Abstract
Microbial contamination remains a significant issue for many industrial, commercial, and medical applications. For instance, microbial surface contamination is detrimental to numerous aspects of food production, infection transfer, and even marine applications. As such, intense scientific interest has focused on improving the antimicrobial properties of surface coatings via both chemical and physical routes. However, there is a lack of synthetic coatings that possess long-term microbiocidal performance. In this study, silver nanoparticle cluster coatings were developed on copper surfaces via an ion-exchange and reduction reaction, followed by a silanization step. The durability of the microbiocidal activity for these develped surfaces was tested against pathogenic bacterial and fungal species, specifically Escherichia coli O157:H7 and Candida auris, over periods of 1- and 7-days. It was observed that more than 90% of E. coli and C. auris were found to be non-viable following the extended exposure times. This facile material fabrication presents as a new surface design for the production of durable microbicidal coatings which can be applied to numerous applications.
1. Introduction
The surface colonization of bacteria and fungi on abiotic substrates is commonly referred to as a biofilm formation and significantly contributes to healthcare and industrial concerns [1,2]. This issue is further impacted by the current rise in antibiotic resistance amongst microbial species, which has caused a significant increase in persistent infections and related deaths [3]. Recent economic projections have estimated that bacterial infections could be responsible for approximately 10 million deaths per annum by 2050 if new antimicrobial surface therapies are not developed [4,5].
In particular, Escherichia coli (E. coli) and Candida auris (C. auris) are common pathogenic microbes responsible for recent outbreaks [6,7]. Shiga toxin-producing E. coli (STEC) O157 has emerged as a public health threat following its initial identification as a pathogen in 1982, which was initiated when an outbreak of illness was associated with the consumption of undercooked ground beef [8]. A 2018 outbreak of the E. coli strain O157:H7 was reported across 36 states, infecting 210 people [6]. C. auris is another emerging pathogen currently responsible for invasive disease in healthcare facilities around the world [7,9] and the fungal infection carries an astonishingly high mortality rate of 60% among infected patients, representing a significant healthcare issue [7,9]. This high mortality rate has occurred due to the simultaneous emergence of multidrug-resistant C. auris isolates across three separate continents [9], raising pressing concerns regarding the identification and detection of invasive candidiasis isolates [10].
Silver nanoparticles (Ag NPs) are highly effective microbiocidal agents against both bacteria and fungi [11]. Ag NPs have been used in forms including solutions, thin-film coatings, or embedding in polymers [12,13,14]. While there have been some reported cases in which bacteria and fungi were found to be resistant against Ag NPs [15,16], recent reviews have shown alternative ways in eradicating bacterial cells via mechanical rupturing with nanostructure modifications [13,17,18,19]. There is a lack of significant research within the ability of these nanostructures to rupture fungal cells, in particular with the combination of Ag NPs and nanostructures not implemented as treatment strategies towards both bacterial and fungal cell surfaces.
In this work, a facile route was used to fabricate Ag NPs coatings on copper (Cu) surfaces, in which Ag NPs would assemble into high-aspect-ratio clusters. These surfaces were assessed for their long-term microbiocidal activity against both Shiga toxin-producing E. coli and C. auris cells. The fabricated surfaces present a new direction in the design of durable microbicidal surface coatings.
