Gluconacetobacter brunescens sp. nov., a Novel Acetic Acid Bacterium Isolated from Pear Vinegar, Producing a Water-Soluble Brown Pigment
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Hr-1-5 and Growth Conditions
2.2. Molecular Identification of Hr-1-5
2.3. Genome Analysis
2.4. Phenotypic Analysis
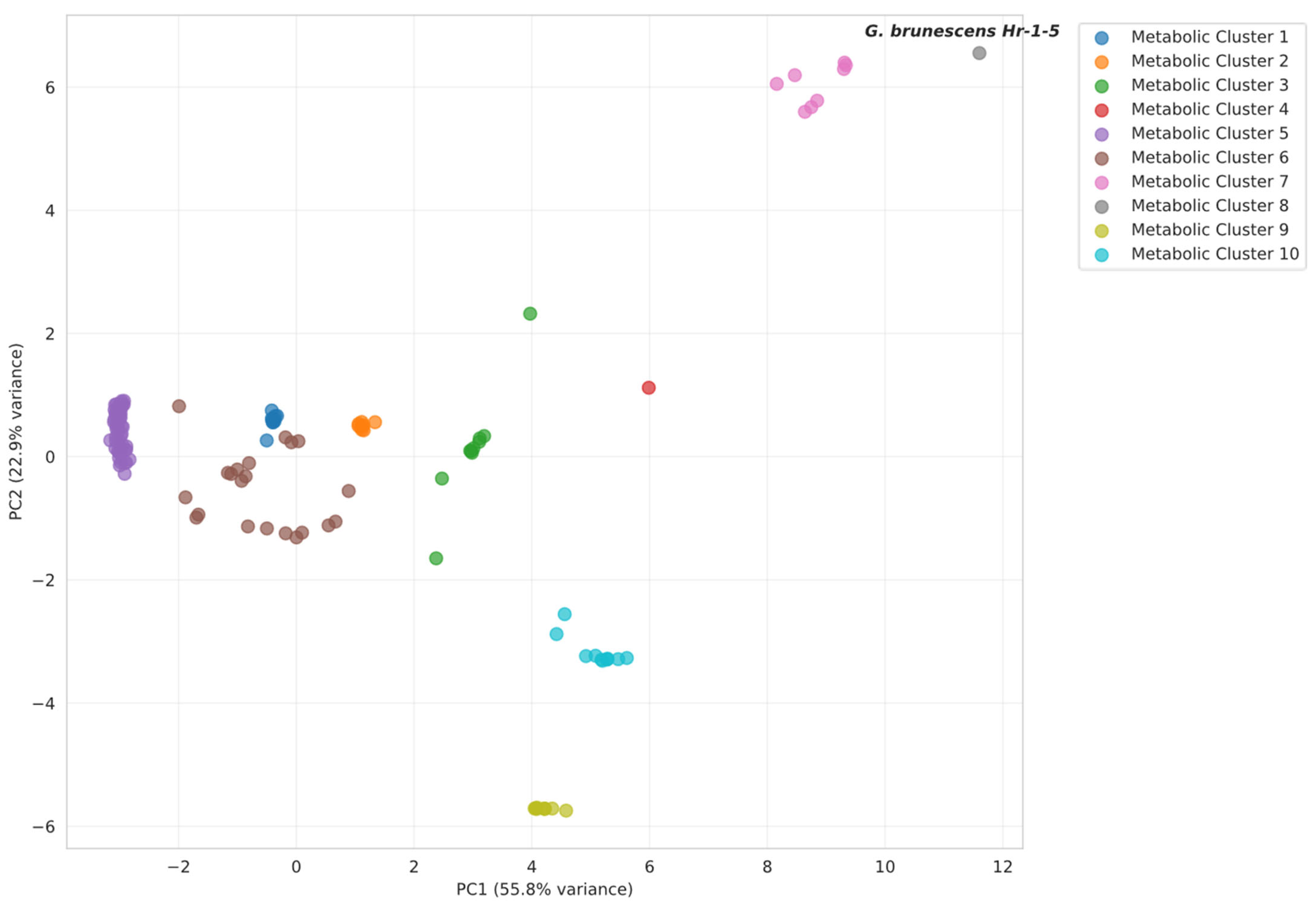
2.5. Metabolic Pathway Analysis
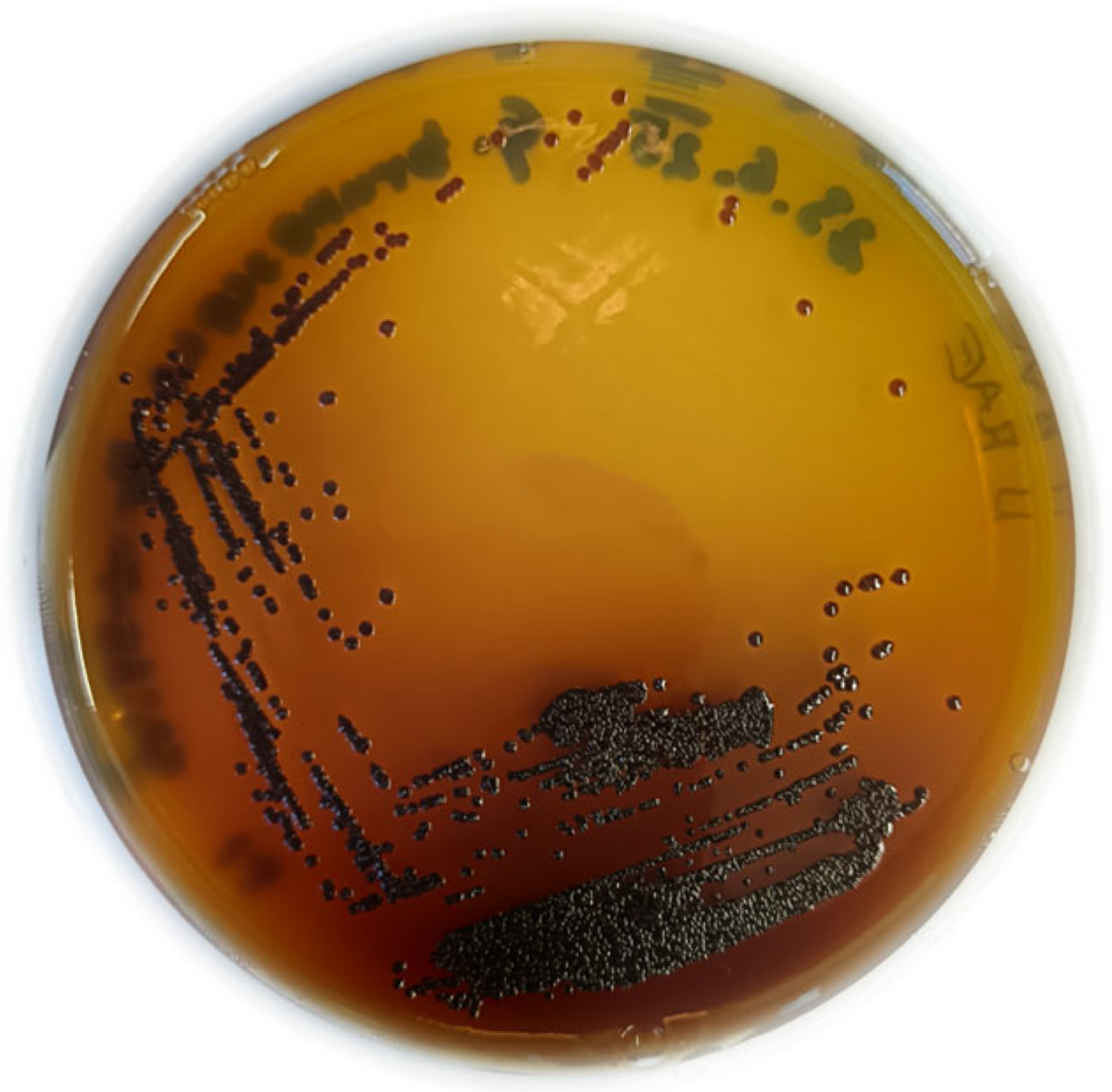
- (a)
- Novacetimonas hansenii (No. 1), Komagataeibacter xylinus (No. 2) and Escherichia coli for negative control, displaying no natural pigment production;
- (b)
- (c)
- Gluconacetobacter tumulisoli (No. 5) for eumelanin production [22];
- (d)
- Gluconacetobacter diazotrophicus (No. 6) for betalain production [17];
- (e)
- Streptomyces scabiei (No. 8) for multiple melanin production [44];
- (f)
- (g)
- Acinetobacter baumannii (No. 11) just for pyomelanin production [48].
