scholar]
- Berkeley, M.J. On a collection of fungi from Cuba. Part II. Bot. J. Linn. Soc. 1869, 10, 341–392. [Google Scholar] [CrossRef]
- Rick, J.E. Pilze aus Rio Grande do Sul (Brazilien). Brotéria 1906, 5, 5–53. [Google Scholar]
- Pfister, D.H. Notes on Caribbean Discomycetes. V. A preliminary annotated checklist of the Caribbean Pezizales. J. Agric. Univ. P R 1974, 58, 358–378. [Google Scholar]
- Zhuang, W.Y. Some new species and new records of discomycetes in China. IV. Mycotaxon 1991, 40, 45–52. [Google Scholar]
- Denison, W.C. Central American Pezizales. III. The genus Phillipsia. Mycologia 1969, 61, 289–304. [Google Scholar] [CrossRef]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.H.; Li, D.Z.; Marhold, K.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017; Regnum Vegetabile 159; Koeltz Botanical Books: Glashütten, Germany, 2018. [Google Scholar]
- Seaver, F.J. The North American Cup-Fungi (Operculates); Lancaster Press: New York, NY, USA, 1928; p. 533. [Google Scholar]
- Spooner, B. The larger cup fungi in Britain, part 4, Sarcoscyphaceae and Sarcosomataceae. Field Mycol. 2002, 3, 9–14. [Google Scholar] [CrossRef]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Ainsworth & Bisby’s Dictionary of the Fungi, 10th ed.; CAB International: Wallingford, UK, 2008. [Google Scholar]
- Wijayawardene, N.N.; Hyde, K.D.; Rajeshkumar, K.C.; Hawksworth, D.L.; Madrid, H.; Kirk, P.M.; Braun, U.; Singh, R.V.; Crous, P.W.; Kukwa, M.; et al. Notes for genera: Ascomycota. Fungal Divers. 2017, 86, 1–594. [Google Scholar]
- Fuckel, L. Symbolae mycologicae. Beiträge zur Kenntniss der Rheinischen Pilze. Jahrb. Nassau. Ver. Naturkd. 1870, 23–24, 1–459. [Google Scholar]
- Krieglsteiner, G.J. On some new, rare and critical Macromycetes in the Federal Republic of Germany. Z. Mykol. 1985, 51, 85–130. [Google Scholar]
- Meléndez-Howell, L.M.; Coute, A.; Mascarell, G.; Bellemère, A. Ultrastructure des asques et des ascospores de Desmazierella acicola (Sarcoscyphaceae, Pezizales, Ascomycetes). Intérêt Systematique et Biologique. Mycotaxon 1998, 68, 53–74. [Google Scholar]
- Castellano, M.A.; Smith, J.E.; O’Dell, T.; Cázares, E.; Nugent, S. Handbook to Strategy 1 Fungal Taxa from the Northwest Forest Plan; U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station: Portland, OR, USA, 1999; pp. 1–195. [Google Scholar]
- Benkert, D. Pithya cupressina und P. vulgaris (Pezizales)–identisch oder nicht? Mycol. Bavarica. 2008, 10, 55–62. [Google Scholar]
- Sammut, C. Pithya cupressina (Ascomycota: Pezizomycetes Sarcoscyphaceae): A new addition to the Maltese mycobiota. Cent. Mediterr. Nat. 2012, 5, 54–55. [Google Scholar]
- Kunca, V. Pithya vulgaris znovuobjavená na Slovensku. Catathelasma 2015, 16, 11–13. [Google Scholar]
- Ortega-López, I.; Valenzuela, R.; Gay-González, A.D.; Lara-Chávez, M.B.N.; López-Villegasy, E.O.; Raymundo, T. La Familia Sarcoscyphaceae (Pezizales, Ascomycota) en México. Acta Bot. Mex. 2019, 126, e1430. [Google Scholar] [CrossRef]
- Index of Fungi 2: 444. Available online: http://sftp.kew.org/pub/data-repositories/LibriFungorum/IXF2/IXF2-444.jpg (accessed on 8 September 2022).
- Saccardo, P.A. Discomyceteae et Phymatosphaeriaceae. Syll. Fung. 1889, 8, 1–1143. [Google Scholar]
- Petrak’s Lists 7: 982. Available online: http://sftp.kew.org/pub/data-repositories/LibriFungorum/Petrak7/Petrak7-982.jpg (accessed on 8 September 2022).
- Döbbeler, P. Octosporella erythrostigma (Pezizales) and Pithyella frullaniae (Helotiales), two remarkable ascomycetes on Frullania dilatate. Feddes Repert. 2004, 115, 5–14. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).