2. Materials and Methods
2.1. Fabrication of Hydrophobized Ag NP Coatings on Cu Surfaces
Cu surfaces were cut into 2 × 2 cm2 squares, pre-cleaned, and washed with HCl (Merck Pty Ltd, VIC, Australia) and MilliQ water (Synergy® UV Millipak Express 20, Merck Pty Ltd, VIC, Australia). The surfaces were then submerged with 0.1 M AgNO3 for 5 min, where Ag NPs formed on the Cu surface via a series of reduction–oxidation reactions. Surfaces were again washed carefully with MilliQ water, then with 100% ethanol (Merck Pty Ltd, VIC, Australia), and with MilliQ water as a last wash, and were blown dry with compressed nitrogen. This study adopted the fabrication method used in the previous work [20]. The Cu surfaces were further submerged with 0.1 M dichloromethyl silane (Sigma–Aldrich, Castle. Hill, NSW, Australia) in dichloromethane (Sigma–Aldrich Pty Ltd, NSW, Australia) for 15 min and were then washed with ethanol and MilliQ water to remove all chemical residues. In this study, pristine ‘copper surfaces’ are noted as ‘Cu surfaces’ and ‘silver nanoparticles embedding on copper surfaces’ were noted as ‘Ag NPs-Cu surfaces’.ImageJ version 1.8.0 (https://imagej.nih.gov/ij/) was implemented to analyze SEM images. Color Threshold and Analyze Particles Plugins were used to determine the dimensions of non-spherical Ag NPs. A total of 90 data points per system were used over three SEM micrographs to determine the distribution of NP clusters.
2.2. Bacterial Strains, Growth Conditions, and Sample Preparation
The antimicrobial efficacy of the surfaces was investigated using the strains E. coli O157:H7 and C. auris, which were obtained from the American Type Culture Collection and SA Pathology Lab, respectively, and were chosen as representatives of two major emerging pathogen outbreaks. The E. coli bacterial cultures were grown on Luria–Bertani (LB) agar (BD Difco, VIC, Australia) and fungal C. auris cultures were grown on Potato Dextrose Agar (PDA) (Sigma-Aldrich Pty Ltd, NSW, Australia) overnight at 37 °C. Bacterial and fungal cells were collected at the logarithmic stage of growth. To determine similar numbers of cells, despite variations in cell densities following collection, the density of the bacterial and fungal suspensions was adjusted to OD 600 = 0.1 at the logarithmic stage of cell growth. To quantify cell numbers in the adjusted bacterial suspensions before attachment experiments, a hemocytometer was used as suggested previously [21]. Pristine Cu and Ag NPs-Cu surfaces were cut into squares of 0.5 × 0.5 cm2. The surfaces were pre-sterilized with 70% ethanol, dried within a sterilized laminar flow cabinet for 24 h and placed in a sterilized 24-well plates (Thermo Fischer Scientific Australia Pty Ltd, VIC, Australia). Bacterial and fungal suspensions were prepared as above with OD600 = 0.1 and 1 ml of each suspension was added into 24-well plates containing the sterilized surfaces. The plates were incubated at 25 °C in dark conditions, to avoid any effects of light on cell viability. The samples were incubated in static conditions for 1 and 7 days without any disturbance from media addition or exchange. This procedure was adopted to avoid any effects of fresh media and shaking on antimicrobial assays. Two technical replicates were done.
2.3. Confocal Laser Scanning Microscopy (CLSM)
The surfaces were washed with 10 mM PBS (pH = 7.4) prior to the CLSM imaging conducted using ZEISS LSM 880 Airyscan upright microscope (Zeiss, Oberkochen, BW, Germany). To assess their viability, adhered cells were stained using a LIVE/DEAD® BacLightTM Viability Kit (including SYTO® 9 and propidium iodide) (Molecular ProbesTM, Invitrogen, Grand Island, NY, USA). In this kit, SYTO® 9 binds to nucleic acids in both intact and damaged cells, while propidium iodide (PI) predominantly enters cells with a damaged membrane considered non-viable. The bacterial and fungal cells on surfaces were stained according to the manufacturer’s protocol [22]. The proportions of live and dead cells on pristine Cu and Ag NPs-Cu surfaces were then evaluated using a ZEISS LSM 880 Airyscan upright microscope (Zeiss, Oberkochen, BW, Germany). To determine the percentage of non-viable cells, CellC Cell Counting Software (https://sites.google.com/site/cellcsoftware/) was used as previously instructed [19]. An analysis was done over 5 representative micrographs over 2 technical replicates. A student t-test (Microsoft Excel) was conducted to compare the antimicrobial performance of these samples.
