Comparative Genomic Hybridization and Transcriptome Sequencing Reveal Genes with Gain in Acute Lymphoblastic Leukemia: JUP Expression Emerges as a Survival-Related Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Cell Line Culturing
2.3. DNA Extraction and Array CGH
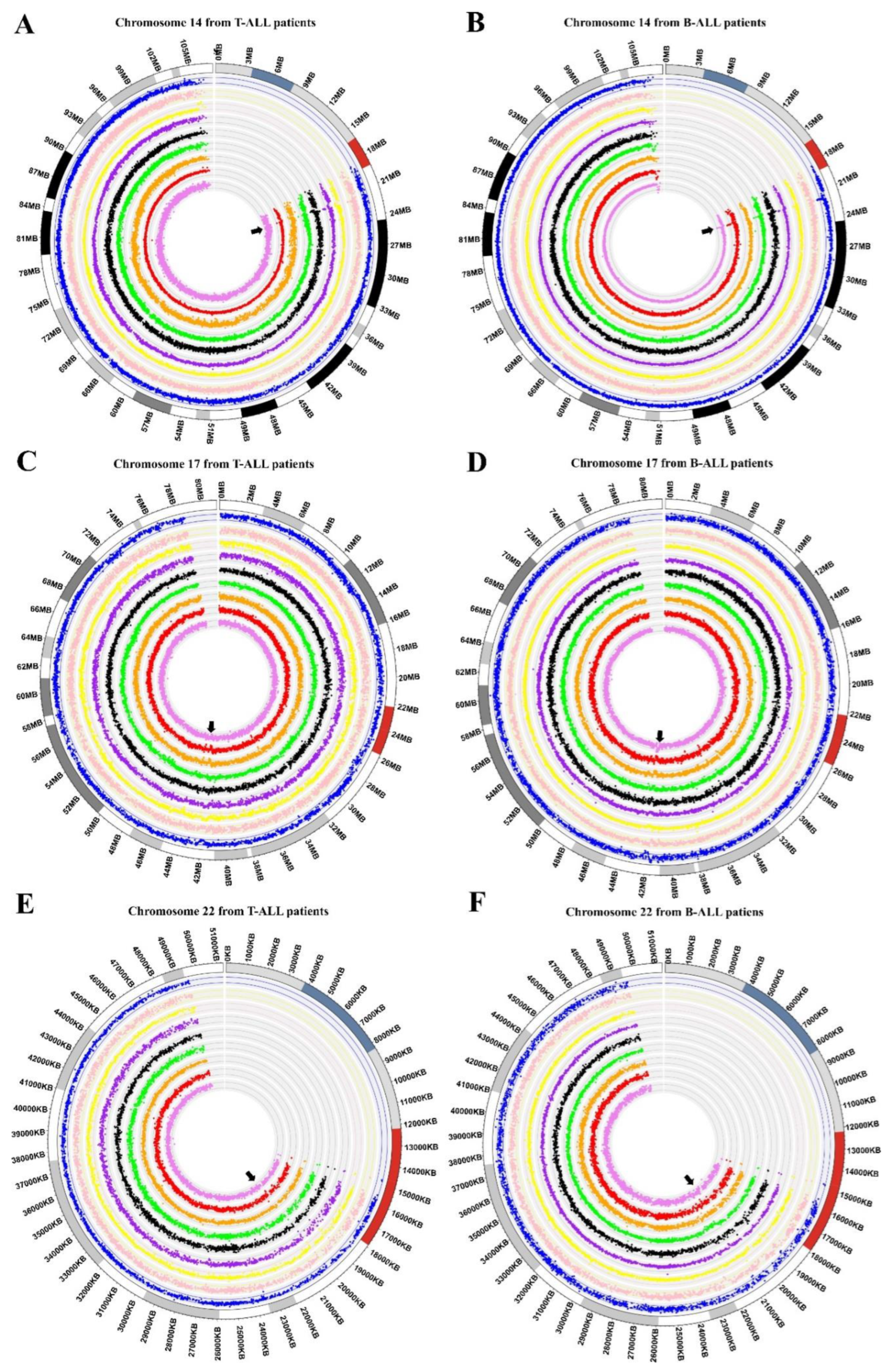
2.4. Circos Plot Representation of Genomic Data
2.5. RNA Dataset Analysis
2.6. Evaluation of JUP Expression by Quantitative PCR
2.7. Expressions of Genes Included in Gain Regions in Patients from the MILE Project
2.8. Tree Plots of Expressions of Genes with Gains in Normal Hematopoiesis versus Leukemia Lineages
2.9. Survival Analysis
3. Results
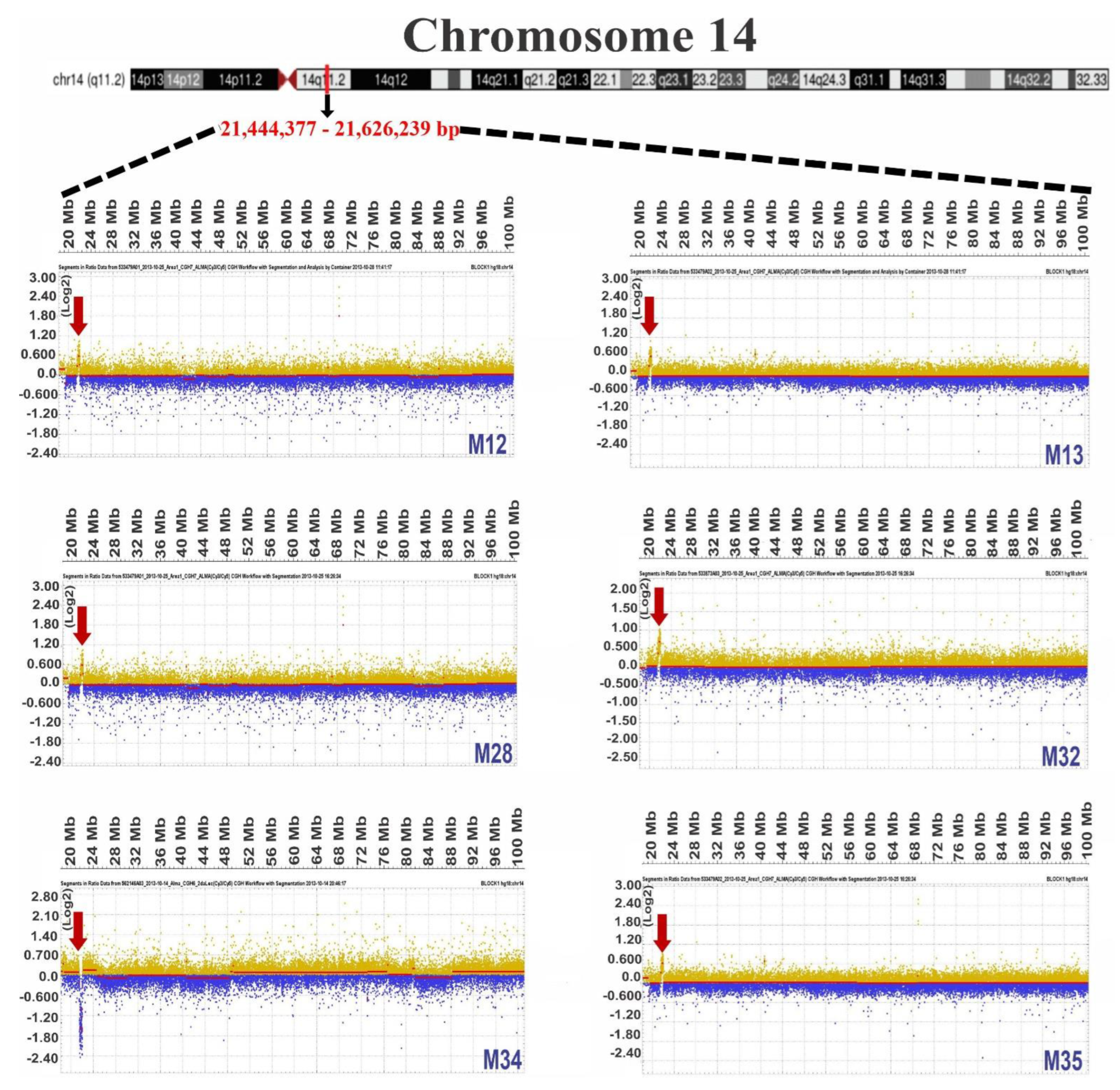
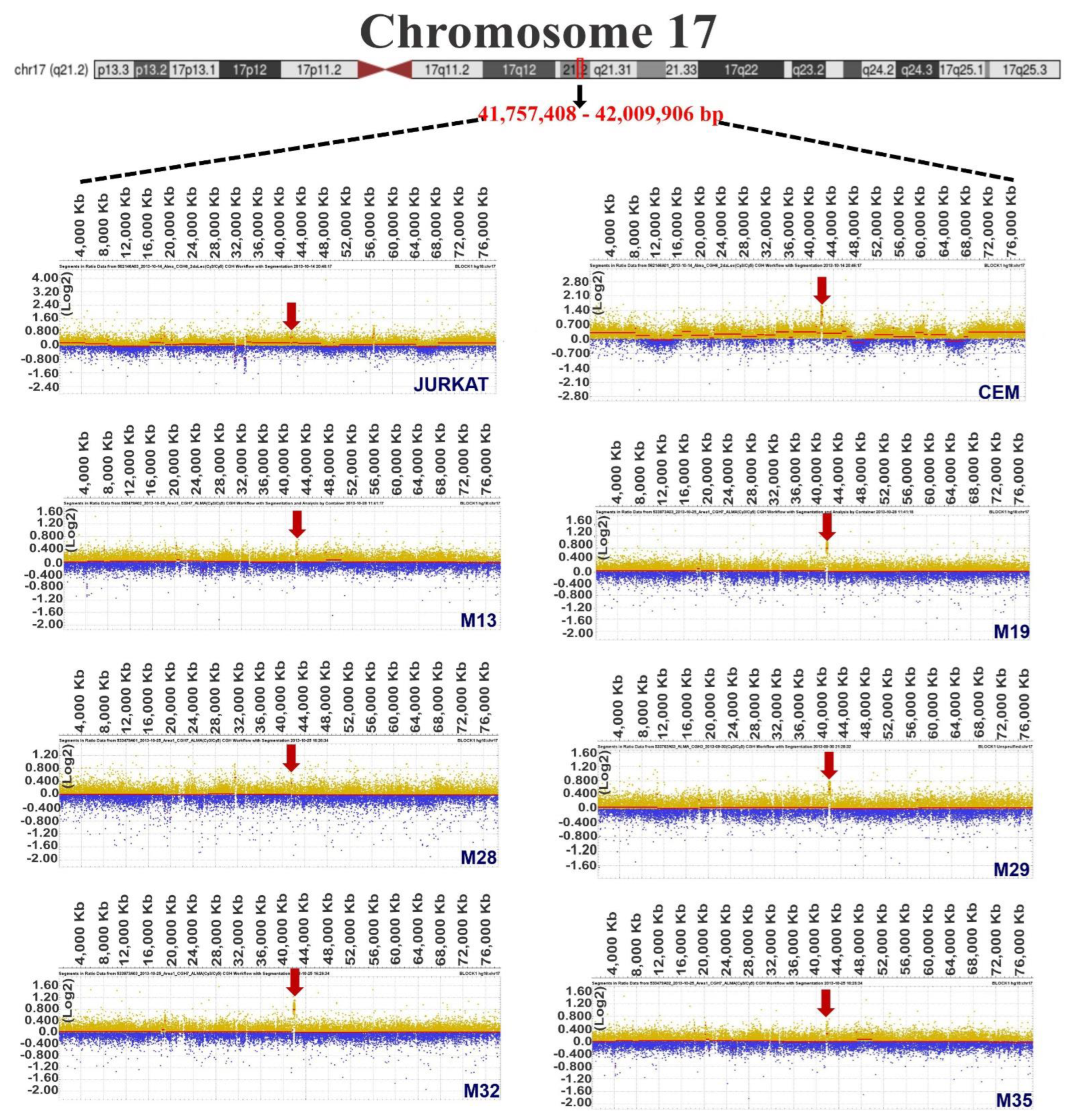
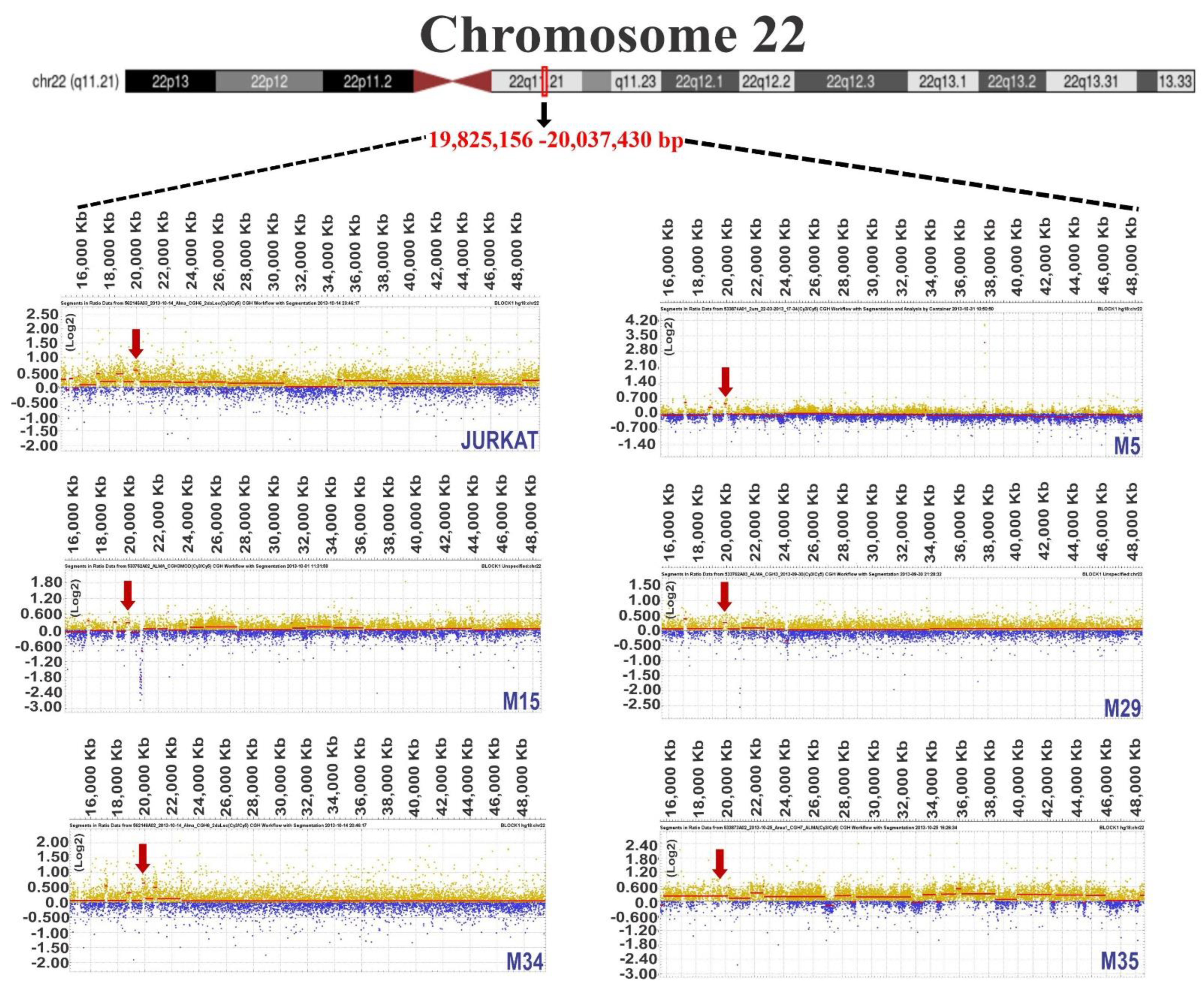
3.1. Chromosome Gains in ALL-Derived Samples
3.2. Common Chromosome Gains in ALL-Derived Samples
3.3. Identification of Genes Located in Regions of Chromosomal Gain
3.4. Expressions at mRNA Level of 22 Genes in JURKAT, CEM, and SUP-B15 Cell Lines
