Abstract
In computer-aided medical diagnosis, deep learning techniques have shown that it is possible to offer performance similar to that of experienced medical specialists in the diagnosis of knee osteoarthritis. In this study, a new deep learning (DL) software, called “MedKnee” is developed to assist physicians in the diagnosis process of knee osteoarthritis according to the Kellgren and Lawrence (KL) score. To accomplish this task, 5000 knee X-ray images obtained from the Osteoarthritis Initiative public dataset (OAI) were divided into train, valid, and test datasets in a ratio of 7:1:2 with a balanced distribution across each KL grade. The pre-trained Xception model is used for transfer learning and then deployed in a Graphical User Interface (GUI) developed with Tkinter and Python. The suggested software was validated on an external public database, Medical Expert, and compared with a rheumatologist’s diagnosis on a local database, with the involvement of a radiologist for arbitration. The MedKnee achieved an accuracy of 95.36% when tested on Medical Expert-I and 94.94% on Medical Expert-II. In the local dataset, the developed tool and the rheumatologist agreed on 23 images out of 30 images (74%). The MedKnee’s satisfactory performance makes it an effective assistant for doctors in the assessment of knee osteoarthritis.
1. Introduction
Knee Osteoarthritis (OA) is one of the most common chronic diseases, and happens when the cartilage in the knee joint breaks down. The resulting friction between the bones of the knee joint can cause discomfort, stiffness, or swelling in the knees. If left untreated, severe osteoarthritis of the knee can result in partial or complete disability. Although there is no known treatment for knee osteoarthritis, if a timely and accurate diagnosis is made there are therapies that can lessen symptoms and halt the disease’s progression. Knee OA affects mainly women, the elderly, and the obese. By 2050, 130 million people worldwide will suffer from knee osteoarthritis [1].
In recent years, Artificial Intelligence (AI) has attracted considerable interest in medical imaging [2,3,4,5,6,7]. Particularly, AI was utilized in the diagnosis of knee osteoarthritis based on MRI and X-ray images [8]. In parallel, Computer-Aided Diagnosis (CAD) is becoming more significant for disease diagnosis, classification, and prediction. The work described in this paper uses X-ray images for automated knee osteoarthritis detection based on a pre-trained deep convolutional neural network (DCNN). The motivation of this study is to develop novel software to assist medical professionals in accurately and speedily diagnosing knee arthritis from X-ray images. To accomplish these goals, a pre-trained Xception model is modified and trained to identify and classify knee osteoarthritis from radiographic images, and then, the automated software, MedKnee, is developed using a graphical user interface (GUI) and the saved Xception model. The model’s performance was analyzed against other CNN models tested on the OAI dataset with comparable sizes. The developed model was validated using an external public database annotated by two experts and a local dataset submitted to a rheumatologist and a radiologist.
2. Related Work
In recent literature, numerous researchers have become interested in disease diagnostics utilizing deep learning and machine learning to detect knee OA according to the Kellgren and Lawrence (KL) grading system. Yang et al. [9] developed an automated KOA diagnostic model that can be used in a mobile device called RefineDet. This model consists of two connected models, the Anchor Refinement Module (ARM) used for region of interest (ROI) localization and the Object Detection Module (ODM) for KOA classification. The Transfer Connection Block (TCB) was introduced to share information between the two models. This model was trained and validated on 2579 X-ray images of the posterior-anterior (PA) view of knees collected from the General Hospital of the People’s Liberation Army in China. In this approach, the images were captured using an iPhone held at a distance of 40 cm. The pre-trained network was built on 2499 train images, 263 validation images, and 941 test images. The method achieved an accuracy of 95.7%.
Dalia et al. [10] aimed to detect knee osteoarthritis from radiographic images using VGG16, Resnet-152, and DenseNet-201. The proposed models were developed and compared using 8892 radiographic knee images from the OAI dataset. The ROI was detected using YOLOv5. The best results were obtained using transfer learning of VGG16 by adding two fully connected (FC) layers with 4096 units, a third FC layer with 1000 units, and a softmax activation function. The VGG16 model achieved an accuracy of 69.8%.
Wahyuningrum et al. [11] developed an end-to-end supervised learning model of a Deep Convolutional Neural Network (DCNN) to classify knee OA using three-fold cross validation. The proposed model consists of five convolution blocks. Each block includes a Rectified Linear Unit (ReLU) layer, a max-pooling layer, a flatten layer, a fully connected layer, and a dropout layer. The model achieved an average accuracy of 77.24% when tested using the three folders containing 527, 501, and 528 images.
Yong et al. [12] proposed an Ordinal Regression Module (ORM) to classify knee OA using VGG, ResNet, ResNext, GoogLeNet, DenseNet, and Mobilenet. The proposed ORM splits the probability space of the scalar output into K classes using K-1 cut-points to perform ordinal regression. The neural networks were optimized using a Cumulative Link (CL) loss function and trained using 4130 X-ray images belonging to the OAI dataset. The best result was obtained using DenseNet-161 with an accuracy of 88.09%.
Ruikar et al. [13] developed an automated computer-aided diagnosis, Osteoarthritis network (OACnet), based on a deep neural network for convincing detection of Knee OA. This model was trained on 4746 radiological images acquired from the OAI dataset. It was built from scratch and with hand-crafted feature engineering (joint space narrowing, bone spur, sclerosis, and deformation). The model achieved an accuracy of 83.74%, improved to 92.7% when combined with hand-crafted features.
