Abstract
The great plasticity and diversity of the Escherichia coli genome, together with the ubiquitous occurrence, make E. coli a bacterium of world-wide concern. Of particular interest are pathogenic strains and strains harboring antimicrobial resistance genes. Overlapping virulence-associated traits between avian-source E. coli and human extraintestinal pathogenic E. coli (ExPEC) suggest zoonotic potential and safety threat of poultry food products. We analyzed whole-genome sequencing (WGS) data of 46 mcr-1-positive E. coli strains isolated from retail raw meat purchased in the Czech Republic. The investigated strains were characterized by their phylogroup—B1 (43%), A (30%), D (11%), E (7%), F (4%), B2 (2%), C (2%), MLST type, and serotype. A total of 30 multilocus sequence types (STs), of which ST744 was the most common (11%), were identified, with O8 and O89 as the most prevalent serogroups. Using the VirulenceFinder tool, 3 to 26 virulence genes were detected in the examined strains and a total of 7 (15%) strains met the pathogenic criteria for ExPEC. Four strains were defined as UPEC (9%) and 18 (39%) E. coli strains could be classified as APEC. The WGS methods and available on-line tools for their evaluation enable a comprehensive approach to the diagnosis of virulent properties of E. coli strains and represent a suitable and comfortable platform for their detection. Our results show that poultry meat may serve as an important reservoir of strains carrying both virulence and antibiotic resistance genes for animal and human populations.
1. Introduction
Humans and warm-blooded animals act as natural reservoirs of a wide range of Gram-negative bacteria, such as numerous strains of Escherichia coli. Evolutionary processes including rearrangement of the existing genes, their loss or, conversely, acquisition of additional genes, result in the high diversity and plasticity of the E. coli genome. Therefore, E. coli can be harmless commensal, but also important pathogen harboring both virulence and antimicrobial resistance genes [1,2]. Pathogenic E. coli are equipped with a wide range of various virulence factors (VFs), including adhesins, invasins, toxins, and several uptake systems for various nutrients, which enable them to survive in inappropriate conditions (e.g., iron-limited environment in the urinary tract) [3,4].
Intestinal pathogenic E. coli can cause mild to life-threatening infections of the gastrointestinal tract in humans and animals. They are equipped with diverse mechanisms of pathogenicity and according to them they may be divided into enterotoxigenic (ETEC), enterohemorrhagic (EHEC), enteroinvasive (EIEC), enteropathogenic (EPEC), and enteroaggregative (EAEC) E. coli [2].
Extraintestinal pathogenic Escherichia coli (ExPEC), first defined by Johnson and Russo in 2002 [5], have a considerable influence on global health status. Based on the host specificity and preferred site of infection, ExPEC strains are classified into four groups—neonatal meningitis E. coli (NMEC), sepsis-associated E. coli (SEPEC), uropathogenic E. coli (UPEC), and avian pathogenic E. coli (APEC). As an extraintestinal pathogen, E. coli is the most common Gram-negative bacterium associated with bloodstream infections in both developed and developing countries. UPEC, which can also cause newborn meningitis and sepsis, is the most common cause of community-acquired urinary tract infections (UTIs) [6,7]. Avian pathogenic E. coli (APEC), which mainly causes respiratory and systemic disease in poultry, is associated with heavy economic losses in poultry industry [8]. Moreover, based on a large genetic overlap between APEC and certain human ExPEC as well as on numerous experimental studies performed in mammalian and avian animal models, APEC is presumed to have a zoonotic potential and represent an external reservoir for extraintestinal infections in humans [7,9,10].
