Abstract
In a recent monograph on the genus Rosellinia, type specimens worldwide were revised and re-classified using a morphological approach. Among them, some came from Pier Andrea Saccardo’s fungarium stored in the Herbarium of the Padova Botanical Garden. In this work, we taxonomically re-examine via a morphological and molecular approach nine different Rosellinia sensu Saccardo types. ITS1 and/or ITS2 sequences were successfully obtained applying Illumina MiSeq technology and phylogenetic analyses were carried out in order to elucidate their current taxonomic position. Only the ITS1 sequence was recovered for Rosellinia areolata, while for R. geophila, only the ITS2 sequence was recovered. We proposed here new combinations for Rosellinia chordicola, R. geophila and R. horridula, while for R. ambigua, R. areolata, R. australis, R. romana and R. somala, we did not suggest taxonomic changes compared to the current ones. The name Rosellinia subsimilis Sacc. is invalid, as it is a later homonym of R. subsimilis P. Karst. & Starbäck. Therefore, we introduced Coniochaeta dakotensis as a nomen novum for R. subsimilis Sacc. This is the first time that these types have been subjected to a molecular study. Our results demonstrate that old types are an important source of DNA sequence data for taxonomic re-examinations.
1. Introduction
The genus Rosellinia (Ascomycota, Xylariales), erected by De Notaris in 1844 [1] and typified by Rosellinia aquila, includes species that form superficial, dark brown to black, ostiolate stromata usually embedded in a mat of hyphae called subiculum, with each stroma containing one or sometimes few perithecia. Perithecia produce unitunicate, cylindrical asci with an amyloid apical ring (apparatus) and unicellular, asymmetrically ellipsoid, brown, often with a germ slit ascospores [2,3]. They occur in temperate and tropical regions as saprobes, some as endophytes and a few as root pathogens of economic important plant species (e.g., Vitis vinifera), growing mainly on deciduous woods of dicotyledonous plants [2]. Rosellinia species have a geniculosporium-like asexual morph and rarely a nodulisporium-like stages [3,4]. Over time, Rosellinia was synonymized with other genera (e.g., Dematophora) and placed in different families until Miller in 1928 [5] considered it as a genus of the family Xylariaceae, a classification now also confirmed by phylogenetic studies [4,6].
In 1882, Pier Andrea Saccardo [7] subdivided Rosellinia into different sections on the basis of stromatal features. Eu-Rosellinia with large, non-setose stromata immersed in a subiculum; Calomastia with large, non-setose stromata without subiculum; Tassiella with large, verrucose, non-setose stromata; Amphisphaerella (Cfr. Anthostomella) with the base of the stromata immersed; Coniomela with small, non-setose gregarious ascomata; Coniochaeta with small, setose, gregarious ascomata; Cucurbitula with erumpent ascomata; and Sphaeropyxis with short stipitate ascomata and globose ascospores. Moreover, he added the sections of lichenicolous species and doubtful species [2,3]. He described new Rosellinia sensu Saccardo species, many of them stored in his personal mycological collection at the Herbarium of the Padova Botanical Garden (PAD). In Saccardo’s fungarium, the genus includes 75 different species, with more or less 35 represented by type specimens [8]. Among Saccardo’s sections, Coniochaeta was raised to generic rank by Cooke in 1887 [9] and is currently accepted as genus of the family Coniochaetaceae (Sordariomycetes, Coniochaetales).
Recently, Petrini revised Rosellinia on the basis of the morphological characters of type specimens worldwide accepting in the genus 142 species and excluding 137 [3]. She subdivided the species in seven morphological groups within three different subgenera (Rosellinia, Calomastia and Corrugata) previously introduced by herself to accommodate species of the two Saccardo’s sections Eu-Rosellinia and Calomastia [2,3]. Species of Rosellinia aquila, R. necatrix and R. buxi groups were placed in the subgenus Rosellinia; species of R. mammaeformis and R. mammoidea groups in the subgenus Calomastia. The Rosellinia emergens group included species of both Rosellinia and Calomastia subgenera, while the species of the subgenus Corrugata were considered distinct from the other morphological groups and placed in the R. thelena group [3]. The multigene phylogeny study of Xylariales published by Wendt et al. [6] suggested that Rosellinia sensu Petrini [3] could be paraphyletic. Indeed, R. necatrix and R. buxi appeared as a sister clade of a clade containing the type species R. aquila. Wittstein et al. [4], through a multigene phylogeny and secondary metabolites study, excluded the species of the Rosellinia necatrix and R. buxi groups from Rosellinia. These species were accommodated in the resurrected genus Dematophora, previously considered a synonym of Rosellinia [4].
In this work, a molecular phylogenetic study, based on the nucleotide sequences of the internal transcribed spacer region (ITS) obtained by applying an Illumina MiSeq technology, was carried out with the aim of defining the current taxonomy of a sub-sample of more than 100-year-old Rosellinia type collections stored in Saccardo’s mycological collection. The molecular study was also coupled with new morphological observations of the type specimens.
2. Materials and Methods
2.1. Specimens Sampling and Morphology
Fungal specimens (indicated in bold in Table 1 and Table 2) were observed with a stereomicroscope Leica EZ4W to sample a small number of dried stromata/ascomata with sterilized tweezers. The material was used for both new morphological and molecular characterizations.

Table 1.
List and details of Coniochaeta and Podosporaceae specimens used in the internal transcribed spacer (ITS) phylogenetic analyses. Newly obtained sequences are reported in bold.

Table 2.
List and details of Xylariaceae and Hypoxylaceae specimens used in the combined ITS-LSU-TUB2 phylogenetic analysis. Newly obtained sequences are reported in bold.
One or two stromata/ascomata were placed on a glass slide, rehydrated in water and smashed up. The characteristics of asci and ascospores, if present, were observed adding 3% lactic acid solution of Cotton Blue, while the presence of an amyloid ascal apical apparatus was tested pre-treating other stromata/ascomata with 10% potassium hydroxide (KOH) and then with Lugol’s solution. Ascospores and asci were observed using an optical microscope Leica DM500 with 400× or 1000× magnifications and photographed with a Leica ICC50W camera integrated in the optical microscope. Stromata/ascomata were photographed with a stereomicroscope Leica EZ4W. They resulted usually collapsed, so their shape and size were recorded when possible. All the measurements were taken using Fiji [10]. Measures of asci and ascospores are indicated as: (minimum–) average minus standard deviation—average—average plus standard deviation (–maximum) of length × (minimum–) average minus standard deviat—average—average plus standard deviation (–maximum) of width. In addition, spore quotient (Q; length/width ratio) = (minimum–) average minus standard deviat—average—average plus standard deviation (–maximum), and average spore quotient (Qav) are reported.
2.2. DNA Extraction, PCR Amplification, Sequencing and Data Analysis
DNA was extracted with the CTAB protocol described in Forin et al. [11]. In order to prepare the libraries for a paired-end sequencing using the Illumina MiSeq technology 2 × 300 bp, ITS1 and ITS2 regions were amplified using a two-step PCR process [12]. The ITS1 region was first amplify using the universal fungal primers ITS1f/ITS1 and ITS2 [13,14], while the ITS2 region with ITS3 and ITS4 [14]. The second PCR was carried out using the products of the first PCR and the same couple of primers used in the first one tagged with different 5 bp identifier tags. The tags are necessary to distinguish the sequences coming from each different type. The second PCR was done in four replicates for each couple of tagged primers. The first PCR was performed in a total volume of 25 μL containing 5 μL of 5X Wonder Taq reaction buffer (EuroClone; 5 mM dNTPs, 15 mM MgCl2), 0.5 μL of bovine serum albumin (BSA, 10 mg/mL), 0.5 μL each of two primers (10 μM), 0.5 μL of Wonder Taq (5 U/μL), 2 μL of genomic DNA and water to reach the final volume. The second PCR was performed without the BSA, using 2 μL of amplicons from the first PCR as template and the primers with tags. The PCR conditions were set as follows: initial denaturation at 95 °C for 3 min; 35 cycles consisting of a denaturation at 95 °C for 30 s, an annealing at 53 °C for ITS1 region and 54 °C for ITS2 region for 40 s and an extension at 72 °C for 45 s; and a final extension at 72 °C for 5 min. The PCRs were purified with the PureLink PCR Purification Kit (Thermo Fisher Scientific, Waltham, MA, United States) and quantified with Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, United States). Amplicons from different samples were mixed in equimolar amount to prepare ITS1 and ITS2 libraries, in accordance with the specifications provided by Fasteris sequencing service (Plan-les-Ouates, Switzerland).