Wahyuningrum et al. [14] developed a CNN-based system using ReseNet, VGGNet, and DenseNet to diagnose knee OA. The proposed system was implemented using Long-Term Memory (LSTM) as a special kind of CNN that can memorize information and store it in complex network elements over a long period. The proposed system was trained and validated using 5148 X-ray images obtained from the OAI dataset. The LSTM combined with VGG16 achieved the highest accuracy of 75.28%.
Both Chen et al. [15] and Wani et Saini. [16] proposed a novel adjustable ordinal loss instead of the cross-entropy loss in the detection of knee OA using VGG, ResNet, DenseNet, and InceptionV3. To develop and compare the proposed models, 8260 knee X-ray images were used in [15] and 1656 X-ray images were applied in [16], all collected from the OAI dataset. The VGG19 model with the proposed ordinal loss in [15,16] obtained the highest knee severity grading accuracy of 70.4%, and 96.7%, respectively. The ordinal loss function-based approach was used also by Jain et al. [17] to develop an automated method of detecting knee osteoarthritis from X-ray images, named High-Resolution Network (HRNet). The developed model was combined with a convolutional mass attention module (CBAM). HRNet is a revolutionary multi-resolution deep CNN consisting of a convolution (2D) layer followed by layers that add up the high-to-low resolution and then merge the multi-resolution in parallel for information exchange. The model was built on 8260 knee X-ray images from the OAI dataset. The method achieved an accuracy of 71.74% and a mean absolute error (MAE) of 0.311.
Yunus et al. [18] tended to apply a specific approach based on Darknet-53 and Alexnet combined with local binary pattern (LBP) to extract deep features and identify knee OA severity from radiological images. The final classification was performed with the support vector machine (SVM) and the K-nearest neighbors (KNN). Then the classified images were localized using a combination of YOLOv2 and an open neural network exchange (ONNX) built in 24 layers for the preparation of the developed model as (i) input layer, (ii) two element-wise Affine layers, (iii) four convolutional layers, (iv) four Batch normalization (BN) layers, (v) three max-pooling layers, and (vi) four activation layers, while YOLO-v2 was built using three convolutional layers, two BN layers, and two ReLU layers. This approach was developed using 3795 X-ray images from the OAI public dataset. The model achieved an accuracy of 90.6%.
Hu et al. [19] developed a novel deep learning architecture, Adversarial Evolving Neural Network (A-ENN), for the longitudinal progression of Knee OA severity over 4 years. The deep learning model was built with Resnet-18 and three classifiers: VGG19, ResNet50, and visual transformer model (Vit). The proposed model was trained and tested on 3294 labeled knee X-ray images belonging to the OAI dataset. The model combined with VGG19 achieved the best accuracy of 64.6%, 63.9%, 63.2%, 61.8%, and 60.2% for progression baseline, 12-month, 24-month, 36-month, and 48-month, respectively.
Raisuddin et al. [20] proposed and evaluated Deep Active Learning (DAL) designed to classify knee OA severity. The proposed model was built with Semi-Supervised Learning (SSL) deep Siamese using the VGG and Consistency Regularization (CR) approach which ensures the model’s stability in front of the input noise. This model was trained and validated using 8953 knee X-ray images from the OAI dataset. The developed DAL achieved a balanced accuracy of 64.13%.
Huu et al. [21] applied the transfer learning of VGG16 for the automated binary classification of KOA severity using a deep Siamese convolution neural network. The proposed model consists of six convolutional layers with a stride of 1, three convolutional layers with a stride of 2, three dropout layers, a Separable Adaptive Max-pooling (SAM) layer, and a fully connected layer. The proposed model was built using 2874 X-ray images collected from the OAI dataset. The updated VGG16 model achieved an accuracy of 89%.
Yifan et al. [22] presented a knee OA classifier using the Transfer learning of ReseNet34 and DenseNet121 combined with a novel learning scheme that splits data into two categories based on reliability. The two models were developed using 8302 X-ray images from the OAI dataset and a hybrid loss function to manipulate the lower reliability sets. Both models, DenseNet121 and ReseNet34, achieved an accuracy of 70.13% and 68.32%, respectively.
More recently, Alshamrani et al. [23] proposed transfer learning models based on sequential CNNs, VGG16, and ResNet-50 to identify normal and abnormal knees from X-ray images. The proposed models were trained using 3836 X-ray images collected from Kaggle. The best developed model was VGG16 which achieved a training accuracy of 99% and a testing accuracy of 92%.
Mohammed et al. [24] suggested a binary classification and a multiclass classification for the severity of KOA from radiographic images. This approach was built using six pre-trained DNN models: ResNet101, MobileNetV2, VGG16, VGG19, InceptionResNetV2, and DenseNet121. The designed models were trained and tested on 9786 knee images taken from OAI. The best-performing model was the pre-trained ResNet101 for three classes and five classes with an accuracy of 89% and 69%, respectively.
Pi et al. [25] presented an ensemble network based on DenseNet-161, EfficientNet-b5, EfficientNet-V2-s, RegNet-Y-8GF, ResNet-101, ResNext-50-32×4d, Wide-ResNet-50-2, and ShuffleNet-V2-×2-0. The proposed method was implemented using 8260 images from the Osteoarthritis Initiative with optimal image sizes for training the various deep learning models. The optimized model ensemble network achieved an accuracy of 76.93%. Table 1 summarizes the binary and multiclass classification techniques utilized in recent publications based on internal validation.

