Abstract
The outbreak of epidemiological diseases creates a major impact on humanity as well as on the world’s economy. The consequence of such infectious diseases affects the survival of mankind. The government has to stand up to the negative influence of these epidemiological diseases and facilitate society with medical resources and economical support. In recent times, COVID-19 has been one of the epidemiological diseases that created lethal effects and a greater slump in the economy. Therefore, the prediction of outbreaks is essential for epidemiological diseases. It may be either frequent or sudden infections in society. The unexpected raise in the application of prediction models in recent years is outstanding. A study on these epidemiological prediction models and their usage from the year 2018 onwards is highlighted in this article. The popularity of various prediction approaches is emphasized and summarized in this article.
1. Introduction
Epidemiology is the scientific analysis of the determinants of these infectious diseases and their spread. Currently, the lethal effects of epidemiological diseases have become more familiar through the COVID-19 pandemic. But such effects should be controlled and rectified by the government. This is possible by predicting the outbreak of the disease as well as finding the determinants of these infectious diseases [1]. The government has to take necessary action to prevent the disease’s spread by providing medical resources to the affected people and forecasting the future effects for executing the preventive measures [2]. The COVID-19 pandemic has caused a terrible impact on the economy and unexpectedly high death rates throughout the world [1]. The unforeseen lockdowns were accomplished. Therefore, there is an increase in the research on epidemiological predictions from 2018 onwards. Owing to such epidemiological scenarios, epidemiological predictions become essential for the prevention of infectious diseases. Other epidemiological diseases like dengue [3], Ebola [4], malaria [5], HIV [6], TB [7], influenza [8], etc., have also had lethal impacts in the world. But the COVID-19 has increased the research in this field. The prediction of death rates, incidence rates, and area-wise outbreaks [3,5,9,10,11] are required to increase the medical facilities for preventing the disease spread, analyzing the expected severity of the disease, and creating timely awareness among the people. The number of research articles in this field is notably high and there has been a boom in this research field of “epidemiological prediction” from 2018 onwards, which is evidenced by nearly 100+ research articles in this paper. This paper is organized as a Scopus analysis of epidemiological prediction models, followed by the existing epidemiological prediction models, a discussion of their significance, and, finally, a summary of the paper.
2. Scopus Analysis on Epidemiological Prediction
The extended literature study on epidemiological prediction of various infectious diseases is discussed in this paper. As per Scopus analysis, there are 11,024 research articles from the year 2016 onwards. In the year 2018, it started increasing due to the COVID-19 infections worldwide. In the years 2021 and 2022, the number of research documents in the epidemiological predictions is significant. The number of research documents from 2016 onwards is shown in Figure 1, where the degree of significance of this research area is an increasing trend in 2021. The boom in this field is due to COVID-19 severity over this period.
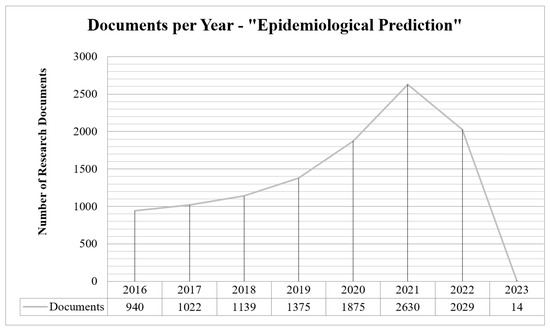
Figure 1.
Number of research articles from 2016 onwards—shows upward trend until 2021.
These research documents are of different categories like journal articles, reviews, conference papers, newsletters, editorials, book chapters, surveys, erratum, and so on. Out of 11,024 documents, around 9634 documents are journal articles, 750 documents are review articles, 286 documents are conference proceedings, 88 are newsletters and the remaining documents are different types, as given in Figure 2.
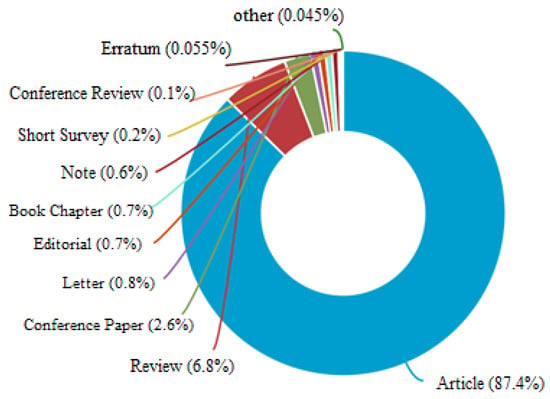
Figure 2.
Percentage of research on “Epidemiological Prediction” based on document type as per Scopus analysis.
These documents are from various research departments like medicine, neuroscience, bioinformatics, computer science, immunology, bioscience, environmental science, and others as shown in Figure 3. The Scopus analysis says that 52.3 percent of research is contributed by the medicine department and 11 percent by the bio-chemistry department. Similarly, the percentage of contributions of various other departments is highlighted in Figure 3.
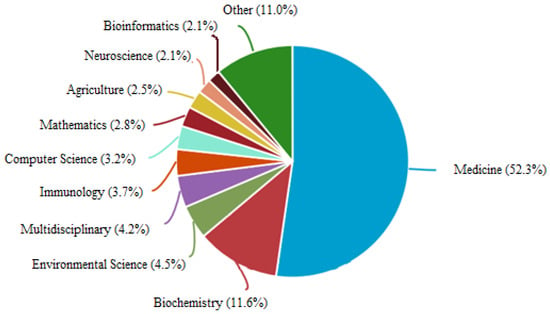
Figure 3.
Percentage of research on “Epidemiological Prediction” based on subject area as per Scopus analysis.
The country-wise analysis report is depicted in Figure 4. Here, the United States ranks first in this research because the infection rate of COVID-19 here was 9.96 crores and the death rate was 11 lakhs to date, which ranks first in the world. Around 3.3 K research documents are from the US. The COVID-19 pandemic is the major reason for this surge in epidemiological research during this period. Next to the US, COVID-19 infections were high in China. Countries like the United Kingdom, Australia, and Canada are also in the first ten rankings, as shown in Figure 4.
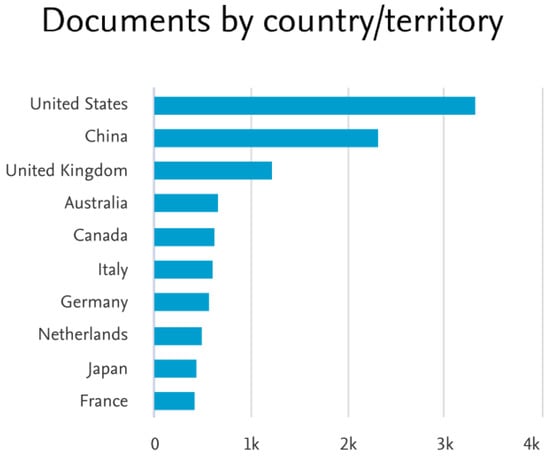
Figure 4.