Forward and reverse fastq files from each library were merged using PEAR v. 0.9.10 [15]. The merged reads were demultiplexed and quality filtered with QIIME v. 1.9.1 [16]. The parameters used are the same reported in Forin et al. [12]. VSEARCH v. 2.3.4 [17] was used to dereplicate the sequences, to filter out chimeric sequences and to cluster the sequences into Operational Taxonomic Units (OTUs). ITS1 and ITS2 regions were extracted using ITSx [18]. To perform the OTUs clustering a 98% similarity threshold was used. OTUs represented by fewer than 10 sequences were discarded, and the Fungi UNITE+INSD dataset v. 8.0 [19] for QIIME was used as reference for the taxonomic assignment. The OTUs were also compared with the sequences of the National Center for Biotechnology Information (NCBI) using a BLASTn search [20], excluding uncultured/environmental sample sequences. The final OTU abundance table was created with VSEARCH, considering an identity value of 98%.
2.3. Phylogenetic Analysis
The sequences used for the phylogenetic analyses are reported in Table 1 and Table 2. Three different datasets were generated according to the final taxonomic assignments and BLAST results: an ITS dataset of Coniochaeta species; an ITS dataset of taxa of the family Podosporaceae; and a combined dataset with ITS, 28S nuclear ribosomal RNA gene (LSU) and β-tubulin gene (TUB2) data of taxa of the families Xylariaceae and Hypoxylaceae. ITS1 and ITS2 sequences, when both identified, of the Saccardo types were combined and used in the phylogenetic analyses.
The sequences were aligned using the online version of MAFFT v. 7 [21] and manually refined with Geneious R11.1.5 (https://www.geneious.com, accessed date: 25 February 2021). Phylogenetic analyses were performed using Maximum likelihood (ML) with RAxML-NG v. 1.0.1 [22] and Bayesian Inference (BI) with MrBayes v. 3.2.6 [23] in the CIPRES science gateway [24]. The best-fit models were estimated by the Bayesian information criterion (BIC) using jModelTest 2 [25] to provide a substitution model for each single alignment. We used the Tamura-Nei model with gamma distribution (TrN + G) for the Coniochaeta ITS dataset and the transition model with gamma distribution (TIM2 + G) for the Podosporaceae ITS dataset. In the combined dataset, for ITS and TUB2 we used the Hasegawa-Kishino-Yano model with proportion of invariable sites and gamma distribution (HKY + I + G) while, for LSU, the Tamura-Nei model with equal base frequencies and proportion of invariable sites (TrNef + I). ML analyses were performed with 1000 bootstrap replicates. BI analyses were performed with two independent Monte Carlo Markov Chains (MCMC) runs, each with four chains of 10 M generations. Trees were sampled every 1000 generations and the first 25% were discarded as burn-in. A majority rule consensus tree of the remaining 10001 trees was calculated to obtain estimates for Bayesian posterior probabilities (BPP). Significance threshold was set ≥0.95 for Bayesian posterior probability (BPP) and ≥70% for ML bootstrap values (MLB).
3. Results
3.1. Phylogenetic Analysis
ML and BI analyses produced trees with congruent topologies. Therefore, the trees obtained from the RAxML-NG analysis with MLB and BPP values are reported (Figure 1, Figure 2 and Figure 3).
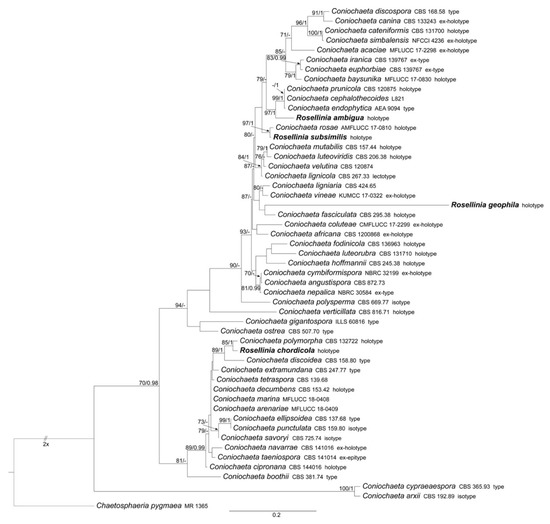
Figure 1.
RAxML phylogram obtained from ITS sequences of selected Coniochaeta species. ChaetoScheme 70. (left) and BPP values ≥0.95 (right) are shown on the branches. Newly obtained sequences are reported in bold.
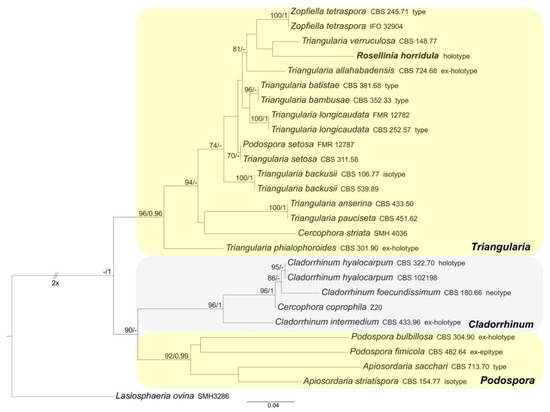
Figure 2.
RAxML phylogram obtained from ITS sequences of selected Triangularia, Cladorrhinum and Podospora (Podosporaceae) species. Lasiosphaeria ovina (Lasiosphaeriaceae) was selected as the outgroup taxon. MLB values ≥70% (left) and BPP values ≥0.95 (right) are shown on the branches. Newly obtained sequence is reported in bold.
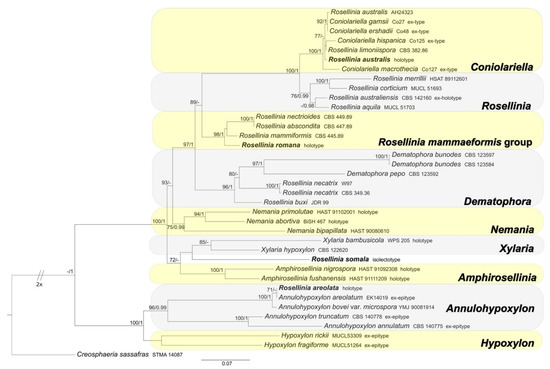
Figure 3.
RAxML phylogram obtained from the combined ITS, LSU and TUB2 sequences of selected species belonging to genera of Xylariaceae and Hypoxylaceae. Creosphaeria sassafras (Lopadostomataceae) was selected as the outgroup taxon. MLB values ≥70% (left) and BPP values ≥0.95 (right) are shown on the branches. Newly obtained sequences are reported in bold.
3.1.1. Coniochaeta
The Coniochaeta dataset includes 52 ITS sequences: four newly generated; 47 Coniochaeta ITS sequences and Chaetosphaeria pygmaea (the outgroup, following Wanasinghe et al. [34]) obtained from GenBank (Table 1). The alignment comprises 563 characters, including indels and missing data. Rosellinia ambigua, R. chordicola, R. geophila and R. subsimilis fall within this genus (Figure 1). The discussion about these Rosellinia types is reported in the taxonomy section.
3.1.2. Podosporaceae
The Podosporaceae dataset includes 27 ITS sequences: one newly generated; 26 Podosporaceae ITS sequences and Lasiosphaeria ovina (the outgroup, following Marin-Felix et al. [27]) obtained from GenBank (Table 1). The alignment comprises 558 characters, including indels and missing data. Triangularia, Cladorrhinum and Podospora form three distinct and well-supported clades (MLB = 96%, BPP = 0.96; MLB = 96%, BPP = 1 and MLB = 92%, BPP = 0.99, respectively). Rosellinia horridula is nested in the Triangularia clade (Figure 2).
3.1.3. Xylariaceae and Hypoxylaceae
The Xylariaceae and Hypoxylaceae combined dataset includes 37 ITS sequences (4 newly generated, 33 obtained from GenBank); 20 28S (LSU) sequences (all obtained from GenBank); 22 TUB2 sequences (all obtained from GenBank). Creosphaeria sassafras was selected as outgroup taxon following Wendt et al. [6]. The alignment comprises 712 (ITS) + 761 (LSU) + 1713 (TUB2) characters, respectively, with a total of 3186 characters, including indels and missing data. Rosellinia australis results included in the Coniolariella clade (MLB = 100%, BPP = 1); R. romana in the Rosellinia mammaeformis sensu Petrini clade (MLB = 98%, BPP = 1), while R. areolata clusters with Annulohypoxylon areolatum sequences in a highly supported clade (MLB = 100%, BPP = 1) (Figure 3). Rosellinia somala occupies an isolate position in the phylogram, sister to the Xylaria clade (Figure 3).