2.6. Production of Colored BC Membranes
3. Results and Discussion
3.1. Morphological, Physiological and Biochemical Tests
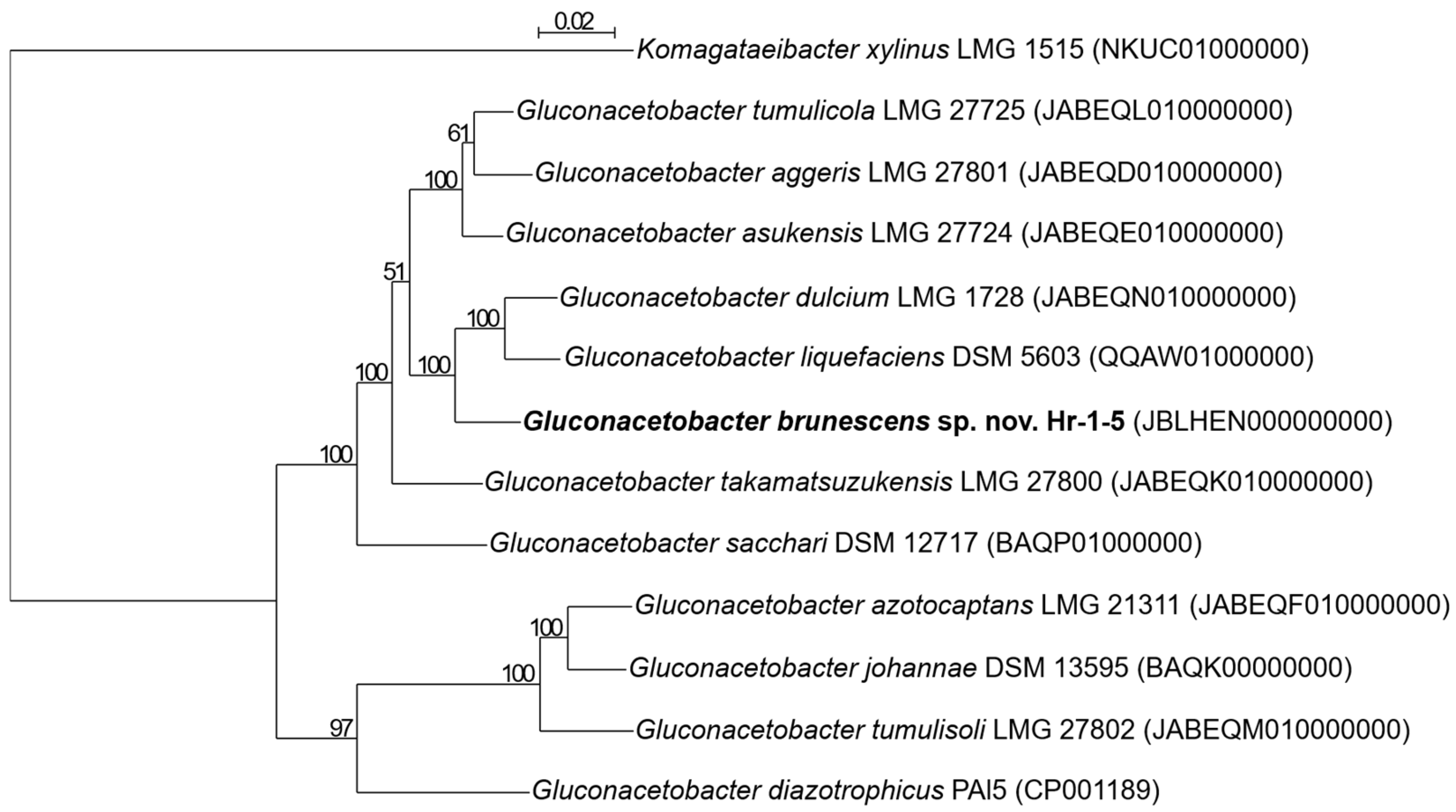
3.2. Phylogenetic Analysis
3.3. Genomic Analysis
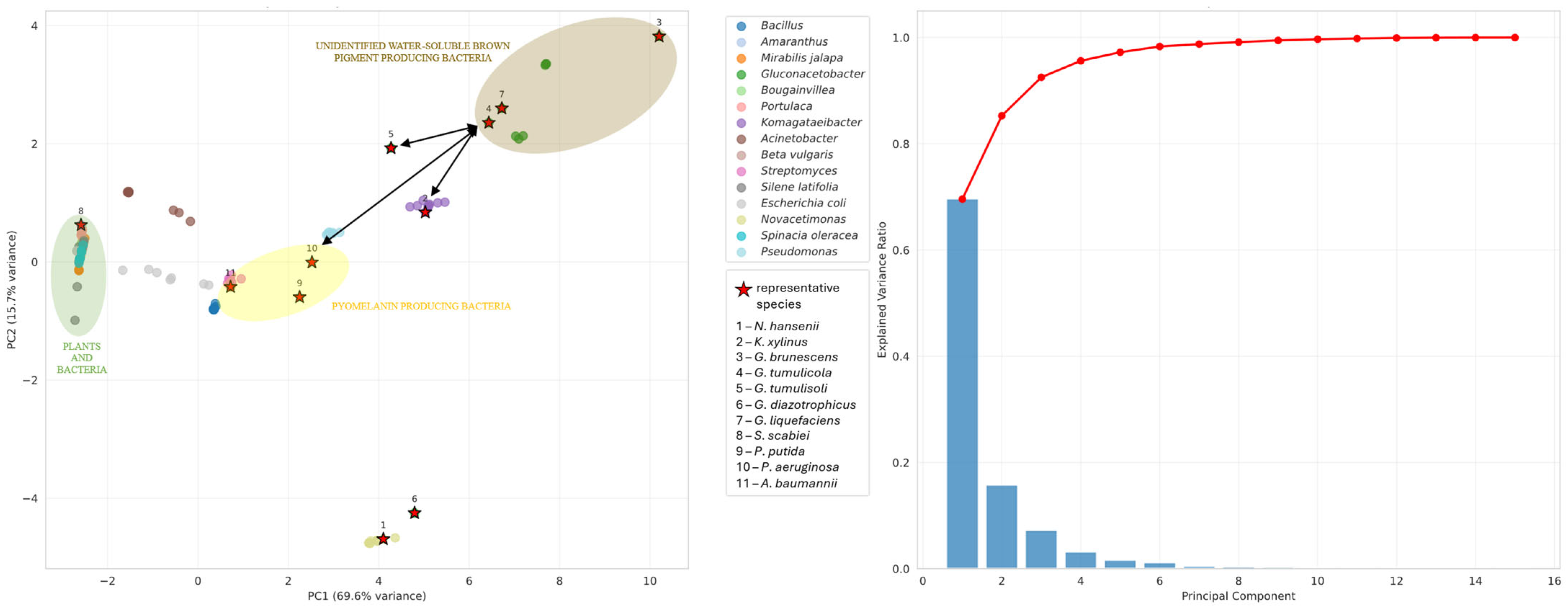
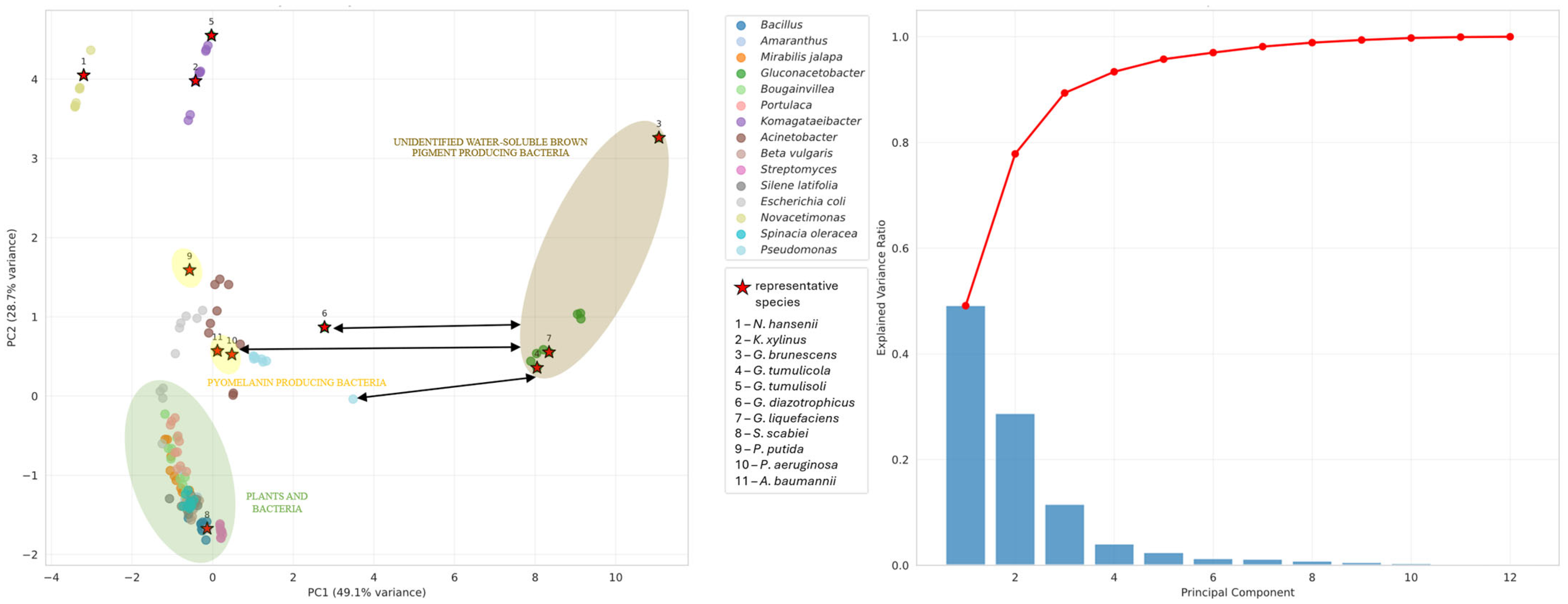
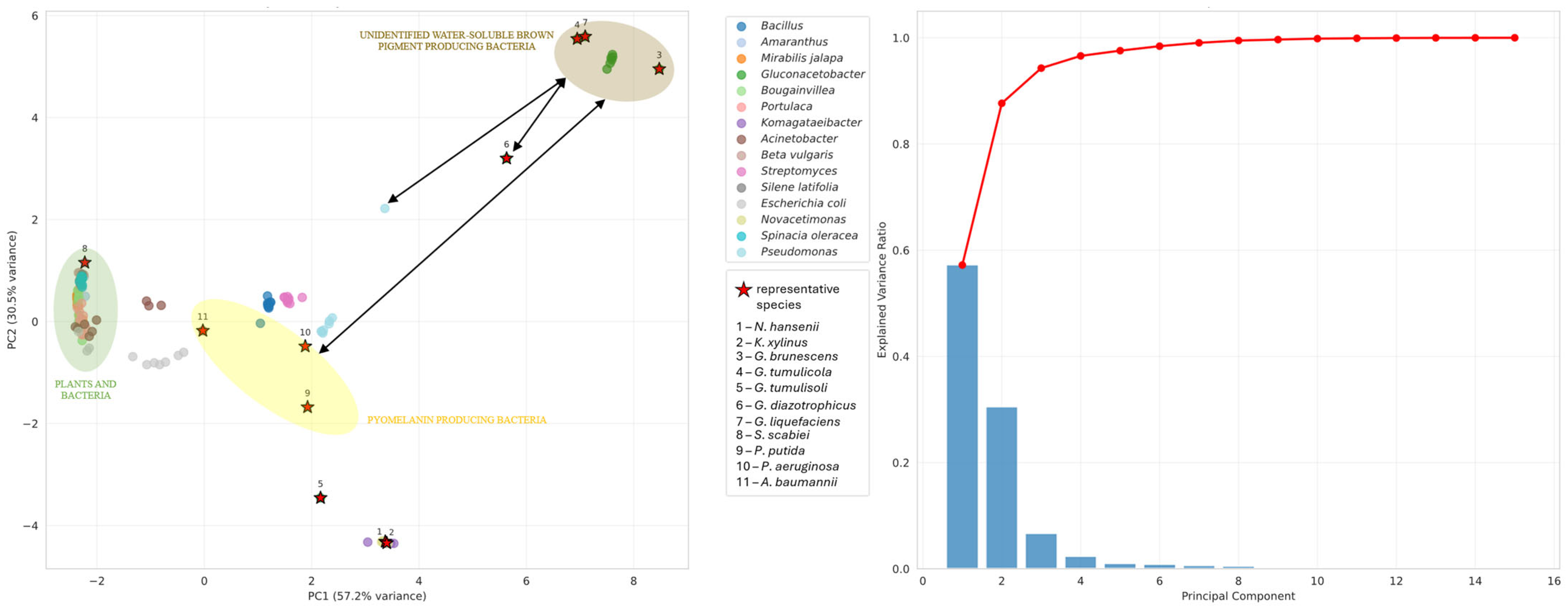
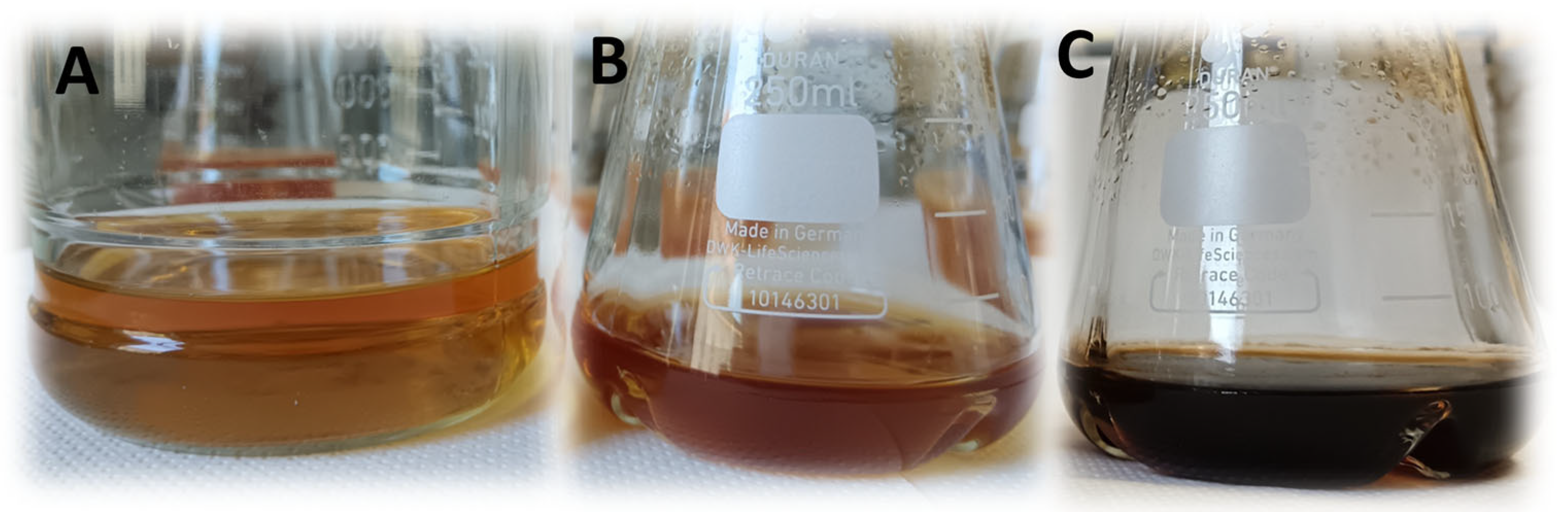
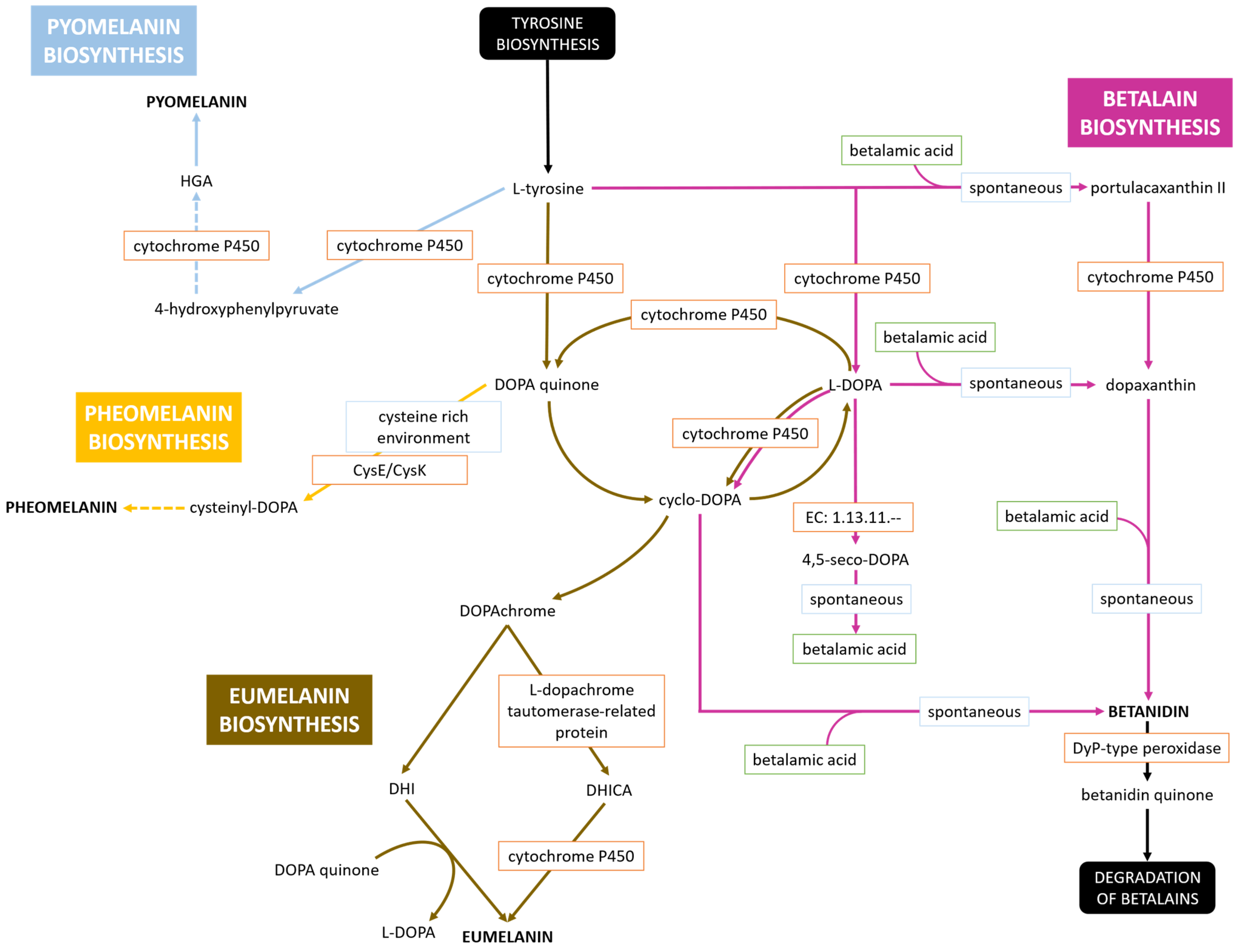
3.4. Pigment Synthesis
- (a)
- The unidentified water-soluble brown pigment of G. brunescens, G. liquefaciens and G. tumulicola is a novel pigment of the Gluconacetobacter group;
- (b)
- The unidentified water-soluble brown pigment is pyomelanin, but the cytochromes P450 of these bacteria may follow a novel, yet undescribed pathway;
- (c)
- G. dulcium requires thorough investigation to determine whether it also produces the water-soluble brown pigment;
- (d)
- All Gluconacetobacter species warrant further study to elucidate their pigment biosynthesis pathways.
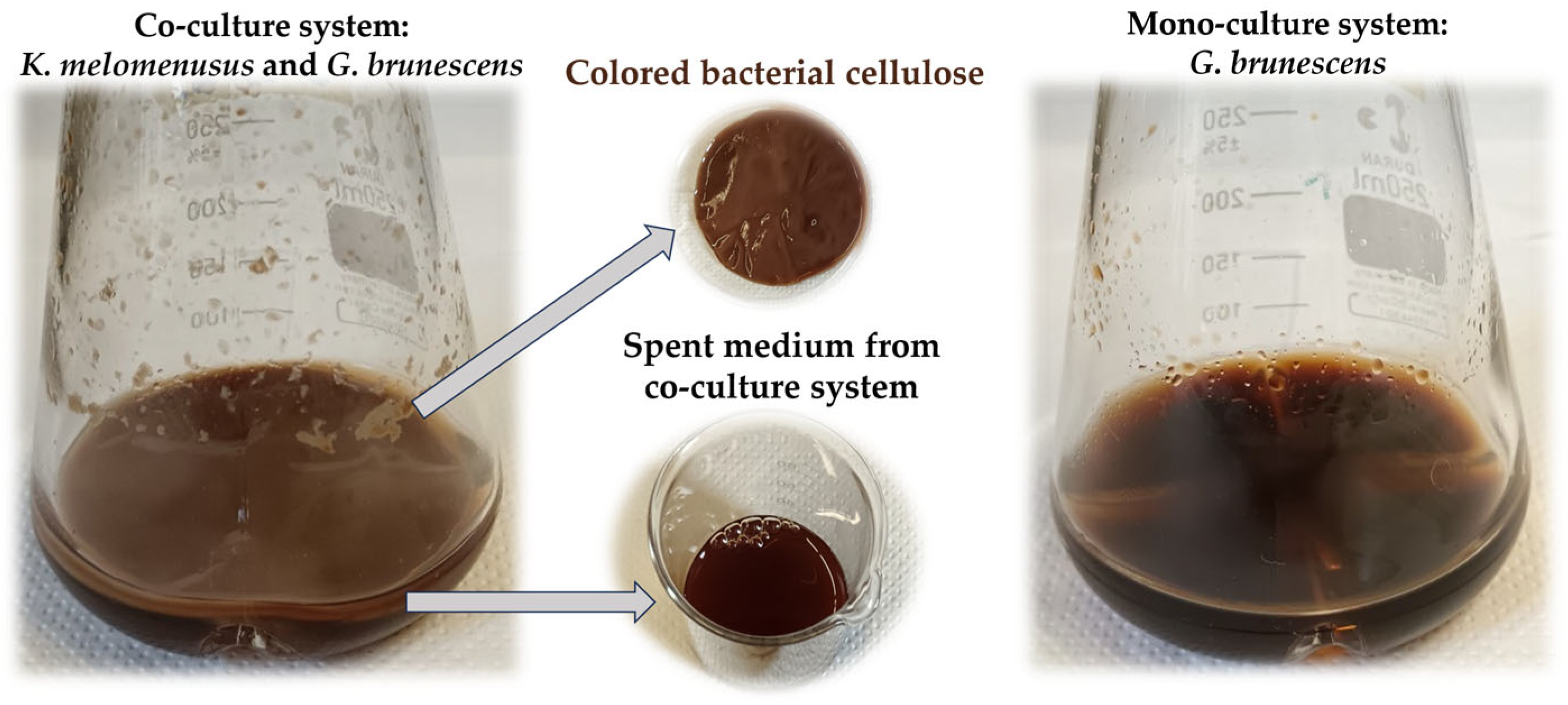
3.5. Bacterial Nanocellulose Synthesis and Co-Cultivation
3.6. Antimicrobial Resistance and Inhibitory Effects
4. Description of Gluconacetobacter brunescens sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Fatty Acid (%) | Hr-1-5 (%) |
|---|---|
| C12:0 | 0.3 |
| C14:0 | 3.5 |
| C16:0 | 12.6 |
| C17:0 | 0.2 |
| C18:0 | 1.9 |
| C16:1 ω7c/C16:1 ω6c | 0.7 |
| C18:1 ω7c | 65.4 |
| C14:0 2OH | 4.9 |
| C16:0 2OH | 5.8 |
| C16:0 3OH | 1.4 |
| C18:0 3OH | 0.5 |
| C14:0 3OH/C16:1 iso I | 0.9 |
| C19:0 cyclo ω8c | 1.1 |
References
- Kersters, K.; Lisdiyanti, P.; Komagata, K.; Swings, J. The Family Acetobacteraceae: The Genera Acetobacter, Acidomonas, Asaia, Gluconacetobacter, Gluconobacter, and Kozakia. In The Prokaryotes; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Jelenko, K.; Cepec, E.; Nascimento, F.X.; Trček, J. Comparative Genomics and Phenotypic Characterization of Gluconacetobacter entanii, a Highly Acetic Acid-Tolerant Bacterium from Vinegars. Foods 2023, 12, 214. [Google Scholar] [CrossRef]
- Brandão, P.R.; Crespo, M.T.B.; Nascimento, F.X. Phylogenomic and Comparative Analyses Support the Reclassification of Several Komagataeibacter Species as Novel Members of the Novacetimonas gen. nov. and Bring New Insights into the Evolution of Cellulose Synthase Genes. Int. J. Syst. Evol. Microbiol. 2022, 72, 005252. [Google Scholar] [CrossRef]
- Gomes, R.J.; Borges, M.d.F.; Rosa, M.d.F.; Castro-Gómez, R.J.H.; Spinosa, W.A. Acetic Acid Bacteria in the Food Industry: Systematics, Characteristics and Applications. Food Technol. Biotechnol. 2018, 56, 139–151. [Google Scholar] [CrossRef]