Number of research on “Epidemiological Prediction” based on country/territory as per Scopus analysis.
3. Existing Epidemiological Prediction Methods
This study highlights different prediction models like mathematical models, statistical models, machine learning models, deep learning models, etc., used in this domain. The essence of these related research works is listed in Table 1 and their significances are highlighted under the following categories: (i) epidemiological disease—the infectious disease concentrated in that article, like COVID, dengue, ebola, malaria, HIV, pneumonia, influenza, scarlet fever, brucellosis, HFMD, etc.; (ii) data variety—the variety of the data used in the work like univariate or multivariate time series, stationary or non-stationary data, and so on; (iii) models explored—the models proposed in that particular research article like ARIMA, LSTM, GRNN, MLP, etc.; (iv) model category—the type of model used in that research such as statistical, machine learning, deep learning, etc.; (v) error forecast—depicts the usage of error forecasting in their research; (vi) parameter tuning—any of the parameters tuned in with those methods are represented; and (vii) novelty—highlighting the significances and contributions of the particular research work. Therefore, this table summarizes these criteria from nearly 80 research articles. Further, some research articles before 2018 are discussed in Table 2. These research studies were on epidemiological diseases like influenza, dengue, pneumonia, etc. But the surge in this research area is mainly due to the lethal effects of COVID-19 after 2018. The necessity of these epidemiological prediction models is discussed further in this article.

Table 1.
Research articles in epidemiological predictions.

Table 2.
Some research articles in epidemiological predictions before 2018.
3.1. Discussions on Epidemiological Prediction Methods
From this study, it is clear that the horizon of every epidemiological prediction is distinct, i.e., it will be disease-specific, geographical location-specific, demographic-specific, and problem-specific. A single model neither suits all the infectious diseases nor all the states/provinces; it varies for different disease-space combinations. The selection of a model for an epidemiological prediction problem plays a vital role in obtaining optimal outcomes. As per the literature, different models are explored and the better model is chosen based on their performances. The model performance always depends on the prediction problem as well as the data.
The prediction problems may be trend extraction, seasonal curve finding, detecting outbreak suspection, outbreak trajectories, peak fluctuations, estimating incidence rate, death rate, recovery rate, daily new cases, treatment requirements, pandemic prediction, outbreak reversal prediction, etc. The margin of these predictions may be either for the long term, like monthly/yearly predictions, or for the short term, like daily/weekly predictions. Therefore, the prediction problems may have different requirements, and the forecast models are chosen based on their characteristics. As mentioned earlier, these models are categorized as mathematical models, statistical models, machine learning models, and deep learning models. The significance of these prediction model categories is shown in Figure 5. The various prediction models under these categories and their significance over this period are also depicted. The negative influences of various epidemiological diseases are shown in Figure 6.
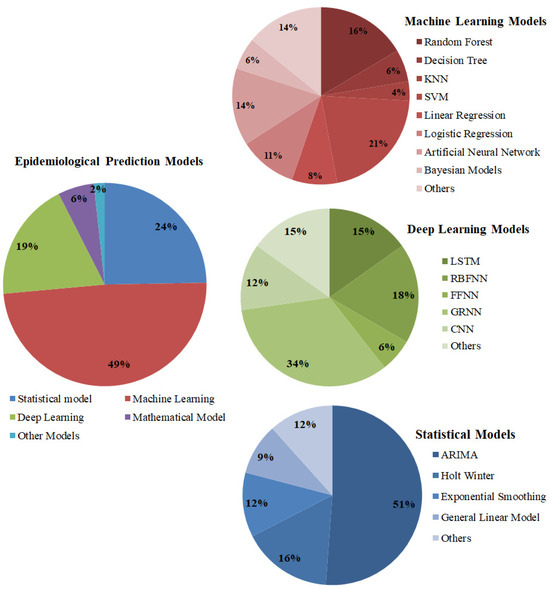
Figure 5.
The significance of epidemiological prediction models from 2018 to 2022.
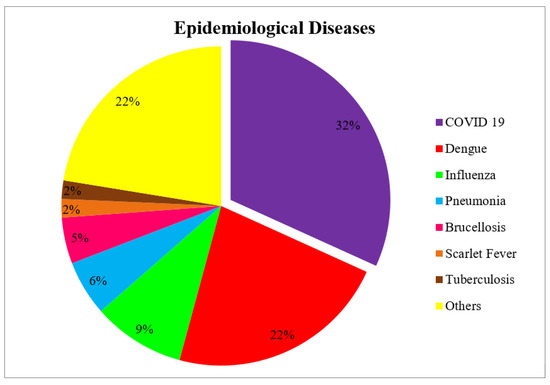
Figure 6.
Epidemiological diseases engaged over this period.
3.1.1. Discussion on Mathematical Models
Mathematical models like Suspected–Exposed–Infectious–Recovery (SEIR), SR, SIR, etc., which are also called compartmental modeling as well as state-space modeling, are used in most of the research works. These models are reliable to use and they can add more states and parameters as required. Some of the limitations are as follows: there is an inconsistency between clinical insights and modeling efforts; the parameter selection must be carried out cautiously; sometimes the influential factors cannot be included; it is complex to provide assumptions on data grouping before modeling; the new evidence are cases may not be taken into account; and treats all infected (I) cases alike, but their clinical insights may differ and they may create a great impact on spreading.
3.1.2. Discussion on Statistical Models
The next commonly used models in the literature are statistical models such as Exponential Smoothing (ES), Auto-Regressive Integrated Moving Average (ARIMA), Holt–Winters (HW), Moving Average, Naïve method, etc. These models use statistical features for modeling the epidemic time series data. These models are the baseline models in this domain. It is easy for everyone to adapt them to the problem domain.
- The ARIMA model is well suited for linear forecasting and provides insights into the linear relationship between the predictors and responses. It is also well suited for stationary time series data. As far as in the literature, the ARIMA model has positive traces in most of the prediction problems.
- Similarly, Exponential Smoothing also has good feedback from various researchers. The distinct nature of ES is that it will not discard past insights; it will gradually decrease them based on their degree of influence. It is good for short-term predictions and the only limitation is that it will lag in tracing out the data transitions.
- The Holt–Winters model is well suited for discovering trends and seasonal patterns. As a whole, these models are used for linear prediction problems either for the short term or the long term with past historical evidence.
The strengths and limitations of these statistical models are as follows:
Strength: The overall strength of these models is well suited for univariate forecasting, providing robust performance for linear trends and seasonal patterns. They are interpretable, require relatively low computational power, and are effective when dealing with stationary data.