Table 1.
Overview of the methods used in recent literature with the average values of the metrics used.
3. Materials and Methods
Although radiography is fast, inexpensive, non-invasive, and easy to use, the quality of the radiographic image requires several treatments to improve contrast and brightness and remove noise. Therefore, before selecting and improving the classification network, image processing was carried out to enhance the identification of knee osteoarthritis severity based on KL grade.
3.1. Dataset Description
OAI is a longitudinal observational study conducted by the US National Institutes of Health (NIH) in men and women over ten years. It consists of 4446 X-ray images of knees labeled by Boston University according to the Kellgren and Lawrence (KL) scoring system. In this study, the knee X-ray images used to train the proposed model belong to the OAI dataset available on Mendeley Data [26]. The images consist of unilateral PA fixed flexion of uniform size with an identical resolution. The raw images are bilateral PA fixed flexion with varied resolutions and sizes, pre-processed before being made available for download on Mendeley. The dataset used is labeled Healthy, Doubtful, Minimal, Moderate, and Severe, with equal size to avoid the problem of an unbalanced dataset. The dataset was augmented into 5000 images and then divided into a training set (70%), a validation set (10%), and a testing set (20%).
3.2. Preprocessing
Histogram equalization is applied in image processing to spread and evenly distribute pixel values. This allows the image quality to be improved by increasing the dispersion of the highest frequency and decreasing the dispersion of other frequencies, allowing the low contrast of the source images to be improved [27]. In this work, we have applied the Contrast Limited Adaptive Histogram Equalization (CLAHE) algorithm used in [21,25]. Then, to improve the performance of the osteoarthritis detection system, it was necessary to use images with variable contrast, brightness, and positions, as they can be captured by the camera used in our work. The original images were resized to 224 × 224, flipped horizontally and vertically, and rotated left and right. Therefore, the image intensity was normalized between 0 and 1, and then the brightness was modified in the range of (0.1,0.7). The preprocessing pipeline is shown in Figure 1.
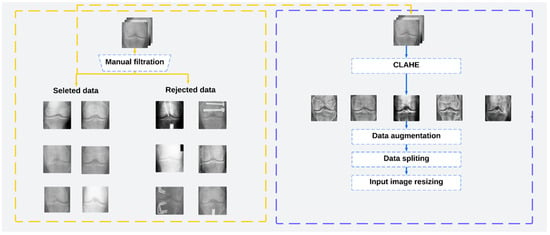
Figure 1.
The preprocessing pipeline used in this work.
3.3. Network Architecture
Xception, also known as the advanced variant of an Inception module, is a deep convolutional neural network based entirely on depthwise separable convolution layers. It was developed in 2017 by the creator of the Keras Library, Francois Chollet [28]. This model moderately surpassed Inception V3 on the ImageNet dataset in 2017. The pre-trained version of the network trained on more than a million images from the ImageNet database. The pre-trained Xception model was selected in this work because it consumes few resources while maintaining acceptable accuracy, and its architecture is very easy to define and modify, making it a prime candidate for medical tasks [29]. It has been utilized in various medical tasks during the past two years, such as the assessment of benign and malignant gastric ulcer lesions based on gastrointestinal endoscopic images [30], the detection of COVID-19 from radiographic images [31], and the detection of knee osteoarthritis [32].
The Xception architecture is easy to define and modify. As shown in Figure 2, it contains 2 convolutional layers, 34 separable convolution layers, 4 max-pooling layers, and a global average pooling layer. Convolutional and separable convolution layers are followed by batch normalization and ReLU layers. The 36 convolutional layers used for extracting network characteristics are arranged in 14 blocks, which are all surrounded by linear residual connections, except for the first and last layers. The convolutional base is succeeded by a logistic regression layer.
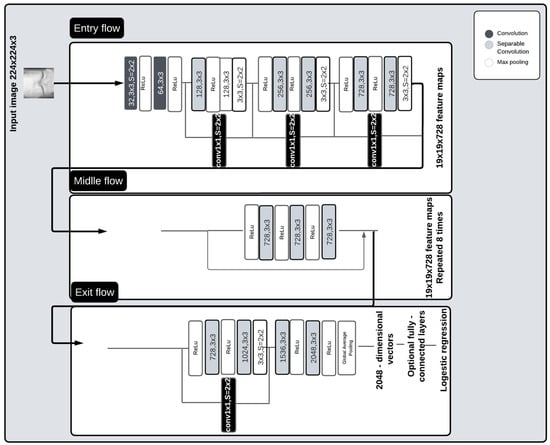
Figure 2.
Pre-trained Xception architecture.
The pre-trained model head in this study has been updated with a global average pooling layer, batch normalization layer, a dropout of 0.2, and a fully connected layer with a softmax activation function. The model is trained using categorical cross-entropy with five outputs corresponding to the Kellgreen and Lawrence (KL) grading scale. The updated network was trained for 200 epochs using a batch size of 16, and an Adam optimizer with an initial learning rate of 0.0001. The best model was saved in a «.H5» file. The network training is conducted with the hyperparameters illustrated in Table 2.

Table 2.
Hyperparameters maintained during training.