Colistin, as the last-line drug for the treatment of life-threatening human infections caused by Gram-negative bacteria, is considered as one of the most critically important antimicrobials. It is used in both human and veterinary medicine, including food-producing animals such as poultry [11,12]. Recent studies have described the plasmid-mediated resistance encoded by mcr genes, detecting ten different variants of the mcr gene of which mcr-1 is predominant [13]. The possibility of spreading these genes through global trade with raw meat is being considered in another recent study, with mcr-1 gene detected in 19% of turkey meat and liver [14]. Considering previous results, we used the whole-genome sequencing (WGS) data to (i) identify virulence-associated genes in mcr-1-positive E. coli isolated from retail poultry meat; (ii) assign the tested E. coli strains to pathotypes, phylogenetic groups, MLST types and serotypes; and (iii) compare and discuss the results obtained by different APEC diagnostic approaches used in silico and, thus, the virulence properties of E. coli strains harboring horizontally transferred genes of antibiotic resistance isolated from retail meat were assessed.
2. Materials and Methods
2.1. Strain Collection
A total of 46 mcr-1-positive E. coli strains isolated from retail raw meat collected between March 2017 and January 2019 in the Czech Republic were analysed in this study. Altogether 45 strains were isolated from turkey meat and liver and 1 strain was obtained from chicken liver. Raw meat and liver samples (20 in total) were purchased from the Czech retail market and originated from the Czech Republic (3), Germany (3), Brazil (5), and Poland (9). More details about sample preparation and detection of colistin-resistant bacteria carrying mcr genes were described previously [14]. In brief, sample cultivation was performed in buffered peptone water under aerobic conditions at 37 °C overnight. After enrichment, the presence of mcr-1 gene was verified by PCR according to Liu et al., 2016 [15]. Positive samples were subsequently inoculated onto Brilliance UTI Clarity agar (Oxoid, Basingstoke, UK) supplemented with colistin sulphate (Discovery Fine Chemicals, Wimborne, UK) at a final concentration of 3.5 mg/L and incubated at 37 °C overnight. Presumptive colonies of E. coli (based on colour and colony morphology) were sub-cultured on Blood agar and were identified by MALDI-TOF MS with the use of Biotyper software (version 3.1, Bruker Daltonics GmbH, Bremen, Germany) with a score above 2.0. Up to 5 colonies from each sample were selected for further characterization.
2.2. Whole-Genome Sequencing
Genomic DNA was extracted using the Blood and Tissue kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). The preparation of DNA libraries and sequencing on the Illumina platform were carried out by Eurofins Genomics (Miseq 2 × 300 bp, n = 6), Macrogen Korea (Hiseq XTM Ten 2 × 300 bp, n = 12), ICM Paris (Nextseq 2 × 150 bp, n = 7) and CEITEC VFU (Miseq 2 × 250 bp, n = 25). The obtained sequence data was assembled using Velvet version 1.1.04 of Ridom SeqSphere+ (version 3.5.0; Ridom GmbH, Münster, Germany).
2.3. Escherichia Genus Strain Phylotyping
Phylogroups were identified by the ClermonTyper at http://clermontyping.iame-research.center/ [16]. The fasta files were downloaded to the above mentioned website. The analysis was accomplished without advanced option.
2.4. Multilocus Sequence Typing
E. coli sequence type (ST) was determined by the Achtman MLST scheme (www.enterobase.warwick.ac.uk/species/e.coli) [17].
2.5. WGS Based Serotyping
Identification of the outer membrane (O) antigen and flagellin protein (H) types was performed using the SerotypeFinder tool from the Centre for Genomic Epidemiology (CGE) website [18].
2.6. Virulence Factors Screening
Virulence genes were identified by VirulenceFinder (https://cge.cbs.dtu.dk/services/VirulenceFinder/). Nucleotide sequences of the selected 8 extra genes were downloaded from the NCBI database (Supplementary Materials, Table S1A). The presence of the investigated genes was analysed using Ridom SeqSphere+ software (version 3.5.0; Ridom GmbH, Münster, Germany). Procedure details of the analysis (default settings) were: required identity to the reference sequence—90%, required percentage aligned to the reference sequence—99%. Extra genes were selected according to ExPEC-defining traits, which are described in Johnson et al., 2003 [19]. APEC diagnostic approaches were used as described in Stromberg et al., 2017) [7], Schouler et al., 2012 [20] and Johnson et al., 2008 [21]. Based on criteria by Spurbeck et al., 2012 [22], the strains were defined as UPEC. Criteria of subpathotypes are shown in Table 1.