2.4. SEM Characterization
Prior to the SEM imaging, bacterial and fungal cells on Cu and Ag NPs-Cu surfaces were fixed using 3% glutaraldehyde and were dehydrated using a series of ethanol concentrations (30%, 50%, 70%, 80%, 90%, 100%). The samples were removed and dried under the laminar flow of a Biosafety Cabinet Class II for 2–4 h. Fixed cells were coated with a thin film of gold prior to imaging. SEM images were taken using a field-emission scanning electron microscope (FE-SEM) (FEI Verios, FEI company, OR, United States) at 5 kV, where imaging of the systems uses methods that have been previously described [23].
2.5. Focussed Ion Beam-Scanning Electron Microscopy (FIB-SEM) Characterization
Surfaces were affixed and dehydrated using the identical dehydration steps described above. A cross-sectional analysis was carried out to show the interaction of E. coli and C. auris with the Ag NP cluster substrates was performed using a FEI Scios Dualbeam FIB-SEM. The cells were first coated with platinum (Pt) using an ion gun at 16 kV/0.15 nA to prevent further damage, followed by a sequential ion slicing at 16 kV/0.15 nA. The imaging was carried out using a standard secondary electron (ETD) and upper in-lens column (T2) detectors at 2 kV/0.1 nA.
3. Results
3.1. Fabrication and Characterisation of Hydrophobized Ag NP-Coated Cu Surfaces
Ag NP clusters were fabricated directly onto Cu surfaces via a single-step electrochemical synthesis. A further surface silanization step was added to create a hydrophobic Ag NP cluster on the Cu surfaces. The fabricated Ag NPs-Cu surfaces were then characterized using SEM and energy dispersive X-ray (EDX) spectroscopy. In Figure 1A,B, the SEM micrographs show that the Ag NPs exhibit sizes that are consistently less than 100 nm in diameter. These Ag NPs were found to form clusters, with wide sizes ranging from 70 nm to 1200 nm and an average size of ~350 nm per cluster. The aspect-ratio of the clusters was estimated using the ratio of length (nm) to width (nm). This measurement revealed that the clusters possess an aspect-ratio (length/width) ranging from 1 to 14. With the addition of silane to the clusters, the surfaces exhibit water contact angles of ~120°, meaning that the surfaces are hydrophobic in nature. Silane coatings are well-known corrosion-inhibitors [24] and therefore provide a strong potential in preventing the corrosion of Ag–Cu surfaces. In Figure 1C, chemical mapping using Energy Dispersive X-ray Analysis (EDX) revealed that the Ag NPs are formed via replacing Cu(0) using a series of electroless galvanic reactions (see Equations (1) and (2)) [25,26]:
Red: −2Ag+ + 2e− → 2Ag0
Ox: Cu → Cu2+ + 2e−
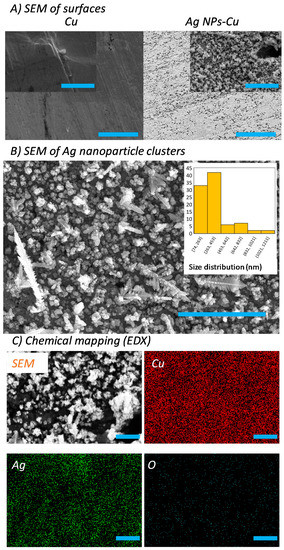
Figure 1.
Surface characterization of pristine Cu and Ag nanoparticle coatings (Ag NPs-Cu). (A) SEM images showing the surface topography of pristine Cu (left) and Ag NPs-Cu (right) surfaces (Scale bar 20 µm, inset 3 µm). (B) High resolution SEM image showing Ag NPs cluster domains (scale bar 4 µm). The inset shows the cluster size distribution. (C) EDX spectroscopy showing the distribution of Cu, Ag, and O elements across the Ag NPs-Cu surfaces (scale bar 1 µm).