3.4. Software Requirements and Libraries
To carry out this work, a machine running the Windows 10 operating system with an i7 processor, 16 GB memory, and a Nvidia Quadro M2200 5.2 graphics processing unit (GPU) is used. Python 3.9.16 (Anaconda3), TensorFlow-GPU 2.10.0, CUDA (Compute Unified Data Architecture) 11.2, and CUDNN 8.1.33 are used to implement the proposed method. Spyder5 was used in these experiments as a framework.
4. Experiment Results and Model Deployment
The saved model was deployed in a graphical user interface (GUI) using a desktop application. Then, to validate our study, the model was tested on the OAI labeled dataset and two other external datasets, including a local dataset.
4.1. Result on OAI Dataset
The proposed automatic system’s performance was assessed through the use of the F1-score, precision, recall, and confusion matrix. The pre-trained model is tested on 1000 images belonging to the OAI dataset, divided into 200 images for each KL grade. The use of batch normalization and drop-out layer has enabled us to avoid overfitting. The model achieved a validation accuracy of 99.39% and a test accuracy of 97.20%.
The test dataset consisted of 1000 knees, 800 knees with OA and 200 knees without OA. As illustrated in Figure 3, the confusion matrix of the Xception model indicates that KL3 was classified without error, followed by KL grade 1, where the method correctly classified 198 knees out of 200. In third place, we find KL4 grade with a re-rating rate of 195 correctly identified knees. Finally, KL2 and KL0 grades have the lowest rate with the correct identification of 190 and 189 knees, respectively. We note the classification of grade KL0 as grade KL 1 in 10 cases due to the minimal difference between the two grades and the difficulty in distinguishing between them.
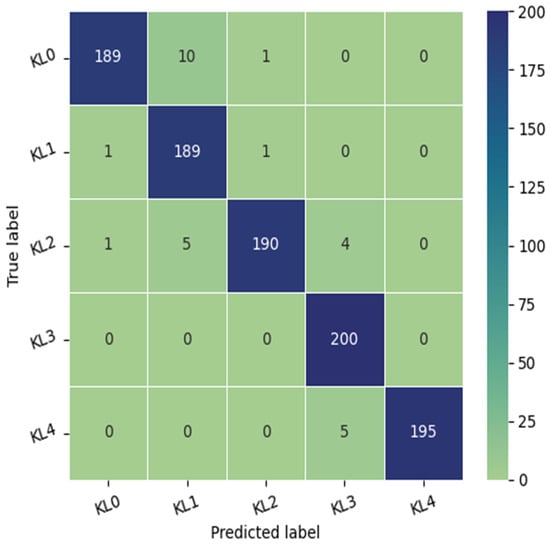
Figure 3.
The confusion matrix of the proposed approach tested on the OAI dataset.
As shown in Table 3, the model perfectly detects grade KL3 with a Recall of 1 and an F1-score of 0.98. KL1 and KL4 were classified with a recall of 0.99 and 0.97, respectively. KL0 and KL2 were identified with an equal precision of 0.99 and an F1-score of 0.97.

Table 3.
Performance of the proposed model in detecting each KL grade of knee osteoarthritis validated on the OAI dataset.
Table 4, compares the proposed model with works deployed on an identical dataset collected from the OAI dataset with a comparable size. The proposed model has outperformed many research studies based on internal validation with a validation accuracy of 99.39% and a test accuracy of 97.20%.

Table 4.
Comparison between the proposed tool, MedKnee, and similar works in KOA’s 5-class classification based on internal validation.
4.2. Graphical User Interface (GUI): MedKnee
A desktop application, the Moroccan KOA diagnosing tool (MedKnee), has been created using TKINTER and FPDF2 Python to simplify the use of the knee osteoarthritis classification implementation. The interface allows the user to enter the patient’s name, date, age, and sex. After entering the correct password, the user can select the left or right knee and obtain an individual diagnosis for each knee (Figure 4).
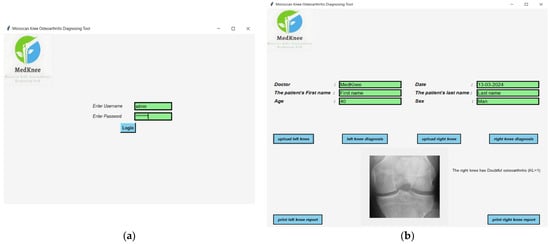
Figure 4.
(a) Login interface; (b) Diagnostic result interface.
As shown in Figure 5, the diagnostic report for each knee can be printed, with a field at the bottom for comments.

Figure 5.
Printable report on knee osteoarthritis diagnosis.
4.3. Results on External Validation
In the first step, to approve our study, Medical Expert-I and Medical Expert-II public datasets were used. Then, a local dataset was employed in the second step to compare the results with a specialist’s diagnosis.
4.3.1. Medical Expert Public Dataset
The database is composed of 1650 digital radiographs of the knee joint collected from various hospitals and diagnostic centers in India [33]. Each X-ray image of the knee is manually labeled by two medical experts according to the Kellgren and Lawrence grades. Both experts are experienced orthopedic surgeons who review between 70 and 100 radiographs per day. To obtain images comparable to those in the Medknee training dataset, preprocessing consistent with that applied to the aforementioned training dataset must be implemented. First, we excluded images with double knees, then we resized the selected images from 362 × 162 to 224 × 162, and finally, we applied contrast-limited adaptive histogram equalization (CLAHE) to improve the local contrast of the images. As shown in Figure 6, the confusion matrix was used to evaluate the performance of our model. The proposed model achieved an accuracy of 95.36% on 1464 images labeled by expert1 and 94.94% on 1463 images annotated by expert2.