3.5. Expressions of 22 Genes in ALL Patients
3.6. Hierarchical Trees in Normal Hematopoiesis and Different Leukemia Lineages of the Gain Genes
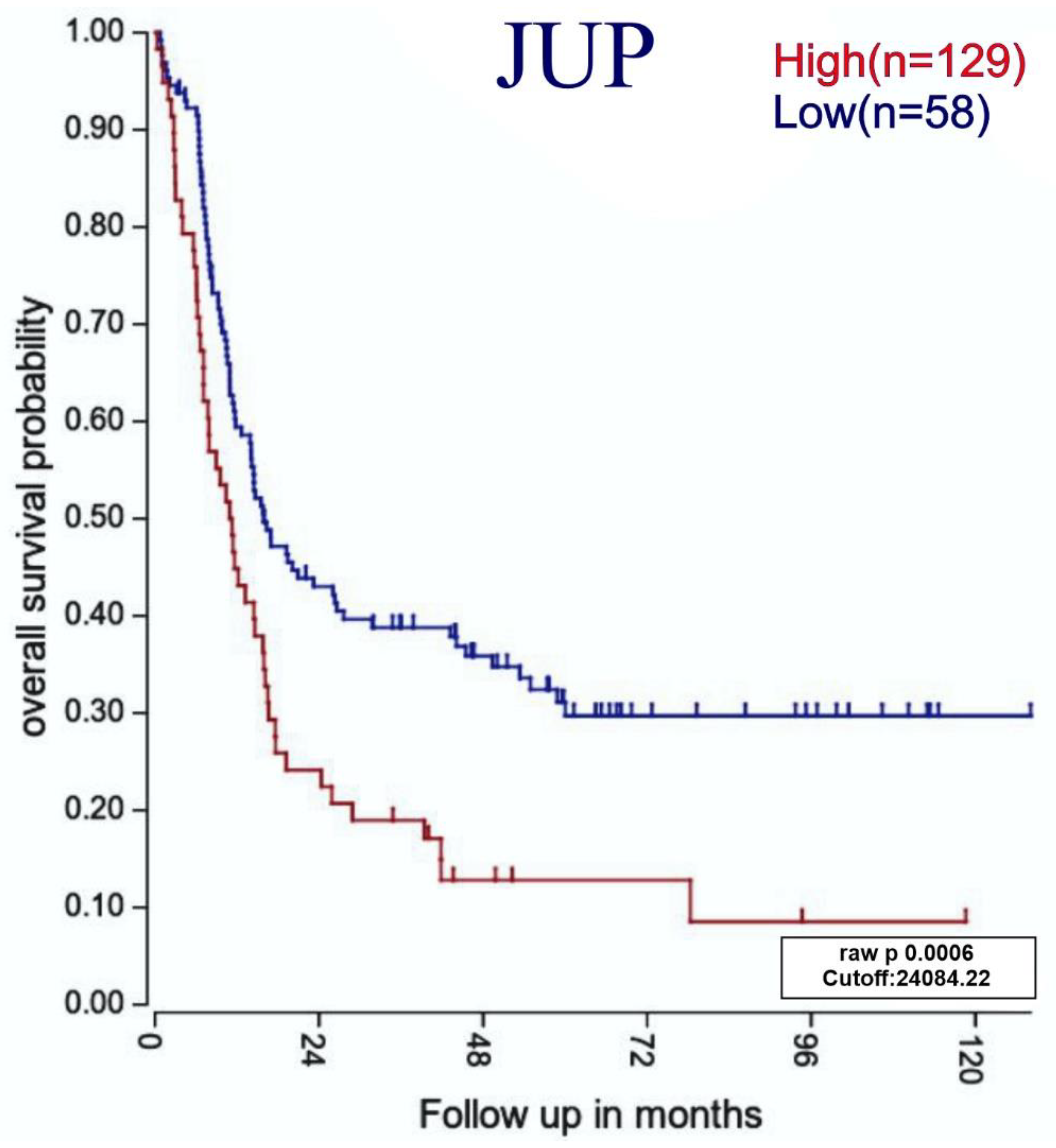
3.7. Relationship between Highly Expressed Genes and ALL Patients with Poor Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corces-Zimmerman, M.R.; Majeti, R. Pre-leukemic evolution of hematopoietic stem cells: The importance of early mutations in leukemogenesis. Leukemia 2014, 28, 2276–2282. [Google Scholar] [CrossRef]
- Marchand, T.; Pinho, S. Leukemic Stem Cells: From Leukemic Niche Biology to Treatment Opportunities. Front. Immunol. 2021, 12, 775128. [Google Scholar] [CrossRef]
- Schmidt, M.P.; Colita, A.; Ivanov, A.V.; Coriu, D.; Miron, I.C. Outcomes of patients with Down syndrome and acute leukemia: A retrospective observational study. Medicine 2021, 100, e27459. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; John, B.M.; Sondhi, V. Acute lymphoblastic leukemia with treatment--naive Fanconi anemia. Indian Pediatr. 2013, 50, 508–510. [Google Scholar] [PubMed]