Galvanic replacement reactions are commonly known to produce Ag NPs via this route [27,28]. Importantly, low level of oxidation was detected on the Ag NPs-Cu surfaces. Therefore we provide a simple method to fabricate Ag NP clusters which have demonstrated several advantages: simplicity, facility, and a low fabrication cost. This galvanic replacement reaction method can be used on an industrial scale, which highlights the utility of this synthetic route [29].
3.2. Microbicidal Performance of Ag NPs-Cu Surfaces over 1- and 7-day Incubations
Two representative species of major global disease outbreaks in recent years, Shiga toxin-producing E. coli O157:H7 and C. auris, were selected to study the relative material and surface interactions, with the use of glass surfaces employed as control substrates (Figures S1 and S2). The data in this study reveal the native cell morphology and viability of the respective bacterial and fungal pathogens incubated on both pristine and Ag NPs-Cu surfaces for periods of 1 and 7 days. To assess their viability, LIVE/DEAD Fluorescent Kits (Molecular ProbesTM, Invitrogen, Grand Island, NY, USA) were used to stain the viable and non-viable cells [22]. SYTO® 9 dye enter the viable cells and propidium iodide would enter the non-viable cells with compromised cell membranes. Control glass surfaces demonstrated over 90% cell viability for both E. coli and C. auris over a 7-day incubation (Figure S2) while pristine Cu surfaces showed slight killing activity against the cells as shown in Figure 1, despite the inherent antimicrobial activity native of Cu surfaces. E. coli specifically formed a well-established biofilm on the pristine Cu surfaces. In comparison, Ag NPs-Cu surfaces inhibited the growth of E. coli and C. auris (Figure 2), with 75% and 98% of non-viability observed respectively, after a 1-day incubation only. Greater results were obtained after a 7-day incubation on the Ag NPs-Cu surfaces with more than 90% and ~100% non-viability observed for E. coli and C. auris respectively. Further investigation of the interactions between bacteria/fungi and Ag NP clusters was carried out using FIB-SEM and clear deformations of E. coli and C. auris were examined and shown by tearing of the cell membrane and presence of multiple hole features in the cell morphology, as shown in Figure 3A,B. Past studies have reported similar examinations [18,30].
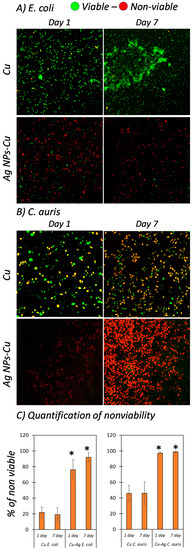
Figure 2.
Assessment of Shiga-toxin-producing E. coli O157:H7 and C. auris on copper and Ag NPs-Cu surfaces over 1- and 7-day incubations. CSLM micrographs showing the viability of (A) E. coli and (B) C. auris on pristine Cu and Ag NPs-Cu surfaces (green indicating viable cells; and red indicating non-viable cells). CLSM images are 150 µm × 150 µm. (C) Quantification of cell viability on Cu and Ag NPs-Cu (* indicating p < 0.05 comparing with Cu surfaces on day 1 and 7, respectively, n = 10).

Figure 3.
Morphology of E. coli and C. auris on control and Ag NPs-Cu surfaces (Scale bar 5 µm). (A) Top-view SEM micrographs showing the cell deformation under the Ag NP clusters (scale bar 5 µm). (B) SEM cross-sections using FIB-SEM reveal the interfacial interaction between cell surfaces and nanostructures (left scale bar 1 µm; right scale bar 4 µm).