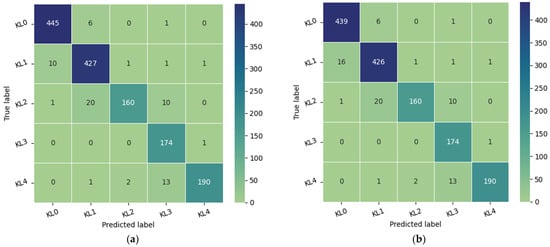
Figure 6.
Model performance: (a) the confusion matrix of the model tested in 1464 images of Medical Expert-I; (b) the confusion matrix of the proposed approach tested on 1463 images of Medical Expert-II.
4.3.2. Local Dataset
In the local dataset, we have selected 60 adult patient files of men and women with knee disorders who were radiographically examined at the Radiology Department of the El Kelaa des Sraghna Provincial Hospital. Each file consists of one or more knee radiographs in Dicom format as presented in Figure 7. The images were acquired using a standard ITALRAY radiology table. After analysis, 30 images with implants and non-posterior-anterior examination views were excluded and 30 left and right Dicom knees were retained to validate this work. To make the validation local dataset, the ROI was selected manually as illustrated in Figure 8. Then, each patient’s file was presented to a rheumatologist with PNG images. In case of disagreement between our diagnostic system and the rheumatologist, a radiologist was consulted for arbitration to make the final decision.
Figure 7.
Example of knee X-ray images collected and filtered from patients with knee symptoms.
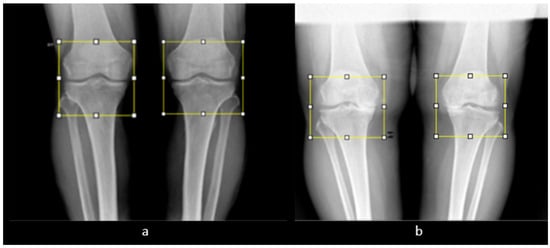
Figure 8.
Manual ROI selection. (a) ROI selection of a man’s knee. (b) ROI selection of a woman’s knee.
Out of the 30 images presented to the doctor, the DL model and the rheumatologist agreed on 23 images (74%). As shown in Table 5, in the seven remaining images where there was a disagreement, the referee confirmed four images, indicating agreement between the referee and the DL model. Three images were not confirmed, of which only one was not confirmed by the referee, indicating agreement between the referee and the rheumatologist. Based on the final decision of the arbitrator physician, our diagnostic model failed in 2 out of 30 images with a test accuracy of 90%.

Table 5.
Arbitration by radiologists to resolve disagreements between software and rheumatologists regarding the local dataset. The bold characters that show that there is an agreement between the result of the developed software diagnostic and the doctors’ diagnostic.
5. Discussion
In this work, an automated approach to classify the severity of knee osteoarthritis from simple radiographic images according to the KL grade is presented and implemented in a desktop application using TKINTER and the DL model based on Xception. The pre-trained Xception was chosen due to its superior performance on the OAI dataset compared to other models used during software development, such as EfficientNetV2M (93.15%) and MobileNetV2 (75.60%). Table 4 gives the multiclass classification values in comparison with similar works. The developed method achieved a validation multiclass accuracy of 99.39% and a test accuracy of 97.20%, which is a high performance compared to [11,12,14]. To validate our work, we tested the model on the Medical expert database and a local database. The model achieved an accuracy of 95.20% on Medical Expert-I and 94.94% on Medical Expert-II. This minimal difference confirms the results obtained in [34]. To validate the results, the model was tested on 30 images and compared with the diagnosis of a rheumatologist. In case of disagreement, a radiologist was consulted. Out of the 30 images, 28 were correctly identified (90%). It is worth noting that when four images were presented twice to the same doctor, the latter failed to give the same diagnosis for two images. However, our software’s diagnosis coincided with the second diagnosis of the doctor for one of the images, which was then revised. Furthermore, it is often challenging to differentiate between images of grades KL-0 and KL-1, which explains the discrepancy in identifying these two grades, but there is no clinical benefit in distinguishing between KL grades 0 and 1. In binary classification for osteoarthritis (OA) (KL < 2), and non-OA (KL ≥ 2), the new software MedKnee tested on a local dataset achieved an accuracy of 100%. Indeed, as shown in Table 5, although only one knee was classified as doubtful (KL = 1) by the rheumatologist, the referee (radiologist) confirmed the software’s diagnosis by validating that the knee in question had minimal osteoarthritis (KL = 2). In 2023, similar software, MediAI-OA [35], was developed using the NASNet DL model, but its accuracy is limited to 83%, which is significantly lower than the performance of our new software, MedKnee.