- Cleenwerck, I.; De Vos, P. Polyphasic Taxonomy of Acetic Acid Bacteria: An Overview of the Currently Applied Methodology. Int. J. Food Microbiol. 2008, 125, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Yukphan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Nakagawa, Y. Subdivision of the Genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: The Proposal of Komagatabacter gen. nov., for Strains Accommodated to the Gluconacetobacter xylinus Group in the α-Proteobacteria. Ann. Microbiol. 2012, 62, 849–859. [Google Scholar] [CrossRef]
- Sombolestani, A.S.; Cleenwerck, I.; Cnockaert, M.; Borremans, W.; Wieme, A.D.; Moutia, Y.; Spaepen, S.; De Vuyst, L.; Vandamme, P. Gluconacetobacter dulcium sp. nov., a Novel Gluconacetobacter Species from Sugar-Rich Environments. Int. J. Syst. Evol. Microbiol. 2021, 71, 004569. [Google Scholar] [CrossRef]
- Filgueiras, L.; Silva, R.; Almeida, I.; Vidal, M.; Baldani, J.I.; Meneses, C.H.S.G. Gluconacetobacter diazotrophicus Mitigates Drought Stress in Oryza sativa L. Plant Soil 2020, 451, 57–73. Plant Soil 2020, 451, 57–73. [Google Scholar] [CrossRef]
- Tazato, N.; Nishijima, M.; Handa, Y.; Kigawa, R.; Sano, C.; Sugiyama, J. Gluconacetobacter tumulicola sp. nov. and Gluconacetobacter asukensis sp. nov., Isolated from the Stone Chamber Interior of the Kitora Tumulus. Int. J. Syst. Evol. Microbiol. 2012, 62, 2032–2038. [Google Scholar] [CrossRef]
- Franke, I.H.; Fegan, M.; Hayward, C.; Leonard, G.; Stackebrandt, E.; Sly, L.I. Description of Gluconacetobacter sacchari sp. nov., a New Species of Acetic Acid Bacterium Isolated from the Leaf Sheath of Sugar Cane and from the Pink Sugar-Cane Mealy Bug. Int. J. Syst. Evol. Microbiol. 1999, 49, 1681–1693. [Google Scholar] [CrossRef]
- Eskin, N.; Vessey, K.; Tian, L. Research Progress and Perspectives of Nitrogen Fixing Bacterium, Gluconacetobacter diazotrophicus, in Monocot Plants. Int. J. Agron. 2014, 2014, 208383. [Google Scholar] [CrossRef]
- Mehnaz, S.; Lazarovits, G. Inoculation Effects of Pseudomonas putida, Gluconacetobacter azotocaptans, and Azospirillum lipoferum on Corn Plant Growth under Greenhouse Conditions. Microb. Ecol. 2006, 51, 326–335. [Google Scholar] [CrossRef]
- Senthilnathan, S.; Rahman, S.S.A.; Pasupathi, S.; Venkatachalam, P.; Karuppiah, S. Stoichiometric Analysis and Production of Bacterial Cellulose by Gluconacetobacter liquefaciens Using Borassus flabellifer L. Jaggery. Appl. Biochem. Biotechnol. 2022, 194, 3645–3667. [Google Scholar] [CrossRef]