The antimicrobial behavior of Ag NPs is known to occur via several mechanisms: (1) physical damage by direct contact [31], (2) the release of silver ions [32,33], and (3) reactive oxygen species production [34,35]. The breadth of information concerning the antibacterial mechanism of Ag NPs is beyond the scope of this article; however, the interested reader is directed towards several important reviews in the field [11,34]. In previous work, Ag NPs have been reported as effective antimicrobial agents against both bacteria (E. coli and Staphylococcus aureus) and fungi (e.g., Candida albicans) [11,12,16,36], but past investigations have failed in showing the long-term effect of silver coatings against both bacteria and fungi over a 7-day incubation, as shown in this study. Furthermore, this study is the first showing the effects of antifungal coating towards C. auris. There are a number of studies demonstrating that high-aspect-ratio nanostructures can be used to capture bacteria and disrupt bacterial membranes [18,37,38,39,40]. Surface nanostructures have been considered to be “promising methods” to stop bacterial adhesion and proliferation; still, there are a number of questions that should be answered before being applied in the actual environments [41]. However, the synergy between both surface nanostructure and chemistry should be considered to be the good candidate in the development of antimicrobial approaches.
4. Conclusions
Both E. coli and C. auris have been reported to be the main cause of recent outbreaks of disease. These two microbes were chosen to investigate the antimicrobial activity of durable Ag NP cluster coated Cu surfaces. Microbiocidal composite Ag NP cluster–Cu coatings were fabricated via an ion-exchange reduction reaction. The durability of microbicidal efficacy of the surfaces was established against pathogenic fungal and bacterial species, specifically E. coli O157:H7 and C. auris, over exposure periods of 1 and 7 days. It was observed that more than ~90% of E. coli and ~100% C. auris were non-viable, after only 7 days of surface immersion. The surfaces reported in this study provide a facile fabrication route and a new design parameter to produce durable microbicidal coatings for numerous applications.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6412/10/1/28/s1, Figure S1: Scanning electron micrographs of E. coli and C. auris on glass surfaces (scale bar 2 µm). The morphology of E. coli and C. auris were found to be intact and no damage on glass substrates, Figure S2: Confocal scanning laser microscopic images of E. coli and C. auris on glass surfaces (scale bar 10 µm). Most of the cells were found to be viable on glass substrates.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “conceptualization, V.K.T. and J.C.; methodology, S.G., A.E., A.E.M., D.C., Y.B.T., R.J.C., P.W., V.K.T., J.C.; formal analysis, S.G., A.E.M., V.K.T., J.C.; writing—original draft preparation, S.G., A.E., V.K.T., J.C.; writing—review and editing, S.G., A.E., A.E.M., D.C., Y.B.T., R.J.C., P.W., V.K.T., J.C.; supervision, V.K.T., J.C. All authors have read and agreed to the published version of the manuscript.
Funding
The work is supported by the School of Science, RMIT University.
Acknowledgments
The authors thank both the Microscopy and Microanalysis Facility (RMMF) and the MicroNano Research Facility (MNRF) at RMIT University for the use of their facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Rodvold, K.A.; McConeghy, K.W. Methicillin-resistant staphylococcus aureus therapy: Past, present, and future. Clin. Infect. Dis. 2014, 58, S20–S27. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, G.; Fleck, F. United Nations Meeting on Antimicrobial Resistance. World Health Organ. Bull. World Health Organ. 2016, 94, 638. [Google Scholar]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; HM Government: London, UK; Welcome Trust: London, UK, 2016.
- CDC. Multistate Outbreak of e. Coli o157:H7 Infections Linked to Chopped Romaine Lettuce; US Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2018.