3.2. Taxonomy
3.2.1. Rosellinia ambigua
Coniochaeta ambigua (Sacc.) Popushoi, Mikoflora plodovykh derevyaev SSSR [Mycoflora of fruit trees of the U.S.S.R.] (Moscow): 90. 1971.
Basionym: Rosellinia ambigua Sacc., Atti Soc. Veneto-Trent. Sci. Nat. 2: 328. 1882.
Sexual stage: Ascomata perithecial, superficial, solitary to gregarious, black, globose with black setae on the surface (41–59 × 4.5–6 μm), papillate, 165–220 µm diam (n = 10); peridium not cephalothecoid. Asci cylindrical without amyloid apical apparatus, 90 × 9.6 μm (n = 1), 8-spored, ascospores obliquely uniseriate. Ascospores broadly-ellipsoidal, with round ends and a convex side giving them also a reniform shape, (7.7–)9.4–10.5–11.7(–13.3) × (5.5–)6.2–7.1–8(–8.8) μm, Q = (1–)1.3–1.5–1.7(–2), Qav = 1.5 (n = 37), hyaline to yellow and brown at maturity, smooth, one-celled, with a straight germ slit as long as the ascospore, one-celled.
Material examined: ITALY, Cansiglio, on Sambucus racemosa, ? October 1879, n. 162, PAD S00027, holotype (Figure 4).
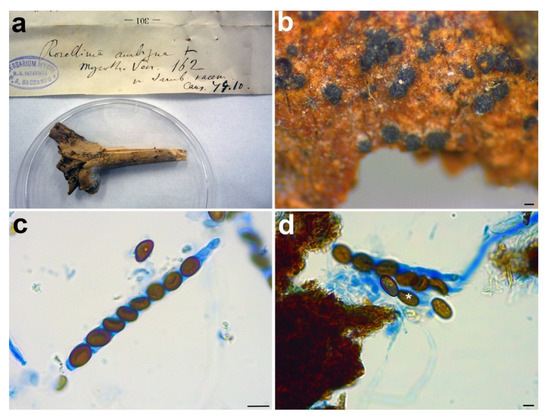
Figure 4.
Rosellinia ambigua. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c) Ascus with ascospores. (d) Ascospores showing germ slit (white asterisk). Scale bars: (b) = 100 μm; (c) = 10 μm; (d) = 5 μm.
Notes: Rosellinia ambigua was moved to the genus Coniochaeta (asexual morph Lecythophora) by Popushoi in 1971 [60]. The genus is characterized by species with perithecial, pyriform to globose ascomata, cylindrical asci without amyloid apical apparatus and ellipsoid to globose, one-celled and brown ascospores often laterally compressed with a germ slit [31,46]. The morphological observation of Rosellinia ambigua fits well with the general phenotypic traits of Coniochaeta reported above (e.g., brown and globose ascospores with a germ slit). In the Coniochaeta phylogram (Figure 1), Rosellinia ambigua clusters in a highly supported clade (MLB = 97%, BPP = 1) where it is sister to a clade (MLB = 99%, BPP = 1) consisting of the ITS sequences of C. cephalothecoides, C. prunicola and C. endophytica. The molecular analysis of the holotype confirms the taxonomic re-classification proposed by Popushoi [60]. Coniochaeta endophytica was described only from its asexual stage [39]. Coniochaeta prunicola has ascospores similar to Rosellinia ambigua (9.2 ± 0.6 × 6.7 ± 0.6 μm) but it differs in the dimension of ascomata (200–250 μm diam), the presence of a long neck above the perithecia and shorter asci (av. 69 × 9.5 μm) [32]. Coniochaeta cephalothecoides and Rosellinia ambigua are morphologically very similar but the former shows a cephalothecoid peridium [61].
3.2.2. Rosellinia areolata
Annulohypoxylon areolatum (Sacc.) Sir & Kuhnert, in Kuhnert, Sir, Lambert, Hyde, Hladki, Romero, Rohde & Stadler, Fungal Diversity 85: 18. 2016.
Basionym: Rosellinia areolata Sacc., Ann. Mycol. 11: 314. 1913.
Synonyms: Annulohypoxylon bovei var. microsporum (J.H. Mill.) Y.M. Ju, J.D. Rogers & H.M. Hsieh (as ‘microspora’), Mycologia 97: 857. 2005.
Hypoxylon bovei var. microsporum J.H. Miller, Monogr. of the World Species of Hypoxylon, p. 95. 1961.
Hypoxylon marginatum var. mammiforme Rehm, Leafl. Philipp. Bot. 8: 2958. 1916.
Hypoxylon chalybeum var. effusum Sacc. apud Sacc. & Trott., Syll. Fung. XXIV, p. 1080. 1928.
Sexual stage: Stromata gregarious, superficial, brown, spherical to spherical compressed, 0.9–1 mm diam (n = 8). Ostioles papillate, with annular disk, 0.4–0.5 mm diam. Asci not found. Ascospores asymmetrically ellipsoidal, with round ends and a convex side, (9.3–)9.6–10.3–10.9(–12) × (3.9–)4.2–4.4–4.7(–5.4) μm, Q = (2–)2.1–2.3–2.5(–2.8), Qav = 2.3 (n = 40), hyaline and brown at maturity, smooth, with a straight germ slit spore-length, one-celled.
Material examined: JAPAN, Mino prov., Kawauye-mura (currently Gifu pref., Nakatsugawa city), on Fagus sp., 30 January 1913, K. Hara, PAD S00028, holotype (Figure 5).
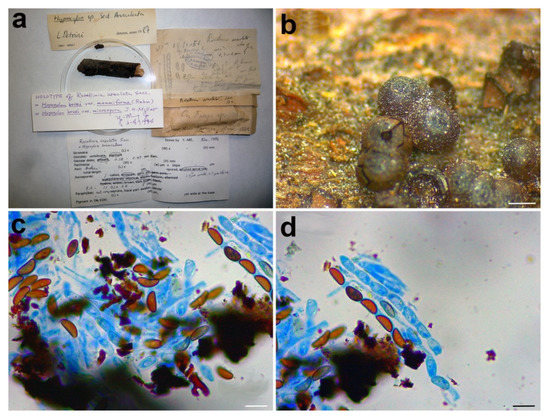
Figure 5.
Rosellinia areolata. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c,d) Ascospores. Scale bars: (b) = 500 μm; (c,d) = 10 μm.
Notes: In 1987, Petrini revised the holotype Rosellinia areolata considering it as a member of the genus Hypoxylon sect. Annulata (Figure 5a). In the monograph about Rosellinia, Petrini revised this species placing it in synonymy with Annulohypoxylon bovei var. microsporum [3]. In 1994 Yu-Ming Ju suggested a synonymy with Hypoxylon bovei var. mammiforme and H. bovei var. microsporum (basionym of Annulohypoxylon bovei var. microsporum). Hypoxylon bovei var. mammiforme does not seem to exist as nomenclatural name and we are convinced that the author intended H. marginatum var. mammiforme (= Annulohypoxylon bovei var. microsporum) (Figure 5a). In 1996, Abe reported a possible synonymy with Hypoxylon truncatum, today Annulohypoxylon truncatum, the type species of the genus Annulohypoxylon. Annulohypoxylon areolatum was recently proposed as new combination for Rosellinia areolata and A. bovei var. microsporum is treated as its synonym [49]. An epitype of Annulohypoxylon areolatum was designated in Kuhnert et al. [49]. The morphological observations of Rosellinia areolata fit with the description of Annulohypoxylon areolatum reported by Kuhnert et al. [49], which was not based on the holotype of R. areolata. Annulohypoxylon species are characterized by the presence of carbonaceous stromata enclosing perithecia, conic-papillate ostioles encircled with an annulate disk, and ascospore perispores with a thickened area visible in 10% KOH at circa 1⁄3 ascospore length when dehiscing [50]. The ITS1 sequence recovered from the holotype clusters with Annulohypoxylon areolatum (MFLUCC 14-1233, ex-epitype) and A. bovei var. microspora (YMJ 90081914) ITS sequences in a highly supported clade (MLB = 100%, BPP = 1) (Figure 3). The ITS1 sequences of Rosellinia australis and Annulohypoxylon areolatum have a nucleotide identity of 99.3%. The molecular analysis confirms the taxonomic reclassification proposed by Kuhnert et al. [49], excluding the synonymy with Annulohypoxylon truncatum suggested by Abe in the label (Figure 5a).