Table 1.
Criteria of ExPEC subpathotypes.
3. Results
3.1. Identification of Escherichia coli Phylogroups, Multilocus Sequence Types, Serotypes
From 20 raw poultry meat and liver samples, 46 mcr-1 positive E. coli strains were selected. These were classified in 7 phylogroups and 30 STs were identified. Phylogenetic analysis revealed that the most of the strains belonged to B1 group 43% (n = 20) with 15 unique STs (ST58, ST86, ST156, ST162, ST224, ST453, ST1079, ST1081, ST1167, ST1196, ST1463, ST1582, ST1589, ST2179, ST7973). Fourteen strains were assigned to group A (30%) with 6 different STs (ST10, ST93, ST744, ST746, ST756, ST5956) and to group D (n = 5; 11%) within which 5 STs were detected (ST38, ST69, ST349, ST1011, ST7233). Three strains belonged to group E (7%) with ST1140 (n = 2) and ST7233 (n = 1). Both strains in group F (4%) belonged to ST354, although they were isolated from different samples. In each of groups B2 and C, only 1 strain was detected and the strains belonged to ST1385 (B2) and ST410 (C). Sequence type 7233 was, as the only one, identified in two different phylogroups (D and E).
O-antigen was identified in 28 (61%) examined strains, whereas serogroups O8 and O89 were the most prevalent (6 and 4 strains). In 13 strains (28%) the O-antigen was not identified. Detailed results are shown in Figure 1 and Supplementary Materials, Table S1B.

Figure 1.
Number of detected virulence genes in E. coli mcr-1-positive strains by phylogroups.
3.2. Virulence Genes Screening
From 3 to 26 virulence genes (VGs) were confirmed in the tested strains (Figure 2; Table S1B). Based on the established criteria of pathotype definition (Table 1), 7 (15%) strains possessed ExPEC characteristics, 4 were defined as UPEC (9%), and 18 (39%) as APEC. Determination of APEC strains differed based on selected criteria. Using the Johnson et al., 2008 [21] approach, 14 strains were recognized as APEC, whereas only 4 strains met APEC criteria using Schouler et al., 2012 [20] method and only 3 strains harbored APEC virulence genes described by Stromberg et al., 2017 [7]. Two strains were confirmed as APEC by 2 different diagnostic approaches simultaneously.
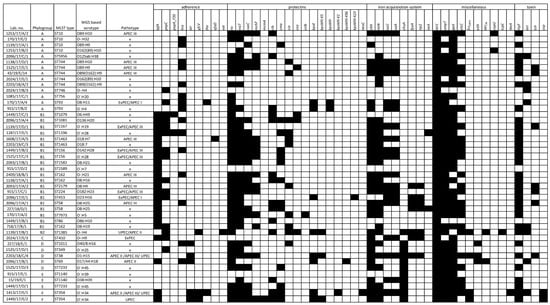
Figure 2.
Characteristics of 46 tested E. coli strains including phylogroup, MLST type, serotype, and virulence genes presence. APEC—avian pathogenic E. coli based on criteria by Stromberg et al., 2017 [7] (APEC I); by Schouler et al., 2012 [20] (APECII), by Johnson et al., 2008 [21] (APECIII); ExPEC—extraintestinal E. coli; UPEC—uropathogenic E. coli; ×—non-pathogenic E. coli; Black boxes represent presence and white boxes absence of respective gene.
Altogether 26 (56%) strains did not fully comply with the characteristics of the selected pathotypes, but they still carried some specific virulence genes (Figure 1, Supplementary Table S1B). Out of these strains, 15 harbored 4 virulence genes detected according to the Johnson et al., 2008 [21] APEC diagnostic approach.