Limitations: ARIMA, Holt–Winters, and Exponential Smoothing are less effective for capturing non-linear relationships, multivariate dynamics, or sudden spikes, which are often encountered in real-world epidemiological data. They also require assumptions of stationarity, which may not hold in disease forecasting scenarios. The other limitations include sudden data instability, fluctuation resistance, unexpected rapid data flooding, minimal historic data, etc.
3.1.3. Discussion on Intelligent Computing Models
The literature evidences that these complexities in prediction problems can be handled by intelligent computing models like machine learning and deep learning models. They can explore non-linear associations among the data, handle non-stationary time series data, and take the rapid epidemic proliferation into account. The various machine learning approaches used for epidemic prediction in the literature are Random Forest, Decision Tree, Support Vector Machine (SVM), Support Vector Regression (SVR), Long- and Short-term Memory (LSTM), Linear Regression, Logistic Regression (LR), Bayesian time series model, etc. These models are good for long-term and short-term predictions. In addition, deep learning models are also used for this problem, some of them are Generalized Regression Neural Networks (GRNN), Convolution Neural Networks, LSTM, Radial Basis Function Neural Networks (RBFNN), Feed Forward Neural Networks (FFNN), Non-Linear Auto-Regressive Neural Networks (NARNN), etc. Among these models, GRNN and LR are most commonly used for medical applications. The various significances of GRNN are as follows: the fastest model that quickly converges; a simple parameter fixing process; possesses great stability; and provides accurate approximations on non-linear data. As this model stores all the samples that cause spatial complexity not in any specific order, the next model in the discussion is LSTM. The researchers widely used this model for long-term predictions and continuous and non-stationary data.
The strengths and limitations of these intelligent computing models are as follows:
Strength: Decision Trees and Random Forests bring the power of non-linearity and are highly useful for capturing complex relationships, while also handling large datasets efficiently. Models like SVM and KNN excel in classification tasks, where they can distinguish between infected and non-infected cases based on multidimensional features. Artificial Neural Networks (ANNs), especially advanced forms like LSTMs and CNNs, are powerful for learning intricate patterns, long-term dependencies, and spatial–temporal features, making them valuable for predicting disease outbreaks.
Limitations: Decision Trees and Random Forests, though flexible, can become prone to overfitting if not properly tuned, and lack interpretability as model complexity increases. SVMs and KNN, although robust, become computationally expensive with large datasets, and KNN is sensitive to the choice of distance metrics and the curse of dimensionality. While ANNs, particularly LSTMs, CNNs, and other deep learning models, are powerful, they require large volumes of data, extensive computational resources, and hyperparameter tuning, making them challenging to implement in resource-constrained settings. Additionally, these deep learning models are often black boxes, posing challenges in interpretation, which is a key requirement in public health decision-making.
3.1.4. Discussion on Practical Applicability of the Prediction Models
In practice, the choice of model for epidemiological disease prediction depends on the data characteristics, the prediction horizon, and the specific requirements of the application.
For short-term forecasts with clear seasonal trends, ARIMA and Exponential Smoothing models are commonly deployed, especially when interpretability and rapid implementation are prioritized. In scenarios with limited data or where linear relationships dominate, General Linear Models, Linear Regression, or Logistic Regression remain practical choices. For complex, non-linear interactions, Decision Trees, Random Forests, and Bayesian Models are suitable due to their flexibility and ability to handle uncertainty. When the goal is to classify disease states or predict outbreaks with high accuracy, SVM, KNN, and ANN-based models like FFNNs and RBFNNs are preferred, especially when sufficient labeled data are available. LSTM and GRNN models are particularly applicable when dealing with time series data with long-range dependencies, while CNNs shine in applications where spatial patterns play a critical role, such as mapping disease spread geographically. Ultimately, model selection should balance complexity, interpretability, computational requirements, and specific epidemiological goals, often leading to a combination of models or hybrid approaches for more robust predictions.
3.1.5. Discussion on Performance These Models Under Epidemiological Context
The performance of various predictive models in epidemiological contexts such as COVID-19, dengue, influenza, pneumonia, brucellosis, scarlet fever, and tuberculosis is influenced by the nature and characteristics of each disease.
Time series models like ARIMA, Exponential Smoothing, and Holt–Winters have been effectively used in diseases with predictable seasonal patterns, such as Influenza and Dengue. These models excel in short-term forecasting where the data follows a linear trend with periodic cycles. However, in cases like COVID-19, where transmission dynamics are influenced by rapidly changing factors like government interventions, mutations, and population behavior, these traditional models tend to struggle. Machine learning models such as Random Forests, Decision Trees, and SVMs have proven more versatile in capturing the complex, non-linear relationships in COVID-19 and Tuberculosis data, where multiple demographic, environmental, and clinical factors interact. Deep learning models, especially LSTMs and CNNs, have shown superior performance in predicting outbreaks by leveraging the temporal and spatial aspects of diseases like scarlet fever and pneumonia. The long memory capabilities of LSTMs make them effective for modeling diseases with varying incubation periods and prolonged transmission cycles.
However, each model’s performance is constrained by data availability, quality, and the inherent complexity of the disease being modeled. For instance, while ARIMA models work well for diseases with consistent patterns, they perform poorly in emerging outbreaks like COVID-19, where data are sparse, non-stationary, and affected by external shocks. Logistic Regression and Decision Trees may perform adequately in classifying disease presence in simpler conditions like brucellosis but struggle with large-scale, noisy data characteristics of more widespread diseases like COVID-19. Deep learning models such as CNNs and LSTMs are powerful for complex scenarios but require extensive data and computational resources, which may not be feasible in all contexts, particularly in low-resource settings typical for diseases like tuberculosis. Additionally, these models often function as black boxes, limiting their interpretability—a critical factor for public health interventions. As such, the choice of model must align with the specific epidemiological context, balancing predictive accuracy with practical considerations such as data availability, computational costs, and the need for interpretability in decision-making.
3.2. Implications for Public Health and Policy
The current study focuses on various epidemiological predictions using versatile models. These predictions can be used for implications of various public health and policy. They are as follows:
- Enhanced Public Health Infrastructure Resilience: Enables government to enhance infrastructure, create contingency plans, and develop mobile healthcare units to face outbreaks.
- Tailored Public Health Interventions: The government can target interventions in areas where outbreaks are most likely to occur, reducing unnecessary disruptions in regions that are less affected.
- Improving Vaccination and Treatment Strategies: Models could help optimize the supply chain for vaccines by predicting where and when they will be needed the most, thus reducing vaccine wastage and improving immunization rates in critical areas.
- Health Equity and Addressing Vulnerable Populations: Predictive models can highlight vulnerable populations, such as those in lower-income or rural areas, who are at greater risk of infection due to limited access to healthcare. Governments and international organizations could leverage these insights to proactively deploy resources and healthcare services to these underserved areas.