3.2.3. Rosellinia australis
Coniolariella limoniispora (Ellis & Everth.) Checa, Arenal & J.D. Rogers, Mycological Research 112: 797. 2008.
Basionym: Rosellinia limoniispora Ellis & Everh., Proc. Acad. Nat. Sci. Phila. 46:326. 1894.
Synonyms: Coniolariella limonispora var. australis Checa, Arenal & J.D. Rogers (as ‘limoniispora’), Mycol. Res. 112: 797. 2008.
Rosellinia australis Sacc. & Trotter, Ann. Mycol. 11: 416. 1913, Nom. illegit., Art. 53.1, preoccupied by Rosellinia australis Speg., Anal. Mus. nac. B. Aires, Ser. 3, 12: 337. 1909. = Rosellinia bonaerensis Speg., Anal. Mus. nac. Hist. nat. B. Aires 6: 258. 1898. fide Petrini 2013.
Sexual stage: Stromata solitary to gregarious, superficial, black with the ostiolar region slightly papillate, globose, 582–904 µm diam (n = 10). Asci cylindrical without amyloid apical apparatus, (113–)113.7–119.4–125.2(–125.4) × (10.5–)10.6–12.1–13.6(–14) μm (n = 5), 8-spored, ascospores obliquely uniseriate. Ascospores citriform with apiculate ends, (13.3–)15.9–17.5–19.2(–20.8) × (7.6–)8.3–9.1–9.9(–10.8) μm, Q = (1.5–)1.7–1.9–2.2(–2.5), Qav = 1.9 (n = 41), dark brown, smooth, with straight and long germ slit, one-celled, monoguttulate.
Material examined: LIBYA, Tripoli, on Nicotiana glauca, 1913, A. Trotter, PAD S00029, holotype (Figure 6).

Figure 6.
Rosellinia australis. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c) Ascus with ascospores; ascospore showing germ slit (white asterisk). Scale bars: (b) = 200 μm; (c) = 20 μm.
Notes: The holotype was morphologically revised by Petrini in 1999, who excluded Rosellinia australis from Rosellinia (Figure 6a) due to the presence of soft stromata, perithecia adhering to the stromatal wall, asci without an amyloid apex and lack of a subiculum [57]. This species is now considered a synonym of Coniolariella limoniispora [51]. Coniolariella was introduced by García et al. [62] to accommodate the single species Coniolariella gamsii, previous placed in the genus Coniochaeta as Coniochaeta gamsii, characterized by stromata solitary or in small groups, globose and dark brown, asci cylindrical, without apical structures and ascospores one-celled, brown, ellipsoidal to citriform, with apiculate ends and a longitudinal germ slit. In a following molecular study, Checa et al. [57] added to Coniolariella the new species C. hispanica and proposed the new combination C. limoniispora for Rosellinia limoniispora. They recognized Coniolariella gamsii, the type species of the genus, and Rosellinia australis as varieties of C. limoniispora. Zare et al. [51] introduced the new species Coniolariella macrothecia and the new combination Coniolariella ershadii for Coniochaeta ershadii. In addition, they did not consider Coniolariella gamsii as a variety of C. limoniispora and synonymized C. limoniispora var. australis under C. limoniispora. In the genus, five different species are recognized [51]. The molecular study places the holotype R. australis in the highly supported Coniolariella clade (MLB = 100%, BPP = 1). Nevertheless, the use of two molecular markers (ITS and LSU, see Table 2) is not sufficient to delimit all the different species. The morphology of Rosellinia australis fits with the morphological description of Coniolariella limoniispora reported by Checa et al. [57], confirming that Rosellinia australis can be considered a synonym of Coniolariella limoniispora.
3.2.4. Rosellinia chordicola
Coniochaeta chordicola (Sacc.) Forin, Fainelli & Vizzini comb. nov. MycoBank MB838853.
Basionym: Rosellinia chordicola Sacc., Michelia 1: 372. 1878.
Sexual stage: Ascomata perithecial, solitary, superficial, black, globose, about 290 µm diam. Asci immature without amyloid apical apparatus. Ascospores broadly-ellipsoidal, with round ends and a convex side giving them also a reniform shape, (8.9–)10.1–11.1–12.1(–14.4) × (6.3–)7.5–8.5–9.6(–10.8) μm, Q = (1.1–)1.2–1.3–1.5(–1.6), Qav = 1.3 (n = 46), brown, smooth with a straight germ slit nearly as long as the ascospore, one-celled.
Material examined: ITALY, Padova, Botanical Garden, on a rope, 1877, PAD S00030, holotype (Figure 7).
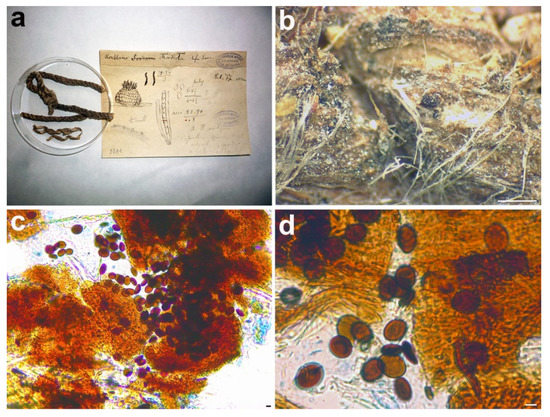
Figure 7.
Rosellinia chordicola. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c,d) Ascospores. Scale bars: (b) = 500 μm; (c,d) = 5 μm.
Notes: Phylogenetically, Rosellinia chordicola in the Coniochaeta genus is closely related to C. polymorpha (CBS 132722, holotype) and C. discoidea (CBS 158.80, type) (Figure 1). Coniochaeta polymorpha was morphologically described only based on its asexual stage [63]; therefore, a comparison between the sexual stages of Saccardo’s type and C. polymorpha was not possible. The ITS sequences of Rosellinia chordicola and C. polymorpha show an identity of 97% with 10 nucleotide differences. Rosellinia chordicola and Coniochaeta discoidea have discoid ascospores with similar dimensions but they differ in the ornamentation of the ascospores. Coniochaeta discoidea has ascospores (8–)9–12 × 8–11 μm characterized by the presence of circular to elongate pits [64]. Our molecular analysis suggests that Rosellinia chordicola should be treated as a distinct species within the genus Coniochaeta.
3.2.5. Rosellinia geophila
Coniochaeta geophila (E. Bommer, M. Rousseau & Sacc.) Forin, Fainelli & Vizzini comb. nov. MycoBank MB838854.
Basionym: Rosellinia geophila E. Bommer, M. Rousseau & Sacc., in Saccardo, Ann. Mycol. 3: 508. 1906.
Sexual stage: Ascomata perithecial, solitary or gregarious, superficial, black, globose with black setae on the surface (66.4–101.2 × 6–8.9 μm) and a slightly papillate ostiolar region, 349–448 µm diam (n = 5). Asci not found. Ascospores ellipsoidal with broadly rounded ends, (18.9–)23–26–29.1(–30.7) × (9.9–)11–12.6–14.2(–16) μm, Q = (1.4–)1.8–2.1–2.4(–2.7), Qav = 2.1 (n = 50), brown, smooth, with a straight germ slit as long as the ascospore, one-celled.
Material examined: BELGIUM, La Panne pr. Furnes, on sandy ground among mosses, November 1900, PAD S00031, holotype (Figure 8).
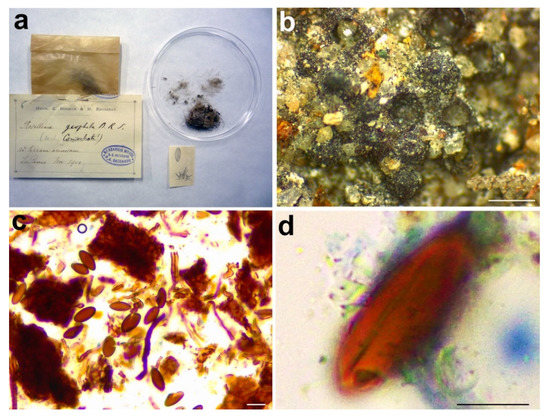
Figure 8.
Rosellinia geophila. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c) Ascospores. (d) Ascospore showing germ slit. Scale bars: (b) = 500 μm; (c) = 20 μm; (d) = 10 μm.