A total of eight E. coli strains were found to be positive for the presence of some kps genes (K2 capsule), of which one strain carried ibeA—gene encoding invasin responsible for neonatal meningitis in humans [23,24]. This E. coli strain (Lab. no. 1413/17/E/1) was isolated from turkey meat originating in the Czech Republic.
None of the tested strains contained specific virulence genes relevant for intestinal E. coli pathotypes, except of eight strains harboring heat-stable enterotoxin astA.
4. Discussion
The strains used in this study were chosen from the set of mcr-1-positive strains isolated from retail raw meat products [14]. Since the emergence of the plasmid-mediated colistin (CT) resistance encoded by the mcr-1 gene in Chinese animal production [15], the worldwide dissemination of this CT resistance has been recorded [25]. The occurrence of Enterobacteriaceae with the mcr-1 gene has been reported in the human population in connection with animal products from different animal species [26,27]. Both of these facts and the role of colistin as the last treatment resort in serious human infections caused by multidrug-resistant bacteria should lead to a great concern around the world [28]. The resistance to antimicrobials can be based on DNA mutation or on horizontal gene transfer. Many genes, coding for resistance to antimicrobials, are inserted into conjugative plasmids, transposons or integrons, called mobile genetic elements. These elements may also carry virulence factor determinants and, therefore, a correlation between virulence and antimicrobial resistance has been documented, at least in some E. coli clones [2].
Virulence potential of E. coli is determined by the occurrence of virulence genes, coding for colonizing factors (fimbriae and adhesins), survival of bacterial cells in unfavorable environments (protectins and siderophores) or causing the host inflammatory response, e. g. toxin production [2]. In the past, different systems have been used to assess the virulence potential of E. coli. Based on recent knowledge, determination of serotypes is not satisfactory, even if some of them are more frequently associated with certain infections than others [29,30]. Out of the serogroups, described as typical of APEC, only O1 (n = 1), O18 (n = 2) and O8 (n = 7) were detected, whereas APEC virulence genes were only confirmed in five of these strains. In the rest of APEC strains, either atypical serogroups O17, O23, O89 and O182 were confirmed or the serogroup was not identified (O-; 28%). Therefore, our findings support the opinion that the association between serogroup and virulence potential is weak.
Phylotyping is a simple method for the assessment of clinical significance of E. coli strains. Based on the presence of four different genes or DNA fragments, E. coli can be classified into eight phylogroups A, B1, B2, C, D, E, F, or cryptic clade I [31]. It has been confirmed that commensal strains mostly fall into A and B1 phylogroups. Intestinal pathogenic E. coli affiliate to groups A, B1, or D. The most of the ExPEC strains belong to groups B2 and D, whereas human pathogenic strains are usually clustered in group B2 [32]. The strains from phylogroup C are considered to be a sister group to those in B1, group E is related to group D and phylogroup F is very similar to B2 [24,33]. In our study, all strains belonging to the phylogroups B2 (n = 1), D (n = 2), and F (n = 2), which are associated with highly virulent E. coli pathotypes [31,34], were determined as pathogenic based on selected criteria in contrast to strains from phylogroup E (3), which did not fully meet the definition of selected pathotypes. On the contrary, the majority of pathogenic strains (9/20; 45%) belonged to B1 or A phylogroup (5/20; 25%). Phylogenetic analysis of all 46 tested strains revealed that the most of E. coli strains belonged to B1 (43%), followed by groups A (30%), D (11%), E (7%), F (4%), B2 (2%), and C (2%). Our data are in accordance with results obtained for 409 E. coli strains from commercial chicken carcasses in Brazil where the most frequent phylogroup was B1 (36.6%), followed by A (31.7%), D (28.1%), and B2 (3.40%) [35]. Other studies aimed at characterization of E. coli isolates from retail poultry meat and eggs described B2 and D as the major phylotypes in ExPEC, while non-ExPEC isolates belonged to groups A and B1 [34,36].