- Cross-border Collaboration and Global Health Governance: These models can also foster better coordination between international organizations like the World Health Organization (WHO) and local governments, leading to synchronized efforts in disease surveillance, resource allocation, and vaccination campaigns.
- Economic Implications and Minimizing Disruption: Governments can use these models to minimize the economic fallout of pandemics by implementing targeted interventions that avoid unnecessary nationwide lockdowns. Accurate predictions can allow businesses to plan for disruptions in advance, enabling smoother supply chain operations and reducing economic losses.
- Monitoring Disease Evolution and Mutation Tracking: Predictive models can help scientists and policymakers anticipate how mutations might spread and affect different populations. This foresight would assist in planning for variant-specific responses, including tailored vaccine development and public health strategies.
- Strengthening Behavioral and Social Policy Interventions: These models can inform behavioral and social interventions, such as public health campaigns to promote hygiene, social distancing, or mask-wearing.
- Environmental and Climate-based Health Planning: As climate change continues to influence the spread of diseases, predictive models can integrate environmental factors, such as temperature, humidity, and precipitation, to forecast outbreaks more accurately.
- Preparedness for Future Pandemics: By forecasting potential outbreaks years in advance, these models enable governments and health organizations to prepare stockpiles of essential medical supplies, develop healthcare infrastructure, and implement public health protocols before a pandemic strikes.
- Reducing Burden on Healthcare Systems: Predictive models can alleviate the burden on healthcare systems by optimizing the allocation of resources during peak outbreak periods.
- Promoting Innovation in Healthcare Technologies: The use of predictive models can drive innovation in healthcare technologies. From AI-powered diagnostics to telemedicine platforms, the ability to forecast disease patterns will inspire the development of new tools that improve patient outcomes and streamline healthcare processes.
3.3. Future Research Directions
In the future, the epidemiological prediction models can focus on improving the efficiency and accuracy of the ML and DL techniques. More sophisticated models can be proposed with hybrid DL architectures, i.e., the combination of models like LSTM, CNN, and GRNN can be explored. These advancements in DL techniques to handle non-linear, complex, and high-dimensional epidemiological data may be critical for improving the predictive outcomes. It can be enhanced with real-time data and can include various environmental factors to predict the outbreaks with greater accuracy and even more for longer intervals.
In the future, the integration of real-time data from diverse resources like satellite images, social media, and health records could provide more granular insights about disease patterns. The researchers can also focus on integrating other data like genomic data and wearable device data into these prediction models. It is also necessary to investigate data privacy concerns and optimize the use of anonymized health data for implementing more scalable and practical models.
Currently, the epidemiological predictions on various diseases like COVID-19, dengue, etc., are concentrated. In the future, more generalized models have to be developed to adapt to novel pathogens and epidemics. It will also handle the unexpected outbreaks of emerging diseases. These future models can be developed to predict outbreaks under various geographies, socio-economic conditions, and climates to enhance the efficiency of the public health response.
4. Conclusions
This review article studies various epidemiological prediction models over the period of 2016 to 2022. It evidences that there is a surge in prediction techniques from 2018 onwards. Around 100 research articles are studied and their significances are summarized. The discussions about these existing research articles are explored in a concise manner. The advantages and limitations of the existing models under different categories are emphasized. Additionally, the performance and limitations of various prediction models in epidemiological contexts are also provided. This article will be one of the supportive resources for researchers of diverse fields. The various possible implications of various epidemiological predictions are discussed. The future research directions in this field are discussed in detail to direct future researchers in this context.
Author Contributions
Conceptualization, G.R. and R.R.; methodology, G.R.; validation, G.R., R.R. and D.S.; formal analysis, G.R. and R.R.; investigation, G.R.; writing—original draft preparation, G.R. and R.R.; writing—review and editing, G.R., R.R. and A.M.; supervision, R.R. and A.A.A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bertozzi, A.L.; Franco, E.; Mohler, G.; Short, M.B.; Sledge, D. The challenges of modeling and forecasting the spread of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 16732–16738. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Kumar, H.; Panigrahi, B.K. Prediction and analysis of COVID-19 positive cases using deep learning models: A descriptive case study of India. Chaos Solitons Fractals 2020, 139, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Basukoski, A. Machine Learning-Based Approaches for Location Based Dengue Prediction: Review. In Proceedings of the Fourth International Congress on information and Communication Technology, Singapore, 4–6 December 2020; pp. 343–352. [Google Scholar]
- Rachah, A.; Torres, D.F.M. Analysis, simulation and optimal control of a SEIR model for Ebola virus with demographic effects. Commun. Fac. Sci. Univ. Ank. Ser. A1 Math. Stat. 2018, 67, 179–197. [Google Scholar]