Notes: Only the ITS2 sequence has been obtained from Rosellinia geophila. The molecular analysis places the holotype close to Coniochaeta fasciculata (CBS 295.38, holotype), only known for its asexual morph [65]. Rosellinia geophila clusters also with Coniochaeta lignaria and C. vineae (Figure 1). C. vineae has ascomata covered by setae and brown ascospores with a straight germ slit, but it differs from R. geophila in having smaller ascomata (170–185 μm diam) and smaller ascospores (6.5–9.5 × 4–6 μm) ovoidal and multi-guttulate [46]. Coniochaeta ligniaria has pointed setae often covering the whole ascomata and brown ellipsoidal ascospores with a germ slit smaller than those of Rosellinia geophila (12–15 × 8–10 μm) [66,67]. The results of the taxonomic assignment coupled with the phylogenetic analysis (Figure 1) suggest that this is a distinct Coniochaeta species confirming the original placement of Rosellinia geophila in the genus Rosellinia sect. Coniochaeta as reported in the original label (Figure 8a).
3.2.6. Rosellinia horridula
Triangularia horridula (Sacc.) Forin, Fainelli & Vizzini comb. nov. MycoBank MB838855.
Basionym: Rosellinia horridula Sacc., Fl. Sard. Comp.: 248. 1884.
Synonym: Podospora horridula (Sacc.) Dennis & S.M. Francis, Trans. Br. Mycol. Soc. 82: 380. 1984.
Sexual stage: Ascomata solitary or gregarious, pyriform to ovate with a short conical neck, superficial or immersed with only the neck protruding, black, covered with flexuous hairs, about 400 µm diam. Asci not found. Ascospores ellipsoidal with an apical end rounded while the other one flattened, inequilateral and slightly curved, (26.2–)29–32.5–36(–41.8) × (12.5–)13.5–15.1–16.7(–17.6) μm, Q = (1.7–)1.9–2.2–2.4(–2.5), Qav = 2.2 (n = 15), dark brown, smooth, one-celled.
Material examined: ITALY, Sardinia, Torralba, on Opuntiae sp., Marcucci, PAD S00032, holotype (Figure 9).
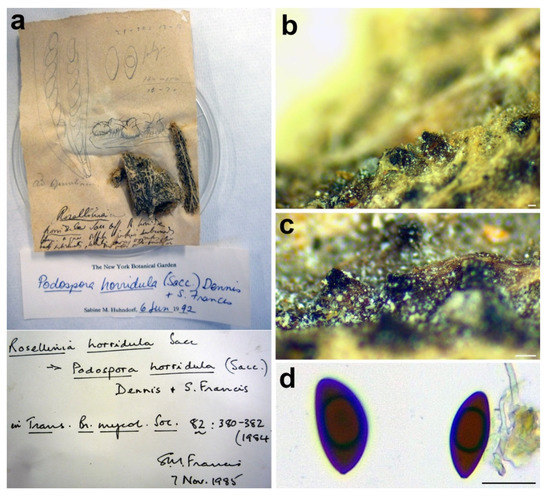
Figure 9.
Rosellinia horridula. (a) Original fungarium specimen. (b,c) Perithecia on natural substrate. (d) Ascospores. Scale bars: (b,c) = 100 μm; (d) = 20 μm.
Notes: The specimen was morphologically revised by S.M. Francis in 1985 and by S.M. Huhndorf in 1992, as reported in the labels associated with the sample (Figure 9a). Rosellinia horridula was redescribed from the holotype as Podospora horridula [68]. Wang et al. [30] introduced the new family Podosporaceae to accommodate three different genera (Podospora, Cladorrhinum and Triangularia) forming a phylogenetic sister lineage of Chaetomiaceae in the Sordariales. Podospora, Cladorrhinum and Triangularia were re-defined, and many species previously identified as Podospora, including the genetic model species P. anserina, were moved to Triangularia [30]. In Podospora sensu stricto, only the type species P. fimicola was maintained and the new combination P. bulbillosa was proposed for Cladorrhinum bulbillosum [30]. Marin-Felix et al. [27] introduced as new combinations the species Podospora striatispora (= Apiosordaria striatispora), P. costaricensis (= Cercophora costaricensis) and P. sacchari (= Apiosordaria sacchari) in Podospora sensu stricto based on a phylogenetic study. The new genera Rhypophila (Naviculisporaceae) and Pseudoechria (Schizotheciaceae) were erected to accommodate different Podospora species. Podospora cochleariformis, P. decipiens, P. myriaspora and P. pleiospora in Rhypophila. Podospora curvicolla, P. longicollis, P. decidua and P. prolifica in Pseudoechria [27]. Our molecular analysis places Rosellinia horridula in the clade Triangularia close to Triangularia verruculosa (Figure 2), from which it differs for the presence of longer and one-celled ascospores (T. verruculosa has (23–)25.5–28.5(–29.5) μm long and two-celled ascospores) [30]. Therefore, a new combination is proposed here for Rosellinia horridula. Triangularia was restricted to the type species of the genus, T. bambusae, together with species characterized by two-celled ascospores that occurred in the same monophyletic clade [30]. Subsequently, a species with one-celled ascospores (Arnium arizonense) has been moved in Triangularia incorporating in the description of the genus also species with one-celled ascospores [27]. Rosellinia horridula has one-celled ascospores and represents the second species with this trait in the genus Triangularia.
3.2.7. Rosellinia romana
Rosellinia glabra (Fuckel) L.E. Petrini, Sydowia 44: 243. 1992.
Basionym: Rosellinia aquila var. glabra Fuckel, Symb. Myc. 149. 1869.
Synonym: Rosellinia romana Sacc., Annales Mycologici 10: 316. 1912.
Sexual stage: Stromata gregarious in small groups, superficial, black, globose, slightly papillate, 675–910 µm diam (n = 10). Asci cylindrical with amyloid apical apparatus, (92.8–)94.4–102.6–110.8(–112.8) × (8.7–)8.9–9.6–10.3(–10.8) μm (n = 6), 8-spored, ascospores obliquely uniseriate. Ascospores ellipsoidal to asymmetrical ellipsoidal with pinched ends, (10.4–)12.2–13.1–14(–14.5) × (4.6–)5.4–5.9–6.3(–7.1) μm, Q = (1.7–)2–2.3–2.5(–3), Qav = 2.2 (n = 48), dark brown, smooth with a straight germ slit nearly as long as the ascospore, one-celled.
Material examined: Italy, Rome, Marino, on Ruscus aculeatus, July 1904, D. Saccardo, PAD S00033, holotype (Figure 10).

Figure 10.
Rosellinia romana. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c) Asci with ascospores; black asterisks indicate the amyloid apical apparatus. Scale bars: (b) = 200 μm; (c) = 20 μm.
Notes: Rosellinia romana was morphologically revised by Petrini in 1987, as reported in the label associated with the specimen (Figure 10a), suggesting a synonymy with R. aquila var. glabra. In 1992 Petrini introduced the new combination Rosellinia glabra for R. aquila var. glabra and, as a consequence, R. romana became a synonym of R. glabra [2,3]. Rosellinia glabra is characterized by large stromata (0.7–0.9 mm diam), semiglobose to cupulate, brown, papillate, gregarious, forming small groups; ascospores (12.5) 16±2 (21) × (5.5) 6.5±0.5 (7.7) μm, ellipsoidal to asymmetrically ellipsoidal with broadly rounded or pinched ends, brown with a germ slit nearly as long as the ascospore, straight and a cellular appendage at one or both spore ends [2,3]. The ITS sequence of R. romana clusters in a highly supported clade (MLB = 98%, BPP = 1) with ITS sequences of Rosellinia species belonging to the R. mammaeformis group introduced by Petrini [3]. This morphological group should include also Rosellinia glabra [3]. However, a molecular comparison between R. romana and R. glabra was not possible because, for the latter, no molecular information is deposited in public databases. As already observed by Petrini [2], the morphologies of R. romana and R. glabra are very similar. In the absence of a molecular confirmation, we agree with the synonymy proposed by Petrini [2,3].
3.2.8. Rosellinia somala
Helicogermslita celastri (S.B. Kale & S.V.S. Kale) Lodha & D. Hawksw., Transactions of the British Mycological Society 81: 91. 1983.
Synonym: Rosellinia somala Bacc., Risultati scientifici della Missione Stefanini Paoli nella Somalia meridionale (Firenze): 195. 1916.
Sexual stage: Stromata solitary or in small groups of two/three, erumpent from bark and eventually almost superficial, black, globose, papillate, 572–793 µm diam (n = 5). Asci cylindrical without amyloid apical apparatus, 140.7–149.5 × 10.2–13.5 μm (n = 2), 8-spored, ascospores obliquely uniseriate. Ascospores ellipsoidal with rounded ends, (14.3–)15.8–17.4–19(–21.7) × (6–)7–7.5–8(–8.7) μm, Q = (1.8–)2.1–2.3–2.6(–3), Qav = 2.3 (n = 48), dark brown, smooth, one-celled, with a helicoid germ slit coiling three times along the entire length of the ascospore.