- German, J. Bloom’s syndrome. XX. The first 100 cancers. Cancer Genet. Cytogenet. 1997, 93, 100–106. [Google Scholar] [CrossRef]
- Bielorai, B.; Fisher, T.; Waldman, D.; Lerenthal, Y.; Nissenkorn, A.; Tohami, T.; Marek, D.; Amariglio, N.; Toren, A. Acute lymphoblastic leukemia in early childhood as the presenting sign of ataxia-telangiectasia variant. Pediatr. Hematol. Oncol. 2013, 30, 574–582. [Google Scholar] [CrossRef]
- Bebeshko, V.G.; Bruslova, K.M.; Pushkareva, T.I.; Tsvyetkova, N.M.; Lyashenko, L.O.; Kuznyetsova, O.Y.; Kuzmenko, V.F.; Gonchar, L.O.; Yaatsemyrskyy, S.M. State of erythroid, granulocyte and platelet branches of hematopoiesis on stages of chemotherapy in children with acute lymphoblastic leukemia, who were exposed to ionizing radiation after the Chornobyl NPP accident. Probl. Radiac. Med. Radiobiol. 2016, 21, 178–190. [Google Scholar] [CrossRef]
- Sehgal, S.; Mujtaba, S.; Gupta, D.; Aggarwal, R.; Marwaha, R.K. High incidence of Epstein Barr virus infection in childhood acute lymphocytic leukemia: A preliminary study. Indian J. Pathol. Microbiol. 2010, 53, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Zubicoa, A.; Heras-Mulero, H.; Tabuenca-Del Barrio, L.; Sagaseta, M. Cytomegalovirus papilitis in a child with acute lymphoblastic leukemia. Enferm. Infecc. Microbiol. Clin. 2020, 38, 246–247. [Google Scholar] [CrossRef]
- Hleihel, R.; Akkouche, A.; Skayneh, H.; Hermine, O.; Bazarbachi, A.; El Hajj, H. Adult T-Cell Leukemia: A Comprehensive Overview on Current and Promising Treatment Modalities. Curr. Oncol. Rep. 2021, 23, 141. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.; Abdul-Hay, M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional de Estadística, INEGI. Estadísticas a Propósito Del Día Mundial Contra El Cáncer. 2021, 105, p. 12. Available online: https://www.inegi.org.mx/contenidos/saladeprensa/aproposito/2021/cancer2021_Nal.pdf (accessed on 17 October 2022).