- Rhee, C.; Kim, H.; Emmanuel, S.A.; Kim, H.-G.; Won, S.; Bae, J.; Bai, S.C.; Koh, S.-C. Probiotic Effects of Mixture of Groenewaldozyma salmanticensis and Gluconacetobacter liquefaciens on Growth and Immune Responses in Paralichthys olivaceus. Lett. Appl. Microbiol. 2020, 70, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gan, L.; He, Z.; Jiang, G.; He, T. Bacterial Pigments as a Promising Alternative to Synthetic Colorants: From Fundamentals to Applications. J. Microbiol. Biotechnol. 2024, 34, 2153–2165. [Google Scholar] [CrossRef]
- Contreras-Llano, L.E.; Guerrero-Rubio, M.A.; Lozada-Ramírez, J.D.; García-Carmona, F.; Gandía-Herrero, F. First Betalain-Producing Bacteria Break the Exclusive Presence of the Pigments in the Plant Kingdom. mBio 2019, 10, e00345-19. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Rubio, M.A.; Sheehan, H.; Brockington, S.F. In Planta Complementation of the Betalain Biosynthetic Pathway with a Bacterial Dioxygenase. PLoS ONE 2025, 20, e0325603. [Google Scholar] [CrossRef]
- Azeredo, H.M.C. Betalains: Properties, Sources, Applications, and Stability—A Review. Int. J. Food Sci. Technol. 2009, 44, 2365–2376. [Google Scholar] [CrossRef]
- Kurian, N.K.; Bhat, S.G. Food, Cosmetic and Biological Applications of Characterized DOPA-Melanin from Vibrio Alginolyticus Strain BTKKS3. Appl. Biol. Chem. 2018, 61, 163–171. [Google Scholar] [CrossRef]
- El-Naggar, N.E.A.; Saber, W.I.A. Natural Melanin: Current Trends, and Future Approaches, with Especial Reference to Microbial Source. Polymers 2022, 14, 1339. [Google Scholar] [CrossRef]
- Song, J.; Ma, Y.; Xie, Z.; Chen, F. Investigation of Eumelanin Biosynthesis in Gluconacetobacter tumulisoli FBFS 97: A Novel Insight into a Bacterial Melanin Producer. Microorganisms 2025, 13, 480. [Google Scholar] [CrossRef]
- Pavan, M.E.; López, N.I.; Pettinari, M.J. Melanin Biosynthesis in Bacteria, Regulation and Production Perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 1357–1370. [Google Scholar] [CrossRef]
- Karničnik, B.; Accetto, T.; Fanedl, L.; Jugović, I.; Trček, J. Isolation and Characterization of Komagataeibacter piraceti sp. nov. and Novacetimonas labruscae sp. nov.: Two Novel Microaerobic Cellulose-Producing Acetic Acid Bacteria from Vinegars. Microorganisms 2025, 13, 456. [Google Scholar] [CrossRef]