- Garcia, K.K.S.; Abrahão, A.A.; Oliveira, A.F.d.M.; Henriques, K.M.d.D.; Pina-Costa, A.d.; Siqueira, A.M.; Ramalho, W.M. Malaria time series in the extra-Amazon region of Brazil: Epidemiological scenario and a two-year prediction model. Malar. J. 2022, 21, 1–11. [Google Scholar] [CrossRef]
- Wang, G.; Wei, W.; Jiang, J.; Ning, C.; Chen, H.; Huang, J.; Liang, B.; Zang, N.; Liao, Y.; Chen, R.; et al. Application of a long short-term memory neural network: A burgeoning method of deep learning in forecasting HIV incidence in Guangxi, China. Epidemiol. Infect. 2019, 147, e194. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, S.; Wang, Z.; Yang, L.; Zhu, Y.; Yuan, J. Temporal trends analysis of tuberculosis morbidity in mainland China from 1997 to 2025 using a new SARIMA-NARNNX hybrid model. Infect. Dis. Res. 2020, 9, e024409. [Google Scholar] [CrossRef]
- Chen, S.; Xu, J.; Wu, Y.; Wang, X.; Fang, S.; Cheng, J.; Ma, H.; Zhang, R.; Liu, Y.; Zhang, L.; et al. Predicting temporal propagation of seasonal influenza using improved Gaussian Process Model. J. Biomed. Inform. 2019, 193, 103144. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, V.; Pal, S. Application of machine learning time series analysis for prediction COVID-19 pandemic. Res. Biomed. Eng. 2022, 38, 35–47. [Google Scholar] [CrossRef]
- Qu, S.; Zhou, M.; Jiao, S.; Zhang, Z.; Xue, K.; Long, J.; Zha, F.; Chen, Y.; Li, J.; Yang, Q.; et al. Optimizing acute stroke outcome prediction models: Comparison of generalized regression neural networks and logistic regressions. PLoS ONE 2022, 17, e0267747. [Google Scholar] [CrossRef]
- Katris, C. A time series-based statistical approach for outbreak spread forecasting: Application of COVID-19 in Greece. Expert Syst. Appl. 2020, 166, 114077. [Google Scholar] [CrossRef]
- Lamia, A.; Fawssaz, A. Detection of pneumonia infection by using deep learning on a mobile platform. Comput. Intell. Neuro Sci. 2022, 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Satrio, C.B.A.; Darmawan, W.; Nadia, B.U.; Hanafiah, N. Time series analysis and forecasting of coronavirus disease in Indonesia using ARIMA model and PROPHET. Procedia Comput. Sci. 2021, 179, 432–524. [Google Scholar]
- Kalantari, M. Forecasting COVID-19 pandemic using optimal singular spectrum analysis. Chaos Solitons Fractals 2020, 142, 110547. [Google Scholar] [CrossRef] [PubMed]
- Ceylan, Z. Short-term prediction of COVID-19 spread using grey rolling model optimized by particle swarm optimization. Appl. Soft Comput. 2021, 109, 107592. [Google Scholar] [CrossRef]
- Triacca, M.; Triacca, U. Forecasting the number of confirmed new cases of COVID-19 in Italy for the period from 19 May to 2 June 2020. Infect. Dis. Model. 2021, 6, 362–369. [Google Scholar] [CrossRef]
- Zhai, M.; Li, W.; Tie, P.; Wang, X.; Xie, T.; Ren, H.; Zhang, Z.; Song, W.; D Quan, M.L.; Chen, L.; et al. Research on the predictive effect of a combined model of Arima and neural networks on human brucellosis in Shanxi Province, China: A time series predictive analysis. BMC Infect. Dis. 2021, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Abbasimehr, H.; Paki, R. Prediction of COVID-19 confirmed cases combining deep learning methods and Bayesian optimization. Chaos Solitons Fractals 2021, 142, 110511. [Google Scholar] [CrossRef]
- Wang, G.; Wu, T.; Wei, W.; Jiang, J.; An, S.; Liang, B.; Ye, L.; Liang, H. Comparison of ARIMA, ES, GRNN and ARIMA–GRNN hybrid models to forecast the second wave of COVID-19 in India and the United States. Epidemiol. Infect. 2021, 149, e240. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Li, D.-C. Selection of key features for PM2.5 prediction using a wavelet model and RBF-LSTM. Appl. Intell. 2021, 51, 2534–2555. [Google Scholar] [CrossRef]
- Suvarna, B.; Nandipati, B.L.; Bhat, M.N. Support Vector Regression fot Predicting COVID-19 Cases. Eur. J. Mol. Clin. Med. 2021, 7, 4882–4893. [Google Scholar]
- González-Pérez, B.; Núñez, C.; Sánchez, J.L.; Valverde, G.; Velasco, J.M. Expert System to Model and Forecast Time Series of Epidemiological Counts with Applications to COVID-19. Mathematics 2021, 9, 1485. [Google Scholar] [CrossRef]
- Watson, G.L.; Xiong, D.; Zhang, L.; Zoller, J.A.; Shamshoian, J.; Sundin, P.; Bufford, T.; Rimoin, A.W.; Suchard, M.A.; Ramirez, C.M. Pandemic velocity: Forecasting COVID-19 in the US with a machine learning & Bayesian time series compartmental model. PLoS Comput. Biol. 2021, 17, e1008837. [Google Scholar]
- Silitonga, P.; Bustamam, A.; Muradi, H.; Mangunwardoyo, W.; Dewi, B.E. Comparison of Dengue Predictive Models Developed Using Artificial Neural Network and Discriminant Analysis with Small Dataset. Appl. Sci. 2021, 11, 943. [Google Scholar] [CrossRef]
- Aiken, E.L.; Nguyen, A.T.; Viboud, C.; Santillana, M. Toward the use of neural networks for influenza prediction at multiple spatial resolutions. Sci. Adv. 2021, 7, eabb1237. [Google Scholar] [CrossRef]
- Mohammadi, Y.; Parsaeian, M.; Farzadfar, F.; Kasaeian, A.; Mehdipour, P.; Sheidaei, A.; Mansouri, A.; Moghaddam, S.S.; Djalalinia, S.; Mahmoudi, M.; et al. Levels and trends of child and adult mortality rates in the Islamic Republic of Iran, 1990-2013; protocol of the NASBOD study. Arch. Iran Med. 2014, 17, 176–181. [Google Scholar] [PubMed]
- Dhamodharavadhani, S.; Rathipriya, R. Computational Intelligence Based Hybrid Hyperparameter Tuned Prediction Techniques for COVID-19 Epidemiological Data. In Understanding COVID-19: The Role of Computational Intelligence. Studies in Computational Intelligence; Springer: Cham, Switzerland, 2021; Volume 963. [Google Scholar]
- Huang, S.W.; Tsai, H.P.; Hung, S.J.; Ko, W.C.; Wang, J.R. Assessing the risk of dengue severity using demographic information and laboratory test results with machine learning. PLoS Neglected Trop. Dis. 2020, 14, e0008960. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zeng, Z.; Wang, K.; Wong, S.-S.; Liang, W.; Zanin, M.; Liu, P.; Cao, X.; Gao, Z.; Mai, Z.; et al. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J. Thorac. Dis. 2020, 12, 165–174. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Peng, Y.; Sun, K. SEIR modeling of the COVID-19 and its dynamics. Non-Linear Dyn. 2020, 101, 1667–1680. [Google Scholar] [CrossRef]