Material examined: SOMALIA, on branch, 1913, G. Paoli, PAD S00034, isolectotype (Figure 11).
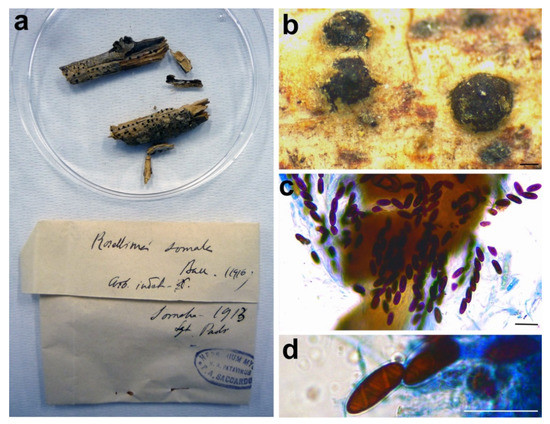
Figure 11.
Rosellinia somala. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c) Ascospores. (d) Ascospore showing helicoid germ slit. Bars: (b) = 200 μm; (c,d) = 20 μm.
Notes: Petrini [3] synonymized Rosellinia somala with Helicogermslita celastri after a morphological examination of the lectotype of R. somala stored in Harvard University (FH). Helicogermslita (Xylariaceae) was originally introduced to accommodate the single species Helicogermslita celastri (formerly Amphisphaerella celastri) [69]. Laessöe and Spooner [70] included in this genus other three previously described species: Helicogermslita fleischhakii (= Sordaria fleischhakii), H. gaudefroyi (= Rosellinia gaudefroyi), H. valdiviensis (= Rosellinia valdiviensis). Petrini [71] introduced three new species (Helicogermslita gisbornia, H. johnstonii, H. mackenziei) and the new combination H. aucklandica for Rosellinia aucklandica. Lee and Crous [72] described the new species Helicogermslita diversa. The main characteristic of the Helicogermslita species is the presence of ascospores with a helical germ slit running along the entire length of the spore [69]. This phenotypic character was observed also in the isolectotype of Rosellinia somala stored in PAD. Unfortunately, there are no molecular data for Helicogermslita species in public databases and we were not able to get original material for comparison, but the isolate position of Rosellinia somala in the phylogram suggests that it does not belong to other Xylariaceae genera (Figure 3). The morphology of Rosellinia somala is similar to those of Helicogermslita celastri reported by Hawksworth and Lodha [69]. In the absence of molecular information, we agree with Petrini who suggested the synonymy of Rosellinia somala with Helicogermslita celastri [3].
3.2.9. Rosellinia subsimilis
Coniochaeta dakotensis Forin, Fainelli & Vizzini nom. nov. MycoBank MB838856 for Rosellinia subsimilis Sacc., Mycologia 12: 199. 1920, non Rosellinia subsimilis P. Karst. & Starbäck, Revue mycol., Toulouse 9 (no. 36): 160. 1887.
Etymology: the specific epithet refers to Dakota, the geographic area where the holotype was collected.
Sexual stage: Ascomata perithecial, gregarious, superficial, black, globose, slightly papillate, 190–250 µm diam (n = 10). Asci cylindrical without amyloid apical apparatus, (84–)90.8–98.4–105.9(–107) × (8–)8.1–8.8–9.5(–10) μm (n = 10), 8-spored, ascospores obliquely uniseriate. Ascospores asymmetrically ellipsoidal with a flattened side and rounded ends, (8–)8.7–10.3–12(–16.3) × (3.7–)4.1–4.8–5.5(–7.5) μm, Q = (1.6–)1.8–2.2–2.5(–2.8), Qav = 2.1 (n = 49), brown, smooth, with a straight germ slit as long as the ascospore, one-celled.
Material examined: USA, North Dakota, Dickey Co., Whitestone Gully, on Crataegus sp., 26 November 1916, J. Brenckle, n. 1188, PAD S00035, holotype (Figure 12).
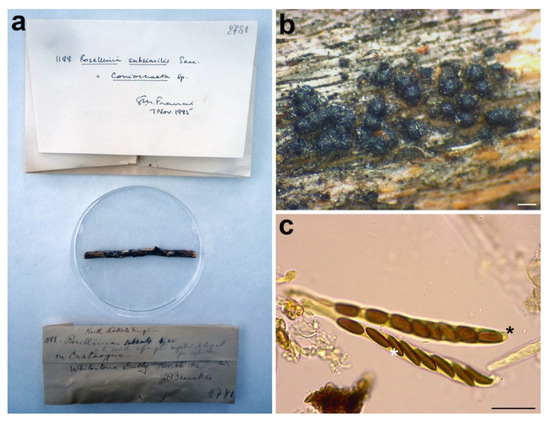
Figure 12.
Rosellinia subsimilis. (a) Original fungarium specimen. (b) Perithecia on natural substrate. (c) Asci without amyloid apical apparatus (black asterisk); ascospores showing germ slit (white asterisk). Bars: (b) = 200 μm; (c) = 20 μm.
Notes: The holotype was morphologically revised in 1985, as reported in the label associated with the sample (Figure 12a), suggesting that the species should be placed in the genus Coniochaeta. Petrini treated Rosellinia subsimilis Sacc. (non R. subsimilis P. Karst. & Starbäck) among the specimens excluded from Rosellinia. She considered R. subsimilis as a Coniochaeta species without giving a new combination or possible synonymies [3]. The ITS sequence of R. subsimilis clusters with the ITS sequence of the holotype Coniochaeta rosae in a highly supported clade (MLB = 97%, BPP = 1). The comparison of the ITS regions reveals an identity of 97.7% between Rosellinia subsimilis and Coniochaeta rosae. Morphologically, the two species are very similar, except that the ascospores of Rosellinia subsimilis are smaller than those of Coniochaeta rosae (14–18 × 4–6 µm, x = 15.8 × 5.2 µm) [34]. In addition, both the species are saprobes of Rosaceae (Coniochaeta rosae was described on a stalk of Rosa hissarica in Uzbekistan) [34]. Based on the morphological and molecular analysis, R. subsimilis Sacc. is considered here as a new species within Coniochaeta. Since the name R. subsimilis Sacc. (1920) is invalid because it is preoccupied by R. subsimilis P. Karst. & Starbäck (1887), we have introduced Coniochaeta dakotensis as a nomen novum for the former.
4. Discussion
The genus Rosellinia has been extensively revised by Petrini considering a specific combination of phenotypic characters. She subdivided the species into seven informal morphological groups, excluding many formerly described species from the genus [2,3]. Among the types taxonomically re-evaluated, some of them come from the Saccardo fungarium stored in the PAD. In this work ITS1 and/or ITS2 sequences were obtained from nine different Rosellinia sensu Saccardo type specimens with the purpose of elucidating the current systematic status of these species. Coupling new morphological observations with molecular phylogenetic analyses, we introduce the new name Coniochaeta dakotensis (for Rosellinia subsimilis Sacc.) and the new nomenclatural combinations Coniochaeta chordicola (formerly R. chordicola), C. geophila (formerly R. geophila) and Triangularia horridula (formerly Podospora horridula). However, for Rosellinia ambigua, R. areolata, R. australis, R. romana and R. somala, we have not suggested any taxonomic change compared to the current one. An exhaustive taxonomic re-evaluation of Rosellinia romana and R. somala was not possible due to the lack of sufficient molecular information deposited in public databases. The absence of a reference database, in term of sequence data, for the species of the genus Helicogermslita has not allowed to have a molecular confirmation on the synonymy of Rosellinia somala with H. celastri proposed by Petrini [3]. Nevertheless, the phenotypic characters of R. somala are congruent with those of the species of the genus Helicogermslita (e.g., helicoid ascospore germ slit), in particular with H. celastri. This also applies to Rosellinia romana, which was placed in synonymy with R. glabra [3]; however, DNA sequences, for the latter, are not available. The morphology is an important component in fungal taxonomy and new species are continuously introduced using this approach [73]. However, molecular information enhances the value of a species description, allowing to integrate them into a modern phylogenetic context. When DNA sequence data for Helicogersmlita species and Rosellinia glabra are available, the current taxonomic position of R. romana and R. somala can be confirmed or changed. Until that moment and relying only on morphological data, we agree with the synonymies proposed by Petrini [3] for these two species. Rosellinia mammaeformis group, with the type of Rosellinia romana, is separated from the clade “Rosellinia sensu stricto” containing the type species R. aquila (Figure 3), suggesting, as also reported in other phylogenetic studies [4,6,56], that Rosellinia sensu Petrini is not a monophyletic clade. It is probable that a molecular study involving type specimens of the Rosellinia aquila, R. emergens, R. mammaeformis, R. mammoidea and R. thelena morphological groups proposed by Petrini would lead to split the genus into different genera. As well as other authors [74,75], we encourage mycologists to always generate molecular data when new species are described or when old fungal species are re-examined in order to make available useful DNA information to the entire mycological community for further studies. Once again, we demonstrate the possibility and the scientific relevance of generating molecular data from fungal type specimens stored in fungaria with the hope that, in the future, greater efforts will be employed for conducting genetic analyses on these important samples.