- Trček, J.; Teuber, M. Genetic and Restriction Analysis of the 16S–23S rDNA Internal Transcribed Spacer Regions of the Acetic Acid Bacteria. FEMS Microbiol. Lett. 2002, 208, 69–75. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Tisza, M.J.; Belford, A.K.; Dominguez-Huerta, G.; Bolduc, B.; Buck, C.B. Cenote-Taker 2 Democratizes Virus Discovery and Sequence Annotation. Virus Evol. 2021, 7, veaa100. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, Better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef] [PubMed]
- Stopnišek, N.; Hedžet, S.; Accetto, T.; Rupnik, M. Insights into Diversity, Host-Range, and Temporal Stability of Bacteroides and Phocaeicola Prophages. BMC Microbiol. 2025, 25, 92. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Škraban, J.; Cleenwerck, I.; Vandamme, P.; Fanedl, L.; Trček, J. Genome Sequences and Description of Novel Exopolysaccharides Producing Species Komagataeibacter pomaceti sp. nov. and Reclassification of Komagataeibacter kombuchae (Dutta and Gachhui 2007) Yamada et al., 2013 as a Later Heterotypic Synonym of Komagataeibacter hansenii (Gosselé et al. 1983) Yamada et al., 2013. Syst. Appl. Microbiol. 2018, 41, 581–592. [Google Scholar] [CrossRef]
- Marič, L.; Cleenwerck, I.; Accetto, T.; Vandamme, P.; Trček, J. Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar. Microorganisms 2020, 8, 1178. [Google Scholar] [CrossRef]
- Tittsler, R.P.; Sandholzer, L.A. The Use of Semi-Solid Agar for the Detection of Bacterial Motility. J. Bacteriol. 1936, 31, 575–580. [Google Scholar] [CrossRef]
- Gorgieva, S.; Jančič, U.; Cepec, E.; Trček, J. Production Efficiency and Properties of Bacterial Cellulose Membranes in a Novel Grape Pomace Hydrolysate by Komagataeibacter melomenusus AV436T and Komagataeibacter xylinus LMG 1518. Int. J. Biol. Macromol. 2023, 244, 125368. [Google Scholar] [CrossRef] [PubMed]
- Cepec, E.; Trček, J. Antimicrobial Resistance of Acetobacter and Komagataeibacter Species Originating from Vinegars. Int. J. Environ. Res. Public Health 2022, 19, 463. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for Taxonomy-Based Analysis of Pathways and Genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The Reactome Pathway Knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- KEGG PATHWAY: Tyrosine Metabolism—Reference Pathway. Available online: https://www.kegg.jp/pathway/map00350 (accessed on 31 July 2025).
- Hearing, V.J. Determination of Melanin Synthetic Pathways. J. Investig. Dermatol. 2011, 131, E8–E11. [Google Scholar] [CrossRef]