- Prasse, B.; Achterberg, M.A.; Ma, L.; Mieghem, P.V. Network-inference-based prediction of the COVID-19 epidemic outbreak in the Chinese province Hubei. Appl. Netw. Sci. 2020, 5, 35. [Google Scholar] [CrossRef]
- ChandraDas, R. Forecasting incidences of COVID-19 using Box-Jenkins method for the period July 12-Septembert 11, 2020: A study on highly affected countries. Chaos Solitons Fractals 2020, 140, 110248. [Google Scholar]
- Petropoulos, F.; Makridakis, S. Forecasting the novel coronavirus COVID-19. PLoS ONE 2020, 15, e0231236. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Caballero, S.; Selles, M.A.; Peydro, M.A.; Perez-Bernabeu, E. An Efficient COVID-19 Prediction Model Validated with the Cases of China, Italy and Spain: Total or Partial Lockdowns? J. Clin. Med. 2020, 9, 1547. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Xianbin, L.; Houquiang, L. Prediction of new coronavirus infection based on a modified SEIR model. medRxiv 2020, 1–13. [Google Scholar] [CrossRef]
- Tsallis, C.; Tirnakli, U. Predicting COVID-19 Peaks Around the World. Front. Phys. 2020, 8, 217. [Google Scholar] [CrossRef]
- Singh, R.K.; Rani, M.; Bhagavathula, A.S.; Sah, R.; Rodriguez-Morales, A.J.; Kalita, H.; Nanda, C.; Sharma, S.; Sharma, Y.D.; Rabaan, A.A.; et al. Prediction of the COVID-19 Pandemic for the Top 15 Affected Countries: Advanced Autoregressive Integrated Moving Average (ARIMA) Model. JMIR Public Health Surviallance 2020, 6, e19115. [Google Scholar] [CrossRef]
- Jia, W.; Han, K.; Song, Y.; Cao, W.; Wang, S.; Yang, S.; Wang, J.; Kou, F.; Tai, P.; Li, J. Extended SIR prediction of the epidemics trend of COVID-19 in Italy and compared with Hunan, China. Front. Med. 2020, 7, 169. [Google Scholar]
- Zhu, G.; Li, J.; Meng, Z.; Yu, Y.; Li, Y.; Tang, X.; Dong, Y.; Sun, G.; Zhou, R.; Wang, H.; et al. Learning from Large-Scale Wearable Device Data for Predicting the Epidemic Trend of COVID-19. Discret. Dyn. Nat. Soc. 2020, 2020, 6152041. [Google Scholar] [CrossRef]
- Chowdhury, R.; Heng, K.; Shawon, M.S.R.; Okonofua, G.G.; Ochoa-Rosales, C.; Gonzalez-Jaramillo, V.; Bhuiya, A.; Reidpath, D.; Prathapan, S.; Shahzad, S.; et al. Dynamic interventions to control COVID-19 pandemic: A multivariate prediction modelling study comparing 16 worldwide countries. Eur. J. Epidemiol. 2020, 35, 389–399. [Google Scholar] [CrossRef]
- Ayyoubzadeh, S.M.; Ayyoubzadeh, S.M.; Zahedi, H.; Ahmadi, M.; Kalhori, S.R.N. Predicting COVID-19 Incidence Through Analysis of Google Trends Data in Iran: Data Mining and Deep Learning Pilot Study. JMIR Public Health Surviallence 2020, 6, e18828. [Google Scholar] [CrossRef]
- Wang, S.; Zha, Y.; Li, W.; Wu, Q.; Li, X.; Niu, M.; Wang, M.; Qiu, X.; Li, H.; Yu, H.; et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020, 56, 2000775. [Google Scholar] [CrossRef]
- Zeroual, A.; Harrou, F.; Dairi, A.; Sun, Y. Deep learning methods for forecasting COVID-19 time-Series data: A Comparative study. Chaos Solitons Fractals 2020, 140, 110121. [Google Scholar] [CrossRef] [PubMed]
- Rahmadani, F.; Lee, H. Hybrid Deep Learning-Based Epidemic Prediction Framework of COVID-19: South Korea Case. Appl. Sci. 2020, 10, 8539. [Google Scholar] [CrossRef]
- Muhammad, L.J.; Algehyne, E.A.; Usman, S.S.; Ahmad, A.; Chakraborty, C.; Mohammed, I.A. Supervised Machine Learning Models for Prediction of COVID-19 Infection using Epidemiology Dataset. SN Comput. Sci. 2020, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, W.; Zhang, S.-Y.; Zhang, K.-Q.; Li, J.; Liu, Y.; Yin, Z.-H. Dental Caries Prediction Based on a Survey of the Oral Health Epidemiology among the Geriatric Residents of Liaoning, China. BioMed Res. Int. 2020, 2020, 5348730. [Google Scholar] [CrossRef]
- Hadi Bagheri, L.T.; Karami, M.; Hosseinkhani, Z.; Najari, H.; Karimi, S.; Cheraghi, Z. Forecasting the monthly incidence rate of brucellosis in west of Iran using time series and data mining from 2010 to 2019. PLoS ONE 2020, 15, e0232910. [Google Scholar] [CrossRef]
- Sulasikin, A.; Nugraha, Y.; Kanggrawan, J.; Suherman, A.L. Forecasting for a data-driven policy using time series methods in handling COVID-19 pandemic in Jakarta. In Proceedings of the 2020 IEEE International Smart Cities Conference (ISC2), Piscataway, NJ, USA, 28 September–1 October 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Pravin, A.; Jacob, T.P.; Nagarajan, G. An intelligent and secure healthcare framework for the prediction and prevention of Dengue virus outbreak using fog computing. Health Technol. 2020, 10, 303–311. [Google Scholar] [CrossRef]
- Sood, S.K.; Kaur, S.; Chahal, K.K. An intelligent framework for monitoring dengue fever risk using LDA-ANFIS. J. Ambient Intell. Smart Environ. 2020, 12, 5–20. [Google Scholar] [CrossRef]
- Zhao, N.; Charland, K.; Carabali, M.; Nsoesie, E.O.; Maheu-Giroux, M.; Rees, E.; Yuan, M.; Garcia Balaguera, C.; Jaramillo Ramirez, G.; Zinszer, K. Machine learning and dengue forecasting: Comparing random forests and artificial neural networks for predicting dengue burdens at the national sub-national scale in Colombia. PLoS Neglected Trop. Dis. 2020, 14, e0008056. [Google Scholar] [CrossRef]
- Mussumeci, E.; Coelho, F.C. Machine-learning forecasting for Dengue epidemics-Comparing LSTM, Random Forest and Lasso regression. medRxiv 2020, 1–17. [Google Scholar] [CrossRef]
- Yang, C.T.; Chen, Y.A.; Chan, Y.W.; Lee, C.L.; Tsan, Y.T.; Chan, W.C.; Liu, P.Y. Influenza-like illness prediction using a long short-term memory deep learning model with multiple Open Data Sources. J. Supercomput. 2020, 76, 9303–9329. [Google Scholar] [CrossRef]
- Harumy, T.H.F.; Chan, H.Y.; Sodhy, G.C. Prediction for Dengue Fever in Indonesia Using Neural Network and Regression Method. J. Phys. Conf. Ser. 2020, 1566, 012019. [Google Scholar] [CrossRef]
- Dourjoy, S.; Rafi, A.; Tumpa, Z.N.; Saifuzzaman, M. A Comparative Study on Prediction of Dengue Fever Using Machine Learning Algorithm. In Advances in Distributed Computing and Machine Learning; Springer: Singapore, 2020; pp. 501–510. [Google Scholar]