Author Contributions
Conceptualization, N.F. and B.B.; Formal analysis, N.F., F.F. and E.E.; Funding acquisition, B.B.; Investigation, N.F., A.V. and F.F.; Methodology, N.F., A.V., F.F. and E.E.; Project administration, N.F., A.V. and B.B.; Resources, N.F., E.E. and B.B.; Supervision, A.V. and B.B.; Validation, A.V.; Visualization, N.F. and A.V.; Writing—original draft, N.F.; Writing—review and editing, N.F., A.V., F.F., E.E. and B.B. All authors have read and agreed to the published version of the manuscript.
Funding
Research was supported by a grant (PANN15T3_00789, Legge 6/2000) from MIUR (Ministero Istruzione Università e Ricerca, Italy) to B.B and DOR (Dotazione Ordinaria della Ricerca) 2018–2020 from the Biology Department (UniPD) to B.B., N.F was the recipient of a post-doc fellowship (prot.704/2018BG) granted by the University of Padova.
Data Availability Statement
New sequence data are available in NCBI GenBank and the accession numbers are reported in Table 1 and Table 2. The Illumina sequencing data are not publicly available because they contain data not involved in this study but currently investigated for other purposes. These data are available upon request to corresponding authors (niccolo.forin@unipd.it, barbara.baldan@unipd.it).
Acknowledgments
The authors are grateful to the curator of the Herbarium of the Padova Botanical Garden Rossella Marcucci for the help to locate the type specimens in the Saccardo collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Notaris, G. Cenni sulla tribù dei pirenomiceti sferiacei e descrizione di alcuni generi spettanti alla medesima. Giornale Botanico Italiano 1844, 1, 322–355. [Google Scholar]
- Petrini, L.E. Rosellinia species of the temperate zones. Sydowia 1992, 44, 169–281. [Google Scholar]
- Petrini, L.E. Rosellinia—A World Monograph. In Bibliotheca Mycologica, 1st ed.; J. Cramer: Stuttgart, DE, USA, 2013; Volume 205. [Google Scholar]
- Wittstein, K.; Cordsmeier, A.; Lambert, C.; Wendt, L.; Sir, E.B.; Weber, J.; Wurzler, N.; Petrini, L.E.; Stadler, M. Identification of Rosellinia species as producers of cyclodepsipeptide PF1022 A and resurrection of the genus Dematophora as inferred from polythetic taxonomy. Stud. Mycol. 2020, 96, 1–16. [Google Scholar] [CrossRef]
- Miller, J. Biologic studies in the Sphaeriales. Mycologia 1928, 20, 305–339. [Google Scholar]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef]
- Saccardo, P.A. Sphaeriaceae, Phaeosporae, Rosellinia. Sylloge Fungorum 1882, 1, 252–277. [Google Scholar]
- Gola, G. L’erbario Micologico di P.A. Saccardo; Antoniana: Padova, Italy, 1930; pp. 214–215. [Google Scholar]
- Cooke, M.C. Synopsis pyrenomycetum. Grevillea 1887, 16, 16–19. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Forin, N.; Nigris, S.; Voyron, S.; Girlanda, M.; Vizzini, A.; Casadoro, G.; Baldan, B. Next generation sequencing of ancient fungal specimens: The case of the Saccardo mycological herbarium. Front. Ecol. Evol. 2018, 6, 129. [Google Scholar] [CrossRef]
- Forin, N.; Vizzini, A.; Nigris, S.; Ercole, E.; Voyron, S.; Girlanda, M.; Baldan, B. Illuminating type collections of nectriaceous fungi in Saccardo’s fungarium. Persoonia 2020, 45, 221–249. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications, 1st ed.; Innis, M.A., Gelfand, D.H., Sninsky, J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; Wit, P.D.; Sa, M.; Amend, A.S.; Jumpponen, A.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 6. [Google Scholar] [CrossRef]
- UNITE Community. Full UNITE+INSD Dataset for Fungi. Version 18.11.2018; 2019. [Google Scholar] [CrossRef]
- Altschul, S. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: A community resource for phylogenetic analyses. In TeraGrid Conference: Extreme Digital Discovery; San Diego Supercomputer Center: San Diego, CA, USA, 2011; Volume 41, pp. 1–8. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Miller, A.N.; Cano-Lira, J.F.; Guarro, J.; García, D.; Stadler, M.; Huhndorf, S.M.; Stchigel, A.M. Re-evaluation of the order Sordariales: Delimitation of Lasiosphaeriaceae s. str., and introduction of the new families Diplogelasinosporaceae, Naviculisporaceae, and Schizotheciaceae. Microorganisms 2020, 8, 1430. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Han, T.; Li, W.; Jia, M.; Xue, L.; Rahman, K.; Qin, L. Geographic and tissue influences on endophytic fungal communities of Taxus chinensis var. mairei in China. Curr. Microbiol. 2013, 66, 40–48. [Google Scholar] [CrossRef]
- Réblová, M.; Winka, K. Phylogeny of Chaetosphaeria and its anamorphs based on morphological and molecular data. Mycolgia 2000, 92, 939–954. [Google Scholar] [CrossRef]
- Wang, X.W.; Bai, F.Y.; Bensch, K.; Meijer, M.; Sun, B.D.; Han, Y.F.; Crous, P.W.; Samson, R.A.; Yang, F.Y.; Houbraken, J. Phylogenetic re-evaluation of Thielavia with the introduction of a new family Podosporaceae. Stud. Mycol. 2019, 93, 155–252. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Gafforov, Y.; Liu, N.; Maharachchikumbura, S.S.N.; Bhat, J.D.; Liu, J.-K.; Promputtha, I.; Hyde, K.D. Combined multi-gene backbone tree for the genus Coniochaeta with two new species from Uzbekistan. Phytotaxa 2018, 336, 43–56. [Google Scholar] [CrossRef]
- Damm, U.; Fourie, P.H.; Crous, P.W. Coniochaeta (Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia 2010, 24, 60–80. [Google Scholar] [CrossRef]
- Dayarathne, M.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chomnunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Jones, G.E.B.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal Diversity Notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Schoch, C.L.; Robbertse, B.; Robert, V.; Vu, D.; Cardinali, G.; Irinyi, L.; Meyer, W.; Nilsson, R.H.; Hughes, K.; Miller, A.N.; et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database Oxford 2014. [Google Scholar] [CrossRef]
- Han, J.; Liu, C.; Li, L.; Zhou, H.; Liu, L.; Bao, L.; Chen, Q.; Song, F.; Zhang, L.; Li, E.; et al. Decalin-containing tetramic acids and 4-Hydroxy-2-Pyridones with antimicrobial and cytotoxic activity from the fungus Coniochaeta cephalothecoides collected in Tibetan Plateau (Medog). J. Org. Chem. 2017, 82, 11474–11486. [Google Scholar] [CrossRef]
- Coronado-Ruiz, C.; Avendaño, R.; Escudero-Leyva, E.; Conejo-Barboza, G.; Chaverri, P.; Chavarría, M. Two new cellulolytic fungal species isolated from a 19th-Century art collection. Sci. Rep. 2018, 8, 7492. [Google Scholar] [CrossRef]
- Perdomo, H.; García, D.; Gené, J.; Cano, J.; Sutton, D.A.; Summerbell, R.; Guarro, J. Phialemoniopsis, a new genus of Sordariomycetes, and new species of Phialemonium and Lecythophora. Mycologia 2013, 105, 398–421. [Google Scholar] [CrossRef]
- Harrington, A.H.; Del Olmo-Ruiz, M.; U’Ren, J.M.; Garcia, K.; Pignatta, D.; Wespe, N.; Sandberg, D.C.; Huang, Y.-L.; Hoffman, M.T.; Arnold, A.E. Coniochaeta endophytica sp. nov., a foliar endophyte associated with healthy photosynthetic tissue of Platycladus orientalis (Cupressaceae). Plant Fungal Syst. 2019, 64, 65–79. [Google Scholar] [CrossRef]