- Arber, D.A.; Borowitz, M.J.; Cessna, M.; Etzell, J.; Foucar, K.; Hasserjian, R.P.; Rizzo, J.D.; Theil, K.; Wang, S.A.; Smith, A.T.; et al. Initial Diagnostic Workup of Acute Leukemia: Guideline from the College of American Pathologists and the American Society of Hematology. Arch. Pathol. Lab. Med. 2017, 141, 1342–1393. [Google Scholar] [CrossRef] [PubMed]
- Motllo, C.; Ribera, J.M.; Morgades, M.; Granada, I.; Montesinos, P.; Mercadal, S.; Gonzalez-Campos, J.; Moreno, M.J.; Barba, P.; Cervera, M.; et al. Frequency and prognostic significance of additional cytogenetic abnormalities to the Philadelphia chromosome in young and older adults with acute lymphoblastic leukemia. Leuk. Lymphoma 2018, 59, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Boby, E.; Nidhi, T.; Jain, A.; Singh, J.; Singh, A.; Chattopadhyay, P.; Bakhshi, S.; Chopra, A.; Palanichamy, J.K. Diagnostic Utility of IGF2BP1 and Its Targets as Potential Biomarkers in ETV6-RUNX1 Positive B-Cell Acute Lymphoblastic Leukemia. Front. Oncol. 2021, 11, 588101. [Google Scholar] [CrossRef] [PubMed]
- Shirai, R.; Osumi, T.; Sato-Otsubo, A.; Nakabayashi, K.; Mori, T.; Yoshida, M.; Yoshida, K.; Kohri, M.; Ishihara, T.; Yasue, S.; et al. Genetic features of B-cell lymphoblastic lymphoma with TCF3-PBX1. Cancer Rep. 2022, 5, e1559. [Google Scholar] [CrossRef]
- Ivanov Ofverholm, I.; Zachariadis, V.; Taylan, F.; Marincevic-Zuniga, Y.; Tran, A.N.; Saft, L.; Nilsson, D.; Syvanen, A.C.; Lonnerholm, G.; Harila-Saari, A.; et al. Overexpression of chromatin remodeling and tyrosine kinase genes in iAMP21-positive acute lymphoblastic leukemia. Leuk. Lymphoma 2020, 61, 604–613. [Google Scholar] [CrossRef]
- Kathiravan, M.; Singh, M.; Bhatia, P.; Trehan, A.; Varma, N.; Sachdeva, M.S.; Bansal, D.; Jain, R.; Naseem, S. Deletion of CDKN2A/B is associated with inferior relapse free survival in pediatric B cell acute lymphoblastic leukemia. Leuk. Lymphoma 2019, 60, 433–441. [Google Scholar] [CrossRef]
- Gonzalez-Gil, C.; Ribera, J.; Ribera, J.M.; Genesca, E. The Yin and Yang-Like Clinical Implications of the CDKN2A/ARF/CDKN2B Gene Cluster in Acute Lymphoblastic Leukemia. Genes 2021, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Mullighan, C.G. Genetic Basis of Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2017, 35, 975–983. [Google Scholar] [CrossRef]
- van Vlierberghe, P.; Meijerink, J.P.; Lee, C.; Ferrando, A.A.; Look, A.T.; van Wering, E.R.; Beverloo, H.B.; Aster, J.C.; Pieters, R. A new recurrent 9q34 duplication in pediatric T-cell acute lymphoblastic leukemia. Leukemia 2006, 20, 1245–1253. [Google Scholar] [CrossRef]
- Noronha, E.P.; Marques, L.V.C.; Andrade, F.G.; Sardou-Cezar, I.; Dos Santos-Bueno, F.V.; Zampier, C.D.P.; Terra-Granado, E.; Pombo-de-Oliveira, M.S. T-lymphoid/myeloid mixed phenotype acute leukemia and early T-cell precursor lymphoblastic leukemia similarities with NOTCH1 mutation as a good prognostic factor. Cancer Manag. Res. 2019, 11, 3933–3943. [Google Scholar] [CrossRef]
- Martelli, A.M.; Paganelli, F.; Fazio, A.; Bazzichetto, C.; Conciatori, F.; McCubrey, J.A. The Key Roles of PTEN in T-Cell Acute Lymphoblastic Leukemia Development, Progression, and Therapeutic Response. Cancers 2019, 11, 629. [Google Scholar] [CrossRef] [PubMed]
- Vermeesch, J.R.; Fiegler, H.; de Leeuw, N.; Szuhai, K.; Schoumans, J.; Ciccone, R.; Speleman, F.; Rauch, A.; Clayton-Smith, J.; Van Ravenswaaij, C.; et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur. J. Hum. Genet. 2007, 15, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, L.G.; Bejjani, B.A. Medical applications of array CGH and the transformation of clinical cytogenetics. Cytogenet. Genome Res. 2006, 115, 303–309. [Google Scholar] [CrossRef]
- Cheung, S.W.; Bi, W. Novel applications of array comparative genomic hybridization in molecular diagnostics. Expert Rev. Mol. Diagn. 2018, 18, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, J.R.; Babicz, M.; Gaworczyk, A.; Lejman, M.; Winnicka, D.; Styka, B.; Jaszczuk, I. Structural and numerical abnormalities resolved in one-step analysis: The most common chromosomal rearrangements detected by comparative genomic hybridization in childhood acute lymphoblastic leukemia. Cancer Genet. Cytogenet. 2010, 200, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. RNA sequencing (RNA-Seq) and its application in ovarian cancer. Gynecol. Oncol. 2019, 152, 194–201. [Google Scholar] [CrossRef]
- Alvarez-Zavala, M.; Riveros-Magana, A.R.; Garcia-Castro, B.; Barrera-Chairez, E.; Rubio-Jurado, B.; Garces-Ruiz, O.M.; Ramos-Solano, M.; Aguilar-Lemarroy, A.; Jave-Suarez, L.F. WNT receptors profile expression in mature blood cells and immature leukemic cells: RYK emerges as a hallmark receptor of acute leukemia. Eur. J. Haematol. 2016, 97, 155–165. [Google Scholar] [CrossRef]
- Forero-Castro, M.; Robledo, C.; Benito, R.; Abaigar, M.; Africa-Martín, A.; Arefi, M.; Fuster, J.L.; De las Heras, N.; Rodríguez, J.N.; Quintero, J.; et al. Genome-Wide DNA Copy Number Analysis of Acute Lymphoblastic Leukemia Identifies New Genetic Markers Associated with Clinical Outcome. PLoS ONE 2016, 11, e0148972. [Google Scholar] [CrossRef] [PubMed]
- Montano, A.; Hernandez-Sanchez, J.; Forero-Castro, M.; Matorra-Miguel, M.; Lumbreras, E.; Miguel, C.; Santos, S.; Ramirez-Maldonado, V.; Fuster, J.L.; de Las Heras, N.; et al. Comprehensive Custom NGS Panel Validation for the Improvement of the Stratification of B-Acute Lymphoblastic Leukemia Patients. J Pers Med 2020, 10, e0148972. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Lemarroy, A.; Zapata-García, J.A.; Jave-Suárez, L.F.; Riveros-Magaña, A. Common chromosome gains and losses in acute lymphoblastic leukemia [RNA-seq]. 2022. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189641 (accessed on 17 October 2022).