- Salami, B.; do Sousa, C.A.M.; Martins, R.; Capinha, C. Dengue importation into Europe: A network connectivity-based approach. PLoS ONE 2020, 15, e0230274. [Google Scholar] [CrossRef]
- Darwish, A.; Rahhal, Y.; Jafar, A. A comparative study on predicting influenza outbreaks using different feature spaces: Application of influenza-like illness data from early warning alert and response system in Syria. BMC Res. Notes 2020, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Poonawala-Lohani, N.; Riddle, P.; Adnan, M.; Wicker, J. A Novel Approach for Time Series Forecasting of Influenza-like Illness Using a Regression Chain Method. Pac. Symp. Biocomput. 2020, 301–312. [Google Scholar] [CrossRef]
- Nordin, N.; Sobri, N.; Ismail, N.; Zulkifli, S.N.; Razak, N.; Mahmud, M. The Classification Performance using Support Vector Machine for ndemic Dengue Cases. J. Phys. Conf. Ser. 2020, 1496, 120–129. [Google Scholar] [CrossRef]
- Adeyinka, D.A.; Muhajarin, N. Time series prediction of under-five mortality rates for Nigeria: Comparative analysis of artificial neural networks, Holt-Winters exponential smoothing and autoregressive integrated moving average models. BMC Med. Res. Methodol. 2020, 20, 292. [Google Scholar] [CrossRef]
- Elsmih, F.E.; Abdelaziz, G.M.M.; Salemalzahrani; Shokeralla, A.A.A. Prediction the daily number of confirmed cases of COVID-19 in Sudan with Arima and holt winter exponential smoothing. Int. J. Dev. Res. 2020, 10, 39408–39413. [Google Scholar]
- Zou, J.-J.; Jiang, G.-F.; Xie, X.-X.; Huang, J.; Yang, X.-B. Application of a combined model with seasonal autoregressive integrated moving average and support vector regression in forecasting hand-foot-mouth disease incidence in Wuhan, China. Medicine 2019, 98, e14195. [Google Scholar] [CrossRef]
- Zhang, C.; Fu, X.; Zhang, Y.; Nie, C.; Li, L.; Cao, H.; Wang, J.; Wang, B.; Yi, S.; Ye, Z. Epidemiological and time series analysis of haemorrhagic fever with renal syndrome from 2004 to 2017 in Shandong Province, China. Sci. Rep. 2019, 9, 14644. [Google Scholar] [CrossRef]
- Chakraborty, T.; Chattopadhyay, S.; Ghosh, I. Forecasting dengue epidemics using a hybrid methodology. Phys. A Stat. Mech. Its Appl. 2019, 527, 121–136. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Wang, Z.; Yuan, J. Seasonality and trend prediction of scarlet fever incidence in mainland China from 2004 to 2018 using a hybrid sarima-NARX model. PeerJ 2019, 7, e6165. [Google Scholar] [CrossRef]
- Srivastava, S.; Soman, S.; Rai, A.; Cheema, A.S. An Online Learning Approach for Dengue Fever Classification. In Proceedings of the IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020. [Google Scholar]
- Devi, C.; Pastor, A.; Oliveira, T.; Neto, F.; Braga-Neto, U.; Bigham, A.; Bigham, A.; Bamshad, M.; Marques, E. Severe Dengue Prognosis Using Human Genome Data and Machine Learning. IEEE Trans. Biomed. Eng. 2019, 66, 2861–2868. [Google Scholar] [CrossRef] [PubMed]
- Mello-Román, J.D.; Mello-Román, J.C.; Gómez-Guerrero, S.; García-Torres, M. Predictive Models for the Medical Diagnosis of Dengue: A Case Study in Paraguay. Comput. Math. Methods Med. 2019, 2019, 7307803. [Google Scholar] [CrossRef] [PubMed]
- Raja, B.; Mallol, R.; Ting, C.Y.; Kamaludin, F.; Ahmad, R.; Ismail, S.; Jayaraj, V.J.; Sundram, B.M. Artificial Intelligence Model as Predictor for Dengue Outbreaks. Malays. J. Public Health Med. 2019, 19, 103–108. [Google Scholar] [CrossRef]
- Ge, Y.; Wang, Q.; Wang, L.; Wu, H.; Peng, C.; Wang, J.; Xu, Y.; Xiong, G.; Zhang, Y.; Yi, Y. Predicting post-stroke pneumonia using deep neural network approaches. Int. J. Med. Inform. 2019, 132, 103986–103996. [Google Scholar] [CrossRef]
- Wu, W.; An, S.Y.; Guan, P.; Huang, D.S.; Zhou, B.S. Time series analysis of human brucellosis in mainland China by using Elman and Jordan Recurrent Neural Networks. BMC Infect. Dis. 2019, 19, 414. [Google Scholar] [CrossRef]
- Iqbal, N.; Islam, M. Machine learning for dengue outbreak prediction: A performance evaluation of different prominent classifiers. Informatica 2019, 43, 1548. [Google Scholar] [CrossRef]
- Tapak, L.; Hamidi, O.; Fathian, M.; Karami, M. Comparative evaluation of time series models for predicting influenza outbreaks: Application of influenza-like illness data from Sentinel Sites of Healthcare Centers in Iran. BMC Res. Notes 2019, 12, 353. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, Q.; Chen, Y.; Xiao, J.; He, J.; Zhang, Y.; Wang, L.; Liu, T.; Ma, W. An ensemble forecast model of dengue in Guangzhou, China using climate and social media surveillance data. Sci. Total Environ. 2019, 647, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Sontisirikit, S.; Iamsirithaworn, S.; Prendinger, H. Prediction of dengue outbreaks based on disease surveillance, meteorological and socio-economic data. BMC Infect. Dis. 2019, 19, 272–280. [Google Scholar] [CrossRef]
- Ruchiraset, A.; Tantrakarnapa, K.A. Time series modeling of pneumonia admissions and its association with air pollution and climate variables in Chiang Mai Province, Thailand. Environ. Sci. Pollut. Res. 2018, 25, 33277–33285. [Google Scholar] [CrossRef]
- Wang, H.; Tian, C.W.; Wang, W.M.; Luo, X.M. Time-series analysis of tuberculosis from 2005 to 2017 in China. Epidemiol. Infect. 2018, 146, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, A.; Ibrahim, M.S.; Usman, M.; Zubair, M.; Khan, S. Chaotic Time Series Prediction using Spatio-Temporal RBF Neural Networks. In Proceedings of the 2018 3rd International Conference on Emerging Trends in Engineering, Sciences and Technology (ICEEST), Karachi, Pakistan, 21–22 December 2018. [Google Scholar]
- Tapak, L.; Shirmohammadi-Khorram, N.; Hamidi, O.; Maryanaji, Z. Predicting the Frequency of Human Brucellosis using Climatic Indices by Three Data Mining Techniques of Radial Basis Function, Multilayer Perceptron and Nearest Neighbor: A Comparative Study. Iran. J. Epidemioligy 2018, 14, 153–165. [Google Scholar]