- Nett, J.E. Candida auris: An emerging pathogen “incognito”? PLoS Pathog. 2019, 15, e1007638. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A. Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. J. Clin. Infect. Dis. 2016, 64, 134–140. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Jackson, B.R.; Vallabhaneni, S.; Ostrosky-Zeichner, L.; Pappas, P.G.; Chiller, T. Thinking beyond the common candida species: Need for species-level identification of candida due to the emergence of multidrug-resistant candida auris. J. Clin. Microbiol. 2017, 55, 3324–3327. [Google Scholar] [CrossRef]
- Prabhu, S.; Poulose, E.K. Silver nanoparticles: Mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int. Nano Lett. 2012, 2, 32. [Google Scholar] [CrossRef]
- Petica, A.; Gavriliu, S.; Lungu, M.; Buruntea, N.; Panzaru, C. Colloidal silver solutions with antimicrobial properties. Mater. Sci. Eng. B 2008, 152, 22–27. [Google Scholar] [CrossRef]
- Elbourne, A.; Truong, V.K.; Cheeseman, S.; Rajapaksha, P.; Gangadoo, S.; Chapman, J.; Crawford, R.J. The use of nanomaterials for the mitigation of pathogenic biofilm formation. In Methods in Microbiology; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Qing, Y.; Cheng, L.; Li, R.; Liu, G.; Zhang, Y.; Tang, X.; Wang, J.; Liu, H.; Qin, Y. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int. J. Nanomed. 2018, 13, 3311–3327. [Google Scholar] [CrossRef] [PubMed]
- Panáček, A.; Kvítek, L.; Smékalová, M.; Večeřová, R.; Kolář, M.; Röderová, M.; Dyčka, F.; Šebela, M.; Prucek, R.; Tomanec, O.; et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat. Nanotechnol. 2018, 13, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.L.; Tajkarimi, M.; Cunningham, Q.; Campbell, A.; Nonga, H.; Harrison, S.H.; Barrick, J.E. Rapid evolution of silver nanoparticle resistance in escherichia coli. Front. Genet. 2015, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Berton, P.; Moraes, C.; Rogers, R.D.; Tufenkji, N. Nanodarts, nanoblades, and nanospikes: Mechano-bactericidal nanostructures and where to find them. Adv. Colloid Interface Sci. 2018, 252, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Elbourne, A.; Chapman, J.; Gelmi, A.; Cozzolino, D.; Crawford, R.J.; Khanh Truong, V. Bacterial-nanostructure interactions: The role of cell elasticity and adhesion forces. J. Colloid Interface Sci. 2019, 546, 192–210. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, P.; Cheeseman, S.; Hombsch, S.; Murdoch, B.J.; Gangadoo, S.; Blanch, E.W.; Truong, Y.B.; Cozzolino, D.; McConville, C.F.; Crawford, R.J.; et al. Antibacterial properties of graphene oxide-copper oxide nanoparticle nanocomposites. ACS Appl. Biol. Mater. 2019, 2, 5687–5696. [Google Scholar] [CrossRef]
- Chapman, J.; Regan, F. Nanofunctionalized superhydrophobic antifouling coatings for environmental sensor applications—Advancing deployment with answers from nature. Adv. Eng. Mater. 2012, 14, B175–B184. [Google Scholar] [CrossRef]
- Liu, S. Chapter 11—How cells grow. In Bioprocess Engineering, 2nd ed.; Liu, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 629–697. [Google Scholar]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.-U.; Egli, T. Assessment and interpretation of bacterial viability by using the live/dead baclight kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283. [Google Scholar] [CrossRef]
- Truong, V.K.; Lapovok, R.; Estrin, Y.S.; Rundell, S.; Wang, J.Y.; Fluke, C.J.; Crawford, R.J.; Ivanova, E.P. The influence of nano-scale surface roughness on bacterial adhesion to ultrafine-grained titanium. Biomaterials 2010, 31, 3674–3683. [Google Scholar] [CrossRef]
- Child, T.F.; van Ooij, W.J. Application of silane technology to prevent corrosion of metals and improve paint adhesion. Trans. IMF 1999, 77, 64–70. [Google Scholar] [CrossRef]