- Kong, D.; Mao, J.H.; Li, H.; Wang, J.Y.; Li, Y.Y.; Wu, X.C.; Re, G.F.; Luo, H.Y.; Kuang, Y.Q.; Wang, K.H. Effects and associated transcriptomic landscape changes of methamphetamine on immune cells. BMC Med. Genom. 2022, 15, 144. [Google Scholar] [CrossRef]
- Ferguson, D.C.; McCorkle, J.R.; Barnett, K.R.; Bonten, E.J.; Bergeron, B.P.; Bhattarai, K.R.; Yang, W.; Smith, C.; Hansen, B.S.; Bajpai, R.; et al. Amino acid stress response genes promote L-asparaginase resistance in pediatric acute lymphoblastic leukemia. Blood Adv. 2022, 6, 3386–3397. [Google Scholar] [CrossRef]
- Diedrich, J.; Savic, D. Genome-wide maps of chromatin state and mRNA expression patterns in leukemic cell lines. 2019. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129066 (accessed on 17 October 2022).
- Huang, Y.H.; Su, T.C.; Wang, C.H.; Wong, S.L.; Chien, Y.H.; Wang, Y.T.; Hwu, W.L.; Lee, N.C. RNA-seq of peripheral blood mononuclear cells of congenital generalized lipodystrophy type 2 patients. Sci. Data 2021, 8, 265. [Google Scholar] [CrossRef]
- Jerez, A.; Hurtado, A.M. RNA-sequencing of Chronic Myelomonocytic Leukemia and healthy donors bone marrow samples. 2020. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102312 (accessed on 17 October 2022).
- Andrews, S. FastQC A Quality Control tool for High Throughput Sequence Data. 2022. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 17 October 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014, 15, 550. [Google Scholar] [CrossRef]
- Gregory, R.; Warnes, B.B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Package ‘gplots’. Repository CRAN. 2020. Available online: https://bio.tools/ggplot2 (accessed on 17 October 2022).
- Haferlach, T.; Kohlmann, A.; Wieczorek, L.; Basso, G.; Kronnie, G.T.; Bene, M.C.; De Vos, J.; Hernandez, J.M.; Hofmann, W.K.; Mills, K.I.; et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010, 28, 2529–2537. [Google Scholar] [CrossRef]
- Liu, W.M. Microarray Innovations in LEukemia (MILE) study. 2009. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13159 (accessed on 17 October 2022).
- Bagger, F.O.; Kinalis, S.; Rapin, N. BloodSpot: A database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 2019, 47, D881–D885. [Google Scholar] [CrossRef]
- Bagger, F.O.; Sasivarevic, D.; Sohi, S.H.; Laursen, L.G.; Pundhir, S.; Sonderby, C.K.; Winther, O.; Rapin, N.; Porse, B.T. BloodSpot: A database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016, 44, D917–D924. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Brennan, S.; Milne, T.A.; Chen, W.Y.; Li, Y.; Hurtz, C.; Kweon, S.M.; Zickl, L.; Shojaee, S.; Neuberg, D.; et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012, 2, 1004–1023. [Google Scholar] [CrossRef] [PubMed]
- Malard, F.; Mohty, M. Acute lymphoblastic leukaemia. Lancet 2020, 395, 1146–1162. [Google Scholar] [CrossRef]
- Krem, M.M.; Press, O.W.; Horwitz, M.S.; Tidwell, T. Mechanisms and clinical applications of chromosomal instability in lymphoid malignancy. Br. J. Haematol. 2015, 171, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Thomas, X.; Heiblig, M. Diagnostic and treatment of adult Philadelphia chromosome-positive acute lymphoblastic leukemia. Int. J. Hematol. Oncol. 2016, 5, 77–90. [Google Scholar] [CrossRef]
- Usvasalo, A.; Raty, R.; Harila-Saari, A.; Koistinen, P.; Savolainen, E.R.; Vettenranta, K.; Knuutila, S.; Elonen, E.; Saarinen-Pihkala, U.M. Acute lymphoblastic leukemias with normal karyotypes are not without genomic aberrations. Cancer Genet. Cytogenet. 2009, 192, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Matteucci, C.; Barba, G.; Varasano, E.; Vitale, A.; Mancini, M.; Testoni, N.; Cuneo, A.; Rege-Cambrin, G.; Elia, L.; La Starza, R.; et al. Rescue of genomic information in adult acute lymphoblastic leukaemia (ALL) with normal/failed cytogenetics: A GIMEMA centralized biological study. Br. J. Haematol. 2010, 149, 70–78. [Google Scholar] [CrossRef]
- Paulsson, K.; Cazier, J.B.; Macdougall, F.; Stevens, J.; Stasevich, I.; Vrcelj, N.; Chaplin, T.; Lillington, D.M.; Lister, T.A.; Young, B.D. Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: Unexpected similarities with pediatric disease. Proc. Natl. Acad. Sci. USA 2008, 105, 6708–6713. [Google Scholar] [CrossRef]