However, several limitations must be noted regarding the proposed approach. First, the study of the OAI dataset did not include lateral radiographs. Therefore, as noted by Ahmed and Mustapha [1], the addition of lateral radiographs would have provided additional information. Secondly, the radiographic images of knee osteoarthritis utilized to train the proposed model are pre-processed and consist of a PA radiograph with fixed flexion and identical resolution and size, whereas raw images require pre-processing before they can be processed. The generalizability of the developed model to external databases is another major limitation. The proposed model is limited to pre-processed images with zooming, resizing, and region of interest (ROI) rearrangement. Since the model was only developed using the pre-treated images from the OAI dataset, its accuracy is not acceptable in the absence of these operations. Nonetheless, the model can be more broadly generalized without requiring significant preprocessing if it is built using a combination of many external databases and local or other institutional datasets, including bilateral and unilateral images of varying sizes and resolutions. Furthermore, the developed application helps physicians to identify knee osteoarthritis with acceptable accuracy using manual localization of the (ROI). The addition of deep learning models, such as YOLO or Faster-RCNN, would be helpful for real-time detection of the ROI. Finally, the implementation of the proposed approach in this study with enhanced computational resources, including Nvidia GeForce, can enhance the degree of accuracy achieved by increasing the number of training epochs and images. In the future, we expect to improve our application by using a large local dataset and introducing automatic ROI selection with the option of adding image segmentation. Nevertheless, the developed software achieved a high level of accuracy and can help physicians predict the exact severity grade of knee OA by analyzing radiographic images.
6. Conclusions and Future Work
This work presents MedKnee, new DL software designed to automatically classify the severity of knee osteoarthritis from radiographic images. The software is developed based on the transfer learning of a pre-trained Xception model and the public OAI dataset. The best DL model achieved a validation accuracy of 99.39% and a test accuracy of 97.20%, moderately better than reported in the recent literature. The developed tool was validated using the Medical expert dataset and a local dataset. The model performed with an accuracy of 95.36% when tested on Medical Expert-I, and 94.94% on Medical Expert-II. On the local dataset, the model was tested on 30 images, and compared with a rheumatologist’s diagnosis. To resolve the disagreement, a radiologist was consulted. The model achieved a multiclass accuracy of 90% and a binary classification accuracy of 100%. It can be concluded that the proposed software “MedKnee” can equip radiologists with the ability to quickly and accurately diagnose and predict knee osteoarthritis from X-ray images. The proposed tool in this study has the potential to be valuable for the early detection and diagnosis of knee osteoarthritis. Future work on this research could include improving the system’s reliability using a large dataset, as well as automatic ROI segmentation and localization. In addition, the deployment of the software in Raspberry Pi can be seen as another advantage for offline diagnostics without the use of a traditional computer.
Author Contributions
Conceptualization, S.T., I.Z. and N.Z.; Methodology, I.Z. and N.Z.; Software, S.T., I.Z. and N.Z.; Validation, I.Z., N.Z., H.D. and M.A.; Formal analysis, I.Z., N.Z. and M.N.N.; Investigation, S.T.; Resources, S.T.; Data curation, S.T.; Supervision, I.Z., N.Z. and M.N.N.; Writing—original draft, S.T.; writing—review and editing, I.Z. and N.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
A local dataset was constructed to validate the software, However, it is not currently available due to ethical and privacy restrictions. The software was additionally validated using a public dataset called Medical Expert: https://www.kaggle.com/datasets/tommyngx/digital-knee-xray?resource=download (accessed on 25 September 2023).
Acknowledgments
The software presented in this article is the result of collaboration between the CPS2E Laboratory of the ENSMR and the radiology department of the El Kelaa Des Sraghna Provincial Hospital. We would like to express our gratitude to the chief of the radiology department at the El Kelaa Des Sraghna provincial hospital and also the representative of the Ministry of Health and Social Protection in the province of El Kelaa Des Sraghna. We would like to thank the radiology team at the El Kelaa Des Sraghna Provincial Hospital for their help in carrying out this investigation.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| CNN | Convolutional neural network |
| CAD | Computer-aided diagnosis |
| DCNN | Deep convolutional neural network |
| DL | Deep Learning |
| KL | Kellgren and Lawrence |
| OA | Osteoarthritis |
| KOA | Knee Osteoarthritis |
| OAI | Osteoarthritis initiative |
| ReLU | Linear rectified unit |
| ROI | Region of interest |
| A-ENN | Adversarial evolving neural network |
| DAL | Deep Active learning |
| LSTM | Long Short-Term Memory |
| ORM | Ordinal regression module |
| HRNet | High-resolution network |
| CBAM | Convolutional mass attention module |
| MCC | Matthews correlation coefficients |
| QWK | Quadratic weighted Cohen’s kappa coefficient |
References
- Ahmed, S.M.; Mstafa, R.J. Identifying Severity Grading of Knee Osteoarthritis from X-ray Images Using an Efficient Mixture of Deep Learning and Machine Learning Models. Diagnostics 2022, 12, 2939. [Google Scholar] [CrossRef] [PubMed]
- Belharar, F.Z.; Zrira, N. DeepRetino: Ophthalmic Disease Classification from Retinal Images using Deep Learning. In Proceedings of the IEEE 9th International Conference on Sciences of Electronics, Technologies of Information and Telecommunications (SETIT), Hammamet, Tunisia, 28–30 May 2022; Available online: https://ieeexplore.ieee.org/document/9875570 (accessed on 25 September 2023).
- Zrira, N.; Benmiloud, I.; Marzouki, K.; Farahat, Z.; Zaimi, I.; El Ghali, B.; El Midaoui, O.; Megdiche, K.; Ngote, N. Automatic and Fast Whole Heart Segmentation for 3D Reconstruction. In Proceedings of the IEEE 9th International Conference on Sciences of Electronics, Technologies of Information and Telecommunications (SETIT), Hammamet, Tunisia, 28–30 May 2022; Available online: https://ieeexplore.ieee.org/document/9875773 (accessed on 25 September 2023).