- Ong, J.; Liu, X.; Rajarethinam, J.; Kok, S.; Liang, S.; Tang, C.; Cook, A.; Ng, L.; Yap, G. Mapping dengue risk in Singapore using Random Forest. PLoS Neglected Trop. Dis. 2018, 12, e0006587. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, Y.; Nishiura, H.; Saitoh, M. Deep Learning for Epidemiological Predictions. In Proceedings of the The 41st International ACM SIGIR Conference on Research & Development in Information Retrieval, Ann Arbor, MI, USA, 8–12 July 2018; pp. 1085–1088. [Google Scholar]
- Thorve, S.; Wilson, M.L.; Lewis, B.L.; Swarup, S.; Kumar, A.; Vullikanti, S.; Marathe, M.V. EpiViewer: An epidemiological application for exploring time series data. BMC Bioinform. 2018, 19, 449. [Google Scholar] [CrossRef] [PubMed]
- Baquero, O.S.; Santana, L.M.R.; Chiaravalloti-Neto, F. Dengue forecasting in São Paulo city with generalized additive models, artificial neural networks and seasonal autoregressive integrated moving average models. PLoS ONE 2018, 13, e0195065. [Google Scholar] [CrossRef]
- Li, H.; Luo, M.; Zheng, J.; Luo, J.; Zeng, R.; Feng, N.; Du, Q.; Fang, J. An artificial neural network prediction model of congenital heart disease based on risk factors. Medicine 2017, 96, e6090. [Google Scholar] [CrossRef]
- Caicedo-Torres, W.; Montes-Grajales, D.; Miranda-Castro, W.; Fennix-Agudelo, M.; Agudelo-Herrera, N. Kernel-Based Machine Learning Models for the Prediction of Dengue and Chikungunya Morbidity in Colombia. In Advances in Computing; Springer: Cham, Switzerland, 2017; pp. 472–484. [Google Scholar]
- He, F.; Hu, Z.-J.; Zhang, W.-C.; Cai, L.; Cai, G.-X.; Aoyagi, K. Construction and evaluation of two computational models for predicting the incidence of influenza in Nagasaki Prefecture, Japan. Sci. Rep. 2017, 7, 7192. [Google Scholar] [CrossRef]
- Goli, S.; Mahjub, H.; Faradmal, J.; Mashayekhi, H.; Soltanian, A.-R. Survival Prediction and Feature Selection in Patients with Breast Cancer Using Support Vector Regression. Comput. Math. Methods Med. 2016, 2016, 2157984. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, D.; Huang, G.; Xia, J.; Wang, X.; Zhang, Y.; Tang, W.; Zhou, H. Time series analysis of temporal trends in the pertussis incidence in mainland China from 2005 to 2016. Sci. Rep. 2016, 6, 32367. [Google Scholar] [CrossRef]
- Caicedo-Torres, W.; Paternina, Á.; Pinzón, H. Machine Learning Models for Early Dengue Severity Prediction. In Advances in Artificial Intelligence—IBERAMIA 2016; Springer: Cham, Switzerland, 2016; pp. 247–258. [Google Scholar]
- Song, Y.; Wang, F.; Wang, B.; Tao, S.; Zhang, H.; Liu, S.; Ramirez, O.; Zeng, Q. Time Series Analyses of Hand, Foot and Mouth Disease Integrating Weather Variables. PLoS ONE 2015, 10, e0117296. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Yang, M.; Zhang, T.; Young, A.A.; Li, X. Comparative Study of Four Time Series Methods in Forecasting Typhoid Fever Incidence in China. PLoS ONE 2013, 8, e63116. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, K.; Gasparrini, A.; Hajat, S.; Smeeth, L.; Armstrong, B. Time series regression studies in environmental epidemiology. Int. J. Epidemiol. 2013, 42, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, X.; Jiang, B.; Yang, W. Forecasting incidence of hemorrhagic fever with renal syndrome in China using ARIMA model. BMC Infect. Dis. 2011, 11, 218. [Google Scholar] [CrossRef] [PubMed]
- Luz, P.M.; Mendes, B.V.M.; Codeço, C.T.; Struchiner, C.J.; Galvani, A.P. Time series analysis of dengue incidence in Rio de Janeiro, Brazil. Am. J. Trop. Med. Hygine 2008, 79, 933–939. [Google Scholar] [CrossRef]
- Medina, D.C.; Findley, S.E.; Guindo, B.; Doumbia, S. Forecasting Non-Stationary Diarrhea, Acute Respiratory Infection, and Malaria Time-Series in Niono, Mali. PLoS ONE 2007, 2, e1181. [Google Scholar] [CrossRef]
- Ture, M.; Kurt, I. Comparison of four different time series methods to forecast hepatitis A virus infection. MevlutTureImranKurt 2006, 31, 41–46. [Google Scholar] [CrossRef]
- Reichert, T.A.; Simonsen, L.; Sharma, A.; Pardo, S.A.; Fedson, D.S.; Miller, M.A. Influenza and the winter increase in mortality in the United States, 1959-1999. Am. J. Epidemiol. 2004, 160, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Burke, H.B.; Goodman, P.H.; Rosen, D.B.; Henson, D.E.; Weinstein, J.N.; Harrell, F.E., Jr.; Marks, J.R.; Winchester, D.P.; Bostwick, D.G. Artificial neural networks improve the accuracy of cancer survival prediction. Cancer 2000, 79, 857–862. [Google Scholar] [CrossRef]
- Tu, J.V. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J. Clin. Epidemiol. 1996, 49, 1225–1231. [Google Scholar] [CrossRef]
- Samaras, L.; García-Barriocanal, E.; Sicilia, M.A. Syndromic surveillance models using web data: The case of scarlet fever in the UK. Inform. Health Soc. Care 2012, 37, 106–124. [Google Scholar] [CrossRef]
- Heckerling, P.S.; Gerber, B.S.; Tape, T.G.; Wigton, R.S. Prediction of community-acquired pneumonia using artificial neural networks. Med. Decis. Mak. 2003, 23, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Liang-liang, M.; Fu-peng, T. Pneumonia incidence rate predictivemodel of nonlinear time series based on Dynamic Learning Rate BP Neural Network. Adv. Intell. Soft Comput. 2010, 739–749. [Google Scholar]
- Imai, C.; Armstrong, B.; Chalabi, Z.; Mangtani, P.; Hashizume, M. Time series regression model for infectious disease and weather. Environ. Res. 2015, 142, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Mansiaux, Y.; Carrat, F. Detection of independent associations in a large epidemiologic dataset: A comparison of random forests, boosted regression trees, conventional and penalized logistic regression for identifying independent factors associated with H1N1pdm influenza infectio. BMC Med. Res. Methodol 2014, 14, 99. [Google Scholar] [CrossRef]
- Mehdipour, P.; Navidi, I.; Parsaeian, M.; Mohammadi, Y.; Moradi, L.M.; Rezaei, D.E.; Nourijelyani, K.; Farzadfar, F. Application of Gaussian Process Regression (GPR) in estimating under-five mortality levels and trends in Iran 1990–2013, study protocol. Arch. Iran. Med. 2014, 17, 189–192. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).