The MLST scheme is another method that is used for E. coli typing. It provides good means for E. coli typing, but there is no direct correlation between STs and their phylogroup affiliation [37]. In our study, 30 different ST types were identified. Out of them, 22 were unique, which supports the finding that E. coli are highly diverse (Figure 1). The most prevalent was ST744 (11%). All these strains were assigned to group A, O89, but were isolated from different samples originating in different countries (Poland, Germany, and Brazil). In three samples, ST162 was detected, which was described by Zhuge et al., 2019 [28] as a highly virulent mcr-positive E. coli clone. Only one of these strains (Lab. no. 2409/18/B/1) fully met the characteristics of APEC by the Johnson et al., 2008 [21] approach.
The developed PCR and sequencing techniques, especially whole genome sequencing methods, enable detailed screening of the occurrence of specific genes and open new possibilities to estimate the relationship between the presence of genes and pathogenicity of E. coli strains [29,38]. The VirulenceFinder software has been updated in 2020 and a broader spectrum of virulence genes encoding, e.g., adhesins, siderophores, toxins, and protectins have been included in multiple variants [39]. Therefore, at least three virulence genes were detected in the tested strains. Genes affecting resistance to host innate immunity (e.g., ompT) or protecting against phagocytosis (e.g., iss) have often been detected, followed by genes employed in the iron acquisition system or transport system, e.g., sitA gene encoding periplasmic-binding protein and iroN encoding the outer membrane receptor fepA belonging to ExPEC siderophores group or iutA gene.
Using WGS data, we focused on the identification of genes by two approaches, which are routinely employed in our laboratory for APEC detection in vitro [20,21]. The method according to Schouler et al., 2012 [20] is based on extensive characterization of 1491 avian E. coli isolates from France, Spain and Belgium. We detected two out of four patterns of VGs-B (iutA+ P(F11)− frzorf4+) and D (iutA− sitA+ aec26+) in four samples from our collection of strains. The method takes into account different strategies of E. coli to invade the host according to genetic equipment of bacterial strain and uses both chromosomally and plasmid located VGs. On the other hand, to identify traits that predict APEC virulence, five plasmid pathogenicity-associated island (PAI) genes were validated for rapid diagnostics according to Johnson et al., 2008 [21]. Fourteen of 46 E. coli isolates carried all of five genes associated with highly pathogenic APEC strains (iutA, hlyF, iss, iroN, ompT). However, four out of five VGs were detected in 37% of strains and three VGs were detected in 4% of strains. The relationship between in vivo virulence and the number of PAI genes in one-day-old chicks was analysed by de Oliveira et al., 2015 [40], who found that 95% of APEC isolates harbored the two, three, or four above mentioned genes, even though isolates with fewer than two VGs were rarely pathogenic. Taking into account these results, the occurrence of APEC strains in our study increased up to 72% (33/46). However, the study of de Oliveira et al., 2015 [40], also showed that two or more VGs are found in approximately 50% of the non-pathogenic isolates. These results suggested that the presence of two or more VGs is necessary but not sufficient to turn E. coli into APEC and, therefore, such interpretation is uncertain.
5. Conclusions
Based on various MLST types, serotypes, and virulence genes detected, high diversity of examined strains was confirmed. Our results show that E. coli strains with plasmid mediated colistin resistance isolated from poultry meat products disposed of various virulence genes. In total, 43% of these strains were assigned to at least one of the assessed pathotype (APEC, 39%; ExPEC, 15%; UPEC, 9%) and may pose a threat of further spreading to the environment, animals, and humans, especially if the hypothesis is taken into account that a combination of resistance and virulence properties may be a kind of advantage for E. coli under a selective pressure of antimicrobials.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/2/308/s1, Table S1: Accession number of genes selected for ExPEC genes screening and detail results of virulence genes findings.