- He, X.; He, R.; Lan, Q.; Duan, F.; Xiao, J.; Song, M.; Zhang, M.; Chen, Y.; Li, Y. A facile fabrication of silver-coated copper nanowires by galvanic replacement. J. Nanomater. 2016, 2016, 8. [Google Scholar] [CrossRef]
- Chen, X.; Ku, S.; Weibel, J.A.; Ximenes, E.; Liu, X.; Ladisch, M.; Garimella, S.V. Enhanced antimicrobial efficacy of bimetallic porous cuo microspheres decorated with ag nanoparticles. ACS Appl. Mater. Interfaces 2017, 9, 39165–39173. [Google Scholar] [CrossRef] [PubMed]
- Brevnov, D.A.; Olson, T.S.; López, G.P.; Atanassov, P. Electroless deposition of silver by galvanic displacement on aluminum alloyed with copper. J. Phys. Chem. B 2004, 108, 17531–17536. [Google Scholar] [CrossRef]
- Chee, S.W.; Tan, S.F.; Baraissov, Z.; Bosman, M.; Mirsaidov, U. Direct observation of the nanoscale kirkendall effect during galvanic replacement reactions. Nat. Commun. 2017, 8, 1224. [Google Scholar] [CrossRef]
- da Silva, A.G.M.; Rodrigues, T.S.; Haigh, S.J.; Camargo, P.H.C. Galvanic replacement reaction: Recent developments for engineering metal nanostructures towards catalytic applications. Chem. Commun. 2017, 53, 7135–7148. [Google Scholar] [CrossRef]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef]
- Chatterjee, T.; Chatterjee, B.K.; Majumdar, D.; Chakrabarti, P. Antibacterial effect of silver nanoparticles and the modeling of bacterial growth kinetics using a modified gompertz model. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2015, 1850, 299–306. [Google Scholar] [CrossRef]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346–2353. [Google Scholar] [CrossRef]
- Xiu, Z.-M.; Zhang, Q.-B.; Puppala, H.L.; Colvin, V.L.; Alvarez, P.J.J. Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 2012, 12, 4271–4275. [Google Scholar] [CrossRef]
- Durán, N.; Durán, M.; de Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 789–799. [Google Scholar] [CrossRef]
- Zhang, W.; Li, Y.; Niu, J.; Chen, Y. Photogeneration of reactive oxygen species on uncoated silver, gold, nickel, and silicon nanoparticles and their antibacterial effects. Langmuir 2013, 29, 4647–4651. [Google Scholar] [CrossRef] [PubMed]
- Lara, H.H.; Romero-Urbina, D.G.; Pierce, C.; Lopez-Ribot, J.L.; Arellano-Jiménez, M.J.; Jose-Yacaman, M. Effect of silver nanoparticles on candida albicans biofilms: An ultrastructural study. J. Nanobiotechnol. 2015, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Masigol, M.; Fattahi, N.; Barua, N.; Lokitz, B.S.; Retterer, S.T.; Platt, T.G.; Hansen, R.R. Identification of critical surface parameters driving lectin-mediated capture of bacteria from solution. Biomacromolecules 2019, 20, 2852–2863. [Google Scholar] [CrossRef] [PubMed]
- Shahali, H.; Hasan, J.; Mathews, A.; Wang, H.; Yan, C.; Tesfamichael, T.; Yarlagadda, P.K.D.V. Multi-biofunctional properties of three species of cicada wings and biomimetic fabrication of nanopatterned titanium pillars. J. Mater. Chem. B 2019, 7, 1300–1310. [Google Scholar] [CrossRef]
- Elbourne, A.; Crawford, R.J.; Ivanova, E.P. Nano-structured antimicrobial surfaces: From nature to synthetic analogues. J. Colloid Interface Sci. 2017, 508, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Elbourne, A.; Dupont, M.F.; Collett, S.; Truong, V.K.; Xu, X.; Vrancken, N.; Baulin, V.; Ivanova, E.P.; Crawford, R.J. Imaging the air-water interface: Characterising biomimetic and natural hydrophobic surfaces using in situ atomic force microscopy. J. Colloid Interface Sci. 2019, 536, 363–371. [Google Scholar] [CrossRef]
- Luan, Y.; Liu, S.; Pihl, M.; van der Mei, H.C.; Liu, J.; Hizal, F.; Choi, C.-H.; Chen, H.; Ren, Y.; Busscher, H.J. Bacterial interactions with nanostructured surfaces. Curr. Opin. Colloid Interface Sci. 2018, 38, 170–189. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).