- Borst, L.; Wesolowska, A.; Joshi, T.; Borup, R.; Nielsen, F.C.; Andersen, M.K.; Jonsson, O.G.; Wehner, P.S.; Wesenberg, F.; Frost, B.M.; et al. Genome-wide analysis of cytogenetic aberrations in ETV6/RUNX1-positive childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2012, 157, 476–482. [Google Scholar] [CrossRef]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150. [Google Scholar] [CrossRef]
- Drews, R.M.; Hernando, B.; Tarabichi, M.; Haase, K.; Lesluyes, T.; Smith, P.S.; Morrill Gavarro, L.; Couturier, D.L.; Liu, L.; Schneider, M.; et al. A pan-cancer compendium of chromosomal instability. Nature 2022, 606, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Simino, J.; Wang, Z.; Bressler, J.; Chouraki, V.; Yang, Q.; Younkin, S.G.; Seshadri, S.; Fornage, M.; Boerwinkle, E.; Mosley, T.H., Jr. Whole exome sequence-based association analyses of plasma amyloid-beta in African and European Americans; the Atherosclerosis Risk in Communities-Neurocognitive Study. PLoS ONE 2017, 12, e0180046. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, A.E.; Postma, D.S.; van Ginneken, B.; Wielputz, M.O.; Schmidt, M.; Becker, N.; Owsijewitsch, M.; Kauczor, H.U.; de Koning, H.J.; Lammers, J.W.; et al. Novel genes for airway wall thickness identified with combined genome-wide association and expression analyses. Am. J. Respir. Crit. Care Med. 2015, 191, 547–556. [Google Scholar] [CrossRef]
- Canning, P.; Cooper, C.D.O.; Krojer, T.; Murray, J.W.; Pike, A.C.W.; Chaikuad, A.; Keates, T.; Thangaratnarajah, C.; Hojzan, V.; Marsden, B.D.; et al. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 2013, 288, 7803–7814. [Google Scholar] [CrossRef] [PubMed]
- Dubey, D.; Wilson, M.R.; Clarkson, B.; Giannini, C.; Gandhi, M.; Cheville, J.; Lennon, V.A.; Eggers, S.; Devine, M.F.; Mandel-Brehm, C.; et al. Expanded Clinical Phenotype, Oncological Associations, and Immunopathologic Insights of Paraneoplastic Kelch-like Protein-11 Encephalitis. JAMA Neurol. 2020, 77, 1420–1429. [Google Scholar] [CrossRef]
- Monoh, K.; Kurihara, T.; Takahashi, Y.; Ichikawa, T.; Kumanishi, T.; Hayashi, S.; Minoshima, S.; Shimizu, N. Structure, expression and chromosomal localization of the gene encoding human 2′, 3′-cyclic-nucleotide 3’-phosphodiesterase. Gene 1993, 129, 297–301. [Google Scholar] [CrossRef]
- Zorniak, M.; Clark, P.A.; Leeper, H.E.; Tipping, M.D.; Francis, D.M.; Kozak, K.R.; Salamat, M.S.; Kuo, J.S. Differential expression of 2′, 3′-cyclic-nucleotide 3′-phosphodiesterase and neural lineage markers correlate with glioblastoma xenograft infiltration and patient survival. Clin. Cancer Res. 2012, 18, 3628–3636. [Google Scholar] [CrossRef]
- Elshourbagy, N.A.; Near, J.C.; Kmetz, P.J.; Wells, T.N.; Groot, P.H.; Saxty, B.A.; Hughes, S.A.; Franklin, M.; Gloger, I.S. Cloning and expression of a human ATP-citrate lyase cDNA. Eur. J. Biochem. 1992, 204, 491–499. [Google Scholar] [CrossRef]
- Basappa, J.; Citir, M.; Zhang, Q.; Wang, H.Y.; Liu, X.; Melnikov, O.; Yahya, H.; Stein, F.; Muller, R.; Traynor-Kaplan, A.; et al. ACLY is the novel signaling target of PIP2/PIP3 and Lyn in acute myeloid leukemia. Heliyon 2020, 6, e03910. [Google Scholar] [CrossRef]
- Shah, S.; Carriveau, W.J.; Li, J.; Campbell, S.L.; Kopinski, P.K.; Lim, H.W.; Daurio, N.; Trefely, S.; Won, K.J.; Wallace, D.C.; et al. Targeting ACLY sensitizes castration-resistant prostate cancer cells to AR antagonism by impinging on an ACLY-AMPK-AR feedback mechanism. Oncotarget 2016, 7, 43713–43730. [Google Scholar] [CrossRef]
- Wen, J.; Min, X.; Shen, M.; Hua, Q.; Han, Y.; Zhao, L.; Liu, L.; Huang, G.; Liu, J.; Zhao, X. ACLY facilitates colon cancer cell metastasis by CTNNB1. J. Exp. Clin. Cancer Res. 2019, 38, 401. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, K.; Gong, D.; Zhang, J.; Li, Q.; Zhao, G.; Lin, P. ACLY: A biomarker of recurrence in breast cancer. Pathol. Res. Pract. 2020, 216, 153076. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Wang, B.; Xu, K.; Nie, L.; Fu, Y.; Wang, Z.; Wang, Q.; Wang, S.; Zou, X. m (6) A Reader HNRNPA2B1 Promotes Esophageal Cancer Progression via Up-Regulation of ACLY and ACC1. Front. Oncol. 2020, 10, 553045. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, K.A.; Wheelock, M.J. Plakoglobin, or an 83-kD homologue distinct from beta-catenin, interacts with E-cadherin and N-cadherin. J. Cell. Biol. 1992, 118, 671–679. [Google Scholar] [CrossRef]