Author Contributions
Conceptualization, R.K., T.G., and I.K.; Investigation, A.K. and M.F.; Methodology, R.K., T.G., M.K., I.K., and M.F.; Writing—original draft, M.K. and I.K.; Writing—review and editing, I.K., R.K., and T.G. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Ministry of Health of the Czech Republic grant no: NV 18-09-00254 and the Ministry of Education, Youth and Sport (MEYS) project no.: CZ.1.05./2.1.00/19.0385 (Prague, Czech Republic).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB34874 (https://www.ebi.ac.uk/ena/data/view/PRJEB34874).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Bielaszewska, M.; Dobrindt, U.; Gärtner, J.; Gallitz, I.; Hacker, J.; Karch, H.; Müller, D.; Schubert, S.; Schmidt, M.A.; Sorsa, L.J.; et al. Aspects of genome plasticity in pathogenic Escherichia coli. Int. J. Med. Microbiol. 2007, 297, 625–639. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Mendonça, N. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 2012, 3, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Wannemuehler, Y.; Johnson, S.J.; Stell, A.L.; Doetkott, C.; Johnson, J.R.; Kim, K.S.; Spanjaard, L.; Nolan, L.K. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 2008, 74, 7043–7050. [Google Scholar] [CrossRef] [PubMed]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-antibiotic Antimicrobial Strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Russo, T.A. Extraintestinal pathogenic Escherichia coli: ‘the other bad E. coli’. J. Lab. Clin. Med. 2002, 139, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Stromberg, Z.R.; Johnson, J.R.; Fairbrother, J.M.; Kilbourne, J.; Van Goor, A.; Curtiss III, R.; Mellata, M. Evaluation of Escherichia coli isolates from healthy chickens to determine their potential risk to poultry and human health. PLoS ONE 2017, 12, e0180599. [Google Scholar] [CrossRef]
- Kabir, S.M.L. Avian Colibacillosis and Salmonellosis: A Closer Look at Epidemiology, Pathogenesis, Diagnosis, Control and Public Health Concerns. Int. J. Environ. Res. Public Health 2010, 7, 89–114. [Google Scholar] [CrossRef]
- Manges, A.R. Escherichia coli and urinary tract infections: The role of poultry-meat. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef]
- Jørgensen, S.L.; Stegger, M.; Kudirkiene, E.; Lilje, B.; Poulsen, L.L.; Ronco, T.; Dos Santos, T.P.; Kiil, K.; Bisgaard, M.; Pedersen, K.; et al. Diversity and Population Overlap between Avian and Human Escherichia coli Belonging to Sequence Type 95. mSphere 2019, 4, e00333-18. [Google Scholar]
- EMA (European Medicines Agency); EFSA (European Food Safety Authority). EMA and EFSA Joint Scientific Opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). EFSA J. 2017, 15, 4666. [Google Scholar] [CrossRef]
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef]
- Gelbíčová, T.; Baráková, A.; Florianová, M.; Jamborová, I.; Zelendová, M.; Pospíšilová, L.; Koláčková, I.; Karpíšková, R. Dissemination and Comparison of Genetic Determinants of mcr-Mediated Colistin Resistance in Enterobacteriaceae via Retailed Raw Meat Products. Front. Microbiol. 2019, 10, 2824. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Wirth, T.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J. Clin. Microbiol. 2018, 53, 2410–2426. [Google Scholar] [CrossRef]
- Johnson, J.R.; Murray, A.C.; Gajewski, A.; Sullivan, M.; Snippes, P.; Kuskowski, M.A.; Smith, K.E. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chem. 2003, 47, 2161–2168. [Google Scholar] [CrossRef]
- Schouler, C.; Schaeffer, B.; Brée, A.; Mora, A.; Dahbi, G.; Biet, F.; Oswald, E.; Mainil, J.; Blanco, J.; Moulin-Schouleur, M. Diagnostic strategy for identifying avian pathogenic Escherichia coli based on four patterns of virulence genes. J. Clin. Microbiol. 2012, 50, 1673–1678. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef]