- Nishijima, M.; Tazato, N.; Handa, Y.; Tomita, J.; Kigawa, R.; Sano, C.; Sugiyama, J. Gluconacetobacter tumulisoli sp. nov., Gluconacetobacter takamatsuzukensis sp. nov. and Gluconacetobacter aggeris sp. nov., Isolated from Takamatsuzuka Tumulus Samples before and during the Dismantling Work in 2007. Int. J. Syst. Evol. Microbiol. 2013, 63, 3981–3988. [Google Scholar] [CrossRef] [PubMed]
- Lerat, S.; Simao-Beaunoir, A.M.; Beaulieu, C. Genetic and Physiological Determinants of Streptomyces Scabies Pathogenicity. Mol. Plant Pathol. 2009, 10, 579–585. [Google Scholar] [CrossRef]
- DeBritto, S.; Gajbar, T.D.; Satapute, P.; Sundaram, L.; Lakshmikantha, R.Y.; Jogaiah, S.; Ito, S.I. Isolation and Characterization of Nutrient Dependent Pyocyanin from Pseudomonas aeruginosa and Its Dye and Agrochemical Properties. Sci. Rep. 2020, 10, 1542. [Google Scholar] [CrossRef]
- Hocquet, D.; Petitjean, M.; Rohmer, L.; Valot, B.; Kulasekara, H.D.; Bedel, E.; Bertrand, X.; Plésiat, P.; Köhler, T.; Pantel, A.; et al. Pyomelanin-Producing Pseudomonas aeruginosa Selected during Chronic Infections Have a Large Chromosomal Deletion Which Confers Resistance to Pyocins. Environ. Microbiol. 2016, 18, 3482–3493. [Google Scholar] [CrossRef] [PubMed]
- Arias-Barrau, E.; Olivera, E.R.; Luengo, J.M.; Fernández, C.; Galán, B.; García, J.L.; Díaz, E.; Miñambres, B. The Homogentisate Pathway: A Central Catabolic Pathway Involved in the Degradation of L-Phenylalanine, L-Tyrosine, and 3-Hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 2004, 186, 5062–5077. [Google Scholar] [CrossRef] [PubMed]
- Coelho-Souza, T.; Martins, N.; Maia, F.; Frases, S.; Bonelli, R.R.; Riley, L.W.; Moreira, B.M. Pyomelanin Production: A Rare Phenotype in Acinetobacter baumannii. J. Med. Microbiol. 2013, 63, 152–154. [Google Scholar] [CrossRef]
- Li, G.; Meng, X.; Zhu, M.; Li, Z. Research Progress of Betalain in Response to Adverse Stresses and Evolutionary Relationship Compared with Anthocyanin. Molecules 2019, 24, 3078. [Google Scholar] [CrossRef]
- Wu, Q.; Fu, X.; Chen, Z.; Wang, H.; Wang, J.; Zhu, Z.; Zhu, G. Composition, Color Stability and Antioxidant Properties of Betalain-Based Extracts from Bracts of Bougainvillea. Molecules 2022, 27, 5120. [Google Scholar] [CrossRef]
- Kamsteeg, J.; van Brederode, J.; van Nigtevecht, G. Genetics of Anthocyanin Formation in Petals of the Red Campion (Silene dioica (L.) Clairv.). Genetica 1979, 51, 5–13. [Google Scholar] [CrossRef]
- Watanabe, S.; Ohtani, Y.; Aoki, W.; Uno, Y.; Sukekiyo, Y.; Kubokawa, S.; Ueda, M. Detection of Betacyanin in Red-Tube Spinach (Spinacia oleracea) and Its Biofortification by Strategic Hydroponics. PLoS ONE 2018, 13, e0203656. [Google Scholar] [CrossRef]