- Jimi, A.; Abouche, H.; Zrira, N.; Benmiloud, I. Automated Skin Lesion Segmentation using VGG-UNet. In Proceedings of the 2022 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM), Istanbul, Turkey, 10–13 November 2022; Available online: https://ieeexplore.ieee.org/document/10068634 (accessed on 25 September 2023).
- Abouche, H.; Jimi, A.; Benmiloud, I. Segmentation and Classification of Dermoscopic Skin Cancer on Green Channel. In Proceedings of the 2022 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM), Istanbul, Turkey, 10–13 November 2022; Available online: https://ieeexplore.ieee.org/document/10068614 (accessed on 25 September 2023).
- Jimi, A.; Abouche, H.; Zrira, N.; Benmiloud, I. Skin Lesion Segmentation Using Attention-Based DenseUNet. In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies, BIOSTEC, Lisbon, Portugal, 16–18 February 2023; Volume 3, pp. 91–100. Available online: https://www.scitepress.org/Link.aspx?doi=10.5220/0011686400003414 (accessed on 25 September 2023).
- Moussaid, A.; Zrira, N.; Benmiloud, I.; Farahat, Z. On the Implementation of a Post-Pandemic Deep Learning Algorithm Based on a Hybrid CT-Scan/X-ray Images Classification Applied to Pneumonia Categories. Healthcare 2023, 11, 662. [Google Scholar] [CrossRef]
- Cigdem, O.; Deniz, C.M. Artificial intelligence in knee osteoarthritis: A comprehensive review for 2022. Osteoarthr. Imaging 2023, 3, 100161. [Google Scholar] [CrossRef]
- Yang, J.; Ji, Q.; Ni, M.; Zhang, G.; Wang, Y. Automatic assessment of knee osteoarthritis severity in portable devices based on deep learning. J. Orthop. Surg. Res. 2022, 17, 540. [Google Scholar] [CrossRef]
- Dalia, Y.; Bharath, A.; Mayya, V.; Sowmya Kamath, S. DeepOA: Clinical Decision Support System for Early Detection and Severity Grading of Knee Osteoarthritis. In Proceedings of the IEEE 5th International Conference on Computer, Communication and Signal Processing (ICCCSP), Chennai, India, 24–25 May 2021; pp. 250–255. Available online: https://ieeexplore.ieee.org/document/9465522 (accessed on 25 September 2023).
- Tri Wahyuningrum, R.; Yasid, A.; Jacob Verkerke, G. Deep Neural Networks for Automatic Classification of Knee Osteoarthritis Severity Based on X-ray Images. In Proceedings of the 8th International Conference on Information Technology ICIT 2020: IoT and Smart City, Xi’an, China, 25–27 December 2020; ACM: New York, NY, USA; pp. 110–114. Available online: https://dl.acm.org/doi/10.1145/3446999.3447020 (accessed on 25 September 2023).
- Yong, C.W.; Teo, K.; Murphy, B.P.; Hum, Y.C.; Tee, Y.K.; Xia, K.; Lai, K.W. Knee osteoarthritis severity classification with ordinal regression module. Multimed. Tools Appl. 2021, 81, 41497–41509. [Google Scholar] [CrossRef]
- Ruikar, D.; Kamble, P.; Ruikar, A.; Houde, K.; Hegadi, R. DNN-Based Knee OA Severity Prediction System: Pathologically Robust Feature Engineering Approach. SN Comput. Sci. 2022, 4, 58. [Google Scholar] [CrossRef]
- Wahyuningrum, R.T.; Anifah, L.; Eddy Purnama, I.K.; Hery Purnomo, M. A New Approach to Classify Knee Osteoarthritis Severity from Radiographic Images based on CNN-LSTM Method. In Proceedings of the IEEE 10th International Conference on Awareness Science and Technology (ICAST), Morioka, Japan, 23–25 October 2019; pp. 1–6. Available online: https://ieeexplore.ieee.org/document/8923284 (accessed on 25 September 2023).
- Chen, P.; Gao, L.; Shi, X.; Allen, K.; Yang, L. Fully automatic knee osteoarthritis severity grading using deep neural networks with a novel ordinal loss. Comput. Med. Imaging Graph. 2019, 75, 84–92. [Google Scholar] [CrossRef]
- Wani, Z.M.; Saini, D.S. Deep Neural Network-based Knee Osteoarthritis Grading Using X-rays. IJRASET 2022, 10, 1293–1299. [Google Scholar] [CrossRef]
- Jain, R.K.; Sharma, P.K.; Gaj, S.; Sur, A.; Ghosh, P. Knee Osteoarthritis Severity Prediction using an Attentive Multi-Scale Deep Convolutional Neural Network. arXiv 2021, arXiv:2106.14292v1. [Google Scholar] [CrossRef]
- Yunus, U.; Amin, J.; Sharif, M.; Yasmin, M.; Kadry, S.; Krishnamoorthy, S. Recognition of Knee Osteoarthritis (KOA) Using YOLOv2 and Classification Based on Convolutional Neural Network. Life 2022, 12, 1126. [Google Scholar] [CrossRef]
- Hu, K.; Wu, W.; Li, W.; Simic, M.; Zomaya, A.; Wang, Z. Adversarial Evolving Neural Network for Longitudinal Knee Osteoarthritis Prediction. IEEE Trans. Med. Imaging 2022, 41, 3207–3217. [Google Scholar] [CrossRef]
- Raisuddin, A.M.; Nguyen, H.H.; Tiulpin, A. Deep Semi-Supervised Active Learning for Knee Osteoarthritis Severity Grading. In Proceedings of the IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; pp. 1–5. Available online: https://ieeexplore.ieee.org/document/9761668 (accessed on 25 September 2023).