- Luong-Gardiol, N.; Siddiqui, I.; Pizzitola, I.; Jeevan-Raj, B.; Charmoy, M.; Huang, Y.; Irmisch, A.; Curtet, S.; Angelov, G.S.; Danilo, M.; et al. gamma-Catenin-Dependent Signals Maintain BCR-ABL1 (+) B Cell Acute Lymphoblastic Leukemia. Cancer Cell 2019, 35, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Huang, X.; Zhang, Y.; Ye, X.; Qian, W. gamma-Catenin Overexpression in AML Patients May Promote Tumor Cell Survival via Activation of the Wnt/beta-Catenin Axis. Onco Targets Ther. 2020, 13, 1265–1276. [Google Scholar] [CrossRef]
- Weiland, F.; Lokman, N.A.; Klingler-Hoffmann, M.; Jobling, T.; Stephens, A.N.; Sundfeldt, K.; Hoffmann, P.; Oehler, M.K. Ovarian Blood Sampling Identifies Junction Plakoglobin as a Novel Biomarker of Early Ovarian Cancer. Front. Oncol. 2020, 10, 1767. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, L.; Qin, Y.; Liu, S.; Qiao, Y.; Wan, X.; Zeng, H.; Tang, X.; Liu, M.; Hou, Y. Effects of differential distributed-JUP on the malignancy of gastric cancer. J. Adv. Res. 2021, 28, 195–208. [Google Scholar] [CrossRef]
- Spethmann, T.; Bockelmann, L.C.; Labitzky, V.; Ahlers, A.K.; Schroder-Schwarz, J.; Bonk, S.; Simon, R.; Sauter, G.; Huland, H.; Kypta, R.; et al. Opposing prognostic relevance of junction plakoglobin in distinct prostate cancer patient subsets. Mol. Oncol. 2021, 15, 1956–1969. [Google Scholar] [CrossRef]
- Zhu, H.J.; Liu, L.; Fan, L.; Zhang, L.N.; Fang, C.; Zou, Z.J.; Li, J.Y.; Xu, W. The BH3-only protein Puma plays an essential role in p53-mediated apoptosis of chronic lymphocytic leukemia cells. Leuk. Lymphoma 2013, 54, 2712–2719. [Google Scholar] [CrossRef]
- Ishiguro, H.; Koga, M.; Horiuchi, Y.; Noguchi, E.; Morikawa, M.; Suzuki, Y.; Arai, M.; Niizato, K.; Iritani, S.; Itokawa, M.; et al. Supportive evidence for reduced expression of GNB1L in schizophrenia. Schizophr. Bull. 2010, 36, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Matsushita, M.; Girirajan, S.; Lisowski, M.; Sun, E.; Sul, Y.; Bernier, R.; Estes, A.; Dawson, G.; Minshew, N.; et al. Evidence for involvement of GNB1L in autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012, 159B, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Nalesnik, M.A.; Tseng, G.; Ding, Y.; Xiang, G.S.; Zheng, Z.L.; Yu, Y.; Marsh, J.W.; Michalopoulos, G.K.; Luo, J.H. Gene deletions and amplifications in human hepatocellular carcinomas: Correlation with hepatocyte growth regulation. Am. J. Pathol. 2012, 180, 1495–1508. [Google Scholar] [CrossRef] [PubMed]
- Hermosilla, V.E.; Hepp, M.I.; Escobar, D.; Farkas, C.; Riffo, E.N.; Castro, A.F.; Pincheira, R. Developmental SALL2 transcription factor: A new player in cancer. Carcinogenesis 2017, 38, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Jiang, L.; Zhong, M.L.; Li, J.F.; Li, B.S.; Peng, L.J.; Dai, Y.T.; Cui, B.W.; Yan, T.Q.; Zhang, W.N.; et al. Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2018, 115, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wang, W.; Tang, Y.; Zhou, D.; Gao, Y.; Zhang, Q.; Zhou, X.; Zhu, H.; Xing, L.; Yu, J. mRNA and methylation profiling of radioresistant esophageal cancer cells: The involvement of Sall2 in acquired aggressive phenotypes. J. Cancer 2017, 8, 646–656. [Google Scholar] [CrossRef]
- Ye, L.; Lin, C.; Wang, X.; Li, Q.; Li, Y.; Wang, M.; Zhao, Z.; Wu, X.; Shi, D.; Xiao, Y.; et al. Epigenetic silencing of SALL2 confers tamoxifen resistance in breast cancer. EMBO Mol. Med. 2019, 11, e10638. [Google Scholar] [CrossRef]
- Alagaratnam, S.; Lind, G.E.; Kraggerud, S.M.; Lothe, R.A.; Skotheim, R.I. The testicular germ cell tumour transcriptome. Int. J. Androl. 2011, 34, e133–e150. [Google Scholar] [CrossRef]
- Suva, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef]
- Chai, L. The role of HSAL (SALL) genes in proliferation and differentiation in normal hematopoiesis and leukemogenesis. Transfusion 2011, 51, 87S–93S. [Google Scholar] [CrossRef]
- Sung, C.K.; Li, D.; Andrews, E.; Drapkin, R.; Benjamin, T. Promoter methylation of the SALL2 tumor suppressor gene in ovarian cancers. Mol. Oncol. 2013, 7, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Imai, A.; Mochizuki, D.; Misawa, Y.; Nakagawa, T.; Endo, S.; Mima, M.; Yamada, S.; Kawasaki, H.; Kanazawa, T.; Misawa, K. SALL2 Is a Novel Prognostic Methylation Marker in Patients with