- Sadowska-Bartosz, I.; Bartosz, G. Biological Properties and Applications of Betalains. Molecules 2021, 26, 2520. [Google Scholar] [CrossRef]
- Trezzini, G.F.; Zrÿd, J.P. Two Betalains from Portulaca grandiflora. Phytochemistry 1991, 30, 1897–1899. [Google Scholar] [CrossRef]
- Christinet, L.; Burdet, F.X.; Zaiko, M.; Hinz, U.; Zrÿd, J.P. Characterization and Functional Identification of a Novel Plant 4,5-Extradiol Dioxygenase Involved in Betalain Pigment Biosynthesis in Portulaca grandiflora. Plant Physiol. 2004, 134, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Trych, U.; Buniowska-Olejnik, M.; Marszałek, K. Bioaccessibility of Betalains in Beetroot (Beta vulgaris L.) Juice under Different High-Pressure Techniques. Molecules 2022, 27, 7093. [Google Scholar] [CrossRef] [PubMed]
- Eyshi, S.; Ghareaghajlou, N.; Afshar Mogaddam, M.R.; Ghasempour, Z. Red Beet Betalains Extraction Process: A Comprehensive Review of Methods, Applications, and Physicochemical Properties. Food Sci. Nutr. 2024, 12, 8540–8558. [Google Scholar] [CrossRef]
- Chang, Y.C.; Chiu, Y.C.; Tsao, N.W.; Chou, Y.L.; Tan, C.M.; Chiang, Y.H.; Liao, P.C.; Lee, Y.C.; Hsieh, L.C.; Wang, S.Y.; et al. Elucidation of the Core Betalain Biosynthesis Pathway in Amaranthus tricolor. Sci. Rep. 2021, 11, 6086, Correction in Sci. Rep. 2021, 11, 15634. [Google Scholar] [CrossRef] [PubMed]
- Sievers, M.; Swings, J. Gluconacetobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–11. [Google Scholar] [CrossRef]
- Raspor, P.; Goranovič, D. Biotechnological Applications of Acetic Acid Bacteria. Crit. Rev. Biotechnol. 2008, 28, 101–124. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Qian, C.; Ma, J.; Liang, J.; Zhang, L.; Liang, X. Comprehensive Deciphering Prophages in Genus Acetobacter on the Ecology, Genomic Features, Toxin–Antitoxin System, and Linkage with CRISPR-Cas System. Front. Microbiol. 2022, 13, 951030. [Google Scholar] [CrossRef]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled Diversity: The Many Roles and Reactions of Bacterial Cytochromes P450 in Secondary Metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef]
- KEGG PATHWAY: Betalain Biosynthesis—Reference Pathway. Available online: https://www.kegg.jp/pathway/map00965 (accessed on 29 July 2025).
- Polturak, G.; Aharoni, A. “La Vie En Rose”: Biosynthesis, Sources, and Applications of Betalain Pigments. Mol. Plant 2018, 11, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Meredith, P.; Sarna, T. The Physical and Chemical Properties of Eumelanin. Pigment. Cell Res. 2006, 19, 572–594. [Google Scholar] [CrossRef]