- Spurbeck, R.R.; Dinh, P.C., Jr.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize, the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef]
- Germon, P.; Chen, Y.H.; He, L.; Blanco, J.E.; Brée, A.; Schouler, C.; Huang, S.H.; Moulin-Schouleur, M. IbeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 2005, 151, 1179–1186. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, H.; Xu, J.; Wang, Y.; Zhang, Q.; Walsh, T.R.; Shao, B.; Wu, C.; Hu, Y.; Yang, L.; et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 2018, 3, 1054–1062. [Google Scholar] [CrossRef]
- Monte, D.F.; Mem, A.; Fernandes, M.R.; Cerdeira, L.; Esposito, F.; Galvão, J.A.; Franco, B.D.G.M.; Lincopan, N.; Landgraf, M. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 2017, 61, e02718-16. [Google Scholar] [CrossRef]
- Mulvey, M.R.; Mataseje, L.F.; Robertson, J.; Nash, J.H.; Boerlin, P.; Toye, B.; Irwin, R.; Melano, R.G. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 2016, 16, 289–290. [Google Scholar] [CrossRef]
- Zhuge, X.; Jiang, M.; Tang, F.; Sun, Y.; Ji, Y.; Xue, F.; Ren, J.; Zhu, W.; Dai, J. Avian-source mcr-1-positive characteristics with E. coli causing human extra-intestinal infections. Vet. Microbiol. 2019, 239, 108483. [Google Scholar] [CrossRef]
- Mainil, J. Escherichia coli virulence factors. Vet. Immun. Immunopathol. 2013, 152, 2–12. [Google Scholar] [CrossRef]
- EFSA BIOHAZ Panel; Koutsoumanis, K.; Allende, A.; Alvarez-Ordonez, A.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Scientific Opinion on the pathogenicity assessment of Shiga toxin-producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020, 18, 5967. [Google Scholar]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Markland, S.M.; LeStrange, K.J.; Sharma, M.; Kniel, K.E. Old Friends in New Places: Exploring the Role of Extraintestinal E. coli in Intestinal Disease and Foodborne Illness. Zoonoses Public Health 2015, 62, 491–496. [Google Scholar] [CrossRef]
- Aslam, M.; Toufeer, M.; Narvaez Bravo, C.; Lai, V.; Rempel, H.; Manges, A.; Diarra, M.S. Characterization of Extraintestinal Pathogenic Escherichia coli isolated from retail poultry meats from Alberta, Canada. Int. J. Food Microbiol. 2014, 177, 49–56. [Google Scholar] [CrossRef]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; Tagliari de Brito, K.C.; Benito Guimarães de Brito, B.; Nakazato, G.; Takayama Kobayashi, R.K. Distribution of ExPEC Virulence Factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli Isolated From Commercialized Chicken Carcasses. Front. Microbiol. 2019, 9, 3254. [Google Scholar] [CrossRef]
- Mitchell, N.M.; Johnson, J.R.; Johnston, B.; Curtis III, R.; Mellata, M. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl. Environ. Microbiol. 2015, 81, 1177–1187. [Google Scholar] [CrossRef]
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 2015, 161 (Pt. 5), 980–988. [Google Scholar] [CrossRef]
- World Health Organization. 1980 World Health Organization (W.H.O.) Scientific Working Group Escherichia coli diarrhoea. Bull. WHO 1980, 58, 23–36. [Google Scholar]
- Tetzschner, A.M.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef]
- De Oliveira, A.L.; Rocha, D.A.; Finkler, F.; de Moraes, L.B.; Barbieri, N.L.; Pavanelo, D.B.; Winkler, C.; Grassotti, T.T.; de Brito, K.C.; de Brito, B.G.; et al. Prevalence of ColV Plasmid-Linked Genes and In Vivo Pathogenicity of Avian Strains of Escherichia coli. Foodborne Pathog. Dis. 2015, 12, 679–685. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).