- Nguyen Huu, P.; Nguyen Thanh, D.; le Thi Hai, T.; Chu Duc, H.; Pham Viet, H.; Nguyen Trong, C. Detection and Classification Knee Osteoarthritis Algorithm using YOLOv3 and VGG16 Models. In Proceedings of the IEEE 7th National Scientific Conference on Applying New Technology in Green Buildings (ATiGB), Da Nang, Vietnam, 11–12 November 2022; pp. 31–36. Available online: https://ieeexplore.ieee.org/document/9984096 (accessed on 25 September 2023).
- Wang, Y.; Bi, Z.; Xie, Y.; Wu, T.; Zeng, X.; Chen, S.; Zhou, D. Learning From Highly Confident Samples for Automatic Knee Osteoarthritis Severity Assessment: Data From the Osteoarthritis Initiative. IEEE J. Biomed. Health Inform. 2022, 26, 1239–1250. [Google Scholar] [CrossRef]
- Alshamrani, H.A.; Rashid, M.; Alshamrani, S.S.; Alshehri, A.H.D. Osteo-NeT: An Automated System for Predicting Knee Osteoarthritis from X-ray Images Using Transfer-Learning-Based Neural Networks Approach. Healthcare 2023, 11, 1206. [Google Scholar] [CrossRef]
- Mohammed, A.S.; Hasanaath, A.A.; Latif, G.; Bashar, A. Knee Osteoarthritis Detection and Severity Classification Using Residual Neural Networks on Preprocessed X-ray Images. Diagnostics 2023, 13, 1380. [Google Scholar] [CrossRef] [PubMed]
- Pi, S.-W.; Lee, B.-D.; Lee, M.S.; Lee, H.J. Ensemble deep-learning networks for automated osteoarthritis grading in knee X-ray images. Sci. Rep. 2023, 13, 22887. [Google Scholar] [CrossRef] [PubMed]
- Chen, P. Knee Osteoarthritis Severity Grading Dataset. Mendeley Data Version 1. 2018. Available online: https://data.mendeley.com/datasets/56rmx5bjcr/1 (accessed on 25 September 2023).
- Brahim, A.; Jennane, R.; Riad, R.; Janvier, T.; Khedher, L.; Toumi, H.; Lespessailles, E. A decision support tool for early detection of knee OsteoArthritis using X-ray imaging and machine learning: Data from the OsteoArthritis Initiative. Comput. Med. Imaging Graph. 2019, 73, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE International Conference on Computer Vision and Pattern Recognition, CVPR, CIVEMSA, Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. Available online: https://ieeexplore.ieee.org/document/8099678 (accessed on 25 September 2023).
- Udas, N.; Beuth, F.; Kowerko, D. Concept Detection in Medical Images using Xception Models. In Proceedings of the Conference and Labs of the Evaluation Forum (CLEF), Ceur Workshop, Thessaloniki, Greece, 22–25 September 2020; Volume 2696. Available online: https://www.researchgate.net/publication/350516158_Concept_Detection_in_Medical_Images_using_Xception_Models_-_TUC_MC_at_ImageCLEFmed_2020 (accessed on 25 September 2023).
- Liu, Y.; Zhang, L.; Hao, Z.; Yang, Z.; Wang, S.; Zhou, X.; Chang, Q. An xception model based on residual attention mechanism for the classification of benign and malignant gastric ulcers. Sci. Rep. 2022, 12, 15365. [Google Scholar] [CrossRef]
- Gülmez, B. A novel deep neural network model based Xception and genetic algorithm for detection of COVID-19 from X-ray images. Ann. Oper. Res. 2023, 328, 617–641. [Google Scholar] [CrossRef] [PubMed]
- El-Ghany, S.A.; Elmogy, M.; El-Aziz, A.A.A. A fully automatic fine-tuned deep learning model for knee osteoarthritis detection and progression analysis. Egypt. Inform. J. 2023, 24, 229–240. [Google Scholar] [CrossRef]
- Gornale, S.S.; Patravali, P.U.; Hiremath, P.S. A Comprehensive Digital Knee X-ray Image Dataset for the Assessment of Osteoarthritis. JSM Biomed Imaging Data Pap. 2020, 6, 1012. Available online: https://www.researchgate.net/publication/351284405_JSM_BIOMEDICAL_IMAGING_DATA_PAPERS_A_Comprehensive_Digital_Knee_X-ray_Image_Dataset_for_the_Assessment_of_Osteoarthritis (accessed on 25 September 2023).
- Gornale, S.S.; Patravali, P.U.; Hiremath, P.S. Detection of Osteoarthritis in Knee Radiographic Images using Artificial Neural Network. Int. J. Innov. Technol. Explor. Eng. IJITEE 2019, 8, 2429–2434. [Google Scholar] [CrossRef]
- Yoon, J.S.; Yon, C.-J.; Lee, D.; Lee, J.J.; Kang, C.H.; Kang, S.-B.; Lee, N.-K.; Chang, C.B. Assessment of a novel deep learning-based software developed for automatic feature extraction and grading of radiographic knee osteoarthritis. BMC Musculoskelet Disord. 2023, 24, 869. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).