- Donoso, R.A.; Ruiz, D.; Gárate-Castro, C.; Villegas, P.; González-Pastor, J.E.; de Lorenzo, V.; González, B.; Pérez-Pantoja, D. Identification of a Self-Sufficient Cytochrome P450 Monooxygenase from Cupriavidus pinatubonensis JMP134 Involved in 2-Hydroxyphenylacetic Acid Catabolism, via Homogentisate Pathway. Microb. Biotechnol. 2021, 14, 1944–1960. [Google Scholar] [CrossRef]
- Ferrer-Sevillano, F.; Fernández-Cañón, J.M. Novel PhacB-Encoded Cytochrome P450 Monooxygenase from Aspergillus nidulans with 3-Hydroxyphenylacetate 6-Hydroxylase and 3,4-Dihydroxyphenylacetate 6-Hydroxylase Activities. Eukaryot. Cell 2007, 6, 514–520. [Google Scholar] [CrossRef]
- Szymczak, I.; Pietrzyk-Brzezińska, A.J.; Duszyński, K.; Ryngajłło, M. Characterization of the Putative Acylated Cellulose Synthase Operon in Komagataeibacter xylinus E25. Int. J. Mol. Sci. 2022, 23, 7851. [Google Scholar] [CrossRef]
- Wijesinghe, V.N.; Choo, W.S. Antimicrobial Betalains. J. Appl. Microbiol. 2022, 133, 3347–3367. [Google Scholar] [CrossRef] [PubMed]
- Zerrad Sidi Mohamed Ben, A.; Sendide, K.; Hassouni, E.M. Antioxidant and Antimicrobial Activities of Melanin Produced by A Pseudomonas balearica Strain. J. Biotechnol. Lett. 2014, 5, 87–94. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karničnik, B.; Jugović, I.; Accetto, T.; Fanedl, L.; Avguštin, G.; Trček, J. Gluconacetobacter brunescens sp. nov., a Novel Acetic Acid Bacterium Isolated from Pear Vinegar, Producing a Water-Soluble Brown Pigment. Microorganisms 2025, 13, 2620. https://doi.org/10.3390/microorganisms13112620
Karničnik B, Jugović I, Accetto T, Fanedl L, Avguštin G, Trček J. Gluconacetobacter brunescens sp. nov., a Novel Acetic Acid Bacterium Isolated from Pear Vinegar, Producing a Water-Soluble Brown Pigment. Microorganisms. 2025; 13(11):2620. https://doi.org/10.3390/microorganisms13112620
Chicago/Turabian StyleKarničnik, Bernarda, Igor Jugović, Tomaž Accetto, Lijana Fanedl, Gorazd Avguštin, and Janja Trček. 2025. "Gluconacetobacter brunescens sp. nov., a Novel Acetic Acid Bacterium Isolated from Pear Vinegar, Producing a Water-Soluble Brown Pigment" Microorganisms 13, no. 11: 2620. https://doi.org/10.3390/microorganisms13112620
APA StyleKarničnik, B., Jugović, I., Accetto, T., Fanedl, L., Avguštin, G., & Trček, J. (2025). Gluconacetobacter brunescens sp. nov., a Novel Acetic Acid Bacterium Isolated from Pear Vinegar, Producing a Water-Soluble Brown Pigment. Microorganisms, 13(11), 2620. https://doi.org/10.3390/microorganisms13112620