- Nasr, S.; Bien, S.; Soudi, M.R.; Alimadadi, N.; Shahzadeh Fazeli, S.A.; Damm, U. Novel Collophorina and Coniochaeta species from Euphorbia polycaulis, an endemic plant in Iran. Mycol. Prog. 2018, 17, 755–771. [Google Scholar] [CrossRef]
- Vázquez-Campos, X.; Kinsela, A.S.; Waite, T.D.; Collins, R.N.; Neilan, B.A. Fodinomyces uranophilus gen. nov. sp. nov. and Coniochaeta fodinicola sp. nov., two uranium mine-inhabiting Ascomycota fungi from northern Australia. Mycologia 2014, 106, 1073–1089. [Google Scholar] [CrossRef]
- Leonhardt, S.; Büttner, E.; Gebauer, A.M.; Hofrichter, M.; Kellner, H. Draft Genome Sequence of the Sordariomycete Lecythophora (Coniochaeta) hoffmannii CBS 245.38. Genome Announc. 2018, 6, e01510-17. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.B.G.; Devadatha, B.; Abdel-Wahab, M.A.; Dayarathne, M.C.; Zhang, S.-N.; Hyde, K.D.; Liu, J.-K.; Bahkali, A.H.; Sarma, V.V.; Tibell, S.; et al. Phylogeny of new marine Dothideomycetes and Sordariomycetes from mangroves and deep-sea sediments. Bot. Mar. 2020, 63, 155–181. [Google Scholar] [CrossRef]
- Friebes, G.; Jaklitsch, W.M.; García, S.; Voglmayr, H. Lopadostoma taeniosporum revisited and a new species of Coniochaeta. Sydowia 2016, 68, 87–97. [Google Scholar] [CrossRef]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Raspé, O.; Karunarathna, S.C.; Wanasinghe, D.N.; Hongsanan, S.; et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.-G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal Diversity Notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Miller, A.N.; Huhndorf, S.M. Using phylogenetic species recognition to delimit species boundaries within Lasiosphaeria. Mycologia 2004, 96, 1106. [Google Scholar] [CrossRef]
- Hsieh, H.-M.; Lin, C.-R.; Fang, M.-J.; Rogers, J.D.; Fournier, J.; Lechat, C.; Ju, Y.-M. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol. Phylogenet. Evol. 2010, 54, 957–969. [Google Scholar] [CrossRef]
- Kuhnert, E.; Sir, E.B.; Lambert, C.; Hyde, K.D.; Hladki, A.I.; Romero, A.I.; Rohde, M.; Stadler, M. Phylogenetic and chemotaxonomic resolution of the genus Annulohypoxylon (Xylariaceae) including four new species. Fungal Divers. 2017, 85, 1–43. [Google Scholar] [CrossRef]
- Hsieh, H.-M.; Ju, Y.-M.; Rogers, J.D. Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 2005, 97, 844–865. [Google Scholar] [CrossRef]
- Zare, R.; Asgari, B.; Gams, W. The species of Coniolariella. Mycologia 2010, 102, 1383–1388. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Camporesi, E.; Tian, Q.; Liu, X.; Chamyuang, S.; Stadler, M.; Hyde, K.D. Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Divers. 2015, 73, 203–238. [Google Scholar] [CrossRef]
- Kuhnert, E.; Fournier, J.; Per, D.; Luangsa-ard, J.J.D.; Stadler, M. New Hypoxylon species from Martinique and new evidence on the molecular phylogeny of Hypoxylon based on ITS rDNA and β-Tubulin data. Fungal Divers. 2014, 64, 181–203. [Google Scholar] [CrossRef]
- Ju, Y.-M.; Hsieh, H.-M.; Ho, M.-C.; Szu, D.-H.; Fang, M.-J. Theissenia rogersii sp. nov. and phylogenetic position of Theissenia. Mycologia 2007, 99, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Vicente, F.; Basilio, A.; Platas, G.; Collado, J.; Bills, G.F.; González Del Val, A.; Martín, J.; Tormo, J.R.; Harris, G.H.; Zink, D.L.; et al. Distribution of the antifungal agents aordarins across filamentous fungi. Mycol. Res. 2009, 113, 754–770. [Google Scholar] [CrossRef]
- Peláez, F.; González, V.; Platas, G.; Sánchez-Ballesteros, J.; Rubio, V. Molecular phylogenetic studies within the Xylariaceae based on ribosomal DNA sequences. Fungal Divers. 2008, 31, 111–134. [Google Scholar]
- Checa, J.; Arenal, F.; Blanco, N.; Rogers, J.D. Coniolariella hispanica sp. nov. and other additions to Coniolariella. Mycol. Res. 2008, 112, 795–801. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Cheewangkoon, R.; van der Bank, M.; Swart, W.J.; Stchigel, A.M.; Cano-Lira, J.F.; Roux, J.; Madrid, H.; et al. Fungal Planet Description Sheets: 154–213. Persoonia 2013, 31, 188–296. [Google Scholar] [CrossRef]
- Sir, E.; Lambert, C.; Wendt, L.; Hladki, A.I.; Romero, A.I.; Stadler, M. A new species of Daldinia (Xylariaceae) from the Argentine subtropical montane forest. Mycosphere 2016, 7, 1378–1388. [Google Scholar] [CrossRef]
- Popuschoi, I.S. Mikoflora Plodorykh Derev’ev SSSR; 1971; p. 90. [Google Scholar]
- Kamiya, S.; Uchiyama, S.; Udagawa, S.-I. Two new species of Coniochaeta with a cephalothecoid peridium wall. Mycoscience 1995, 36, 377–383. [Google Scholar] [CrossRef]
- García, D.; Stchigel, A.M.; Cano, J.; Calduch, M.; Hawksworth, D.L.; Guarro, J. Molecular phylogeny of Coniochaetales. Mycol. Res. 2006, 110, 1271–1289. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Gené, J.; Ahmad, S.; Cano, J.; Al-Sweih, N.; Joseph, L.; Chandy, R.; Guarro, J. Coniochaeta polymorpha, a new species from endotracheal aspirate of a preterm neonate, and transfer of Lecythophora species to Coniochaeta. Antonie Van Leeuwenhoek 2013, 104, 243–252. [Google Scholar] [CrossRef] [PubMed]
- García, D.; Stchigel, A.M.; Guarro, J. A new species of Poroconiochaeta from Russian soils. Mycologia 2003, 95, 525–529. [Google Scholar] [CrossRef]
- Weber, E. The Lecythophora-Coniochaeta complex I. Morphological studies on Lecythophora species isolated from Picea abies. Nova Hedwigia 2002, 74, 159–185. [Google Scholar] [CrossRef]
- Mahoney, D.P.; LaFavre, J.S. Coniochaeta extramundana, with a synopsis of other Coniochaeta species. Mycologia 1981, 73, 931–952. [Google Scholar] [CrossRef]
- Checa, J.; Barrasa, J.M.; Moreno, G.; Fort, F.; Guarro, J. The genus Coniochaeta (Sacc.) Cooke (Coniochaetaceae, Ascomycotina) in Spain. Cryptog. Mycol. 1988, 9, 1–34. [Google Scholar]
- Francis, S.M.; Sparrow, J.R. Podospora horridula. Trans. Br. Mycol. Soc. 1984, 82, 380–382. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lodha, B.C. Helicogermslita, a new stromatic xylariaceous genus with a spiral germ slit from India. Trans. Br. Mycol. Soc. 1983, 81, 91–96. [Google Scholar] [CrossRef]
- Laessöe, T.; Spooner, B.M. Rosellinia & Astrocystis (Xylariaceae): New species and generic concepts. Kew Bull. 1993, 49, 1–70. [Google Scholar] [CrossRef]
- Petrini, L.E. Rosellinia and related genera in New Zealand. N. Zeal. J. Bot. 2003, 41, 71–138. [Google Scholar] [CrossRef]
- Lee, S.; Crous, P.W. A new species of Helicogermslita from South Africa. Sydowia 2003, 55, 109–114. [Google Scholar]
- Li, Q.; Kang, J.; Hyde, K.D. Two new Rosellinia species from southwest China. Mycotaxon 2015, 130, 563–567. [Google Scholar] [CrossRef]
- Seifert, K.A.; Rossman, A.Y. How to describe a new fungal species. IMA Fungus 2010, 1, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 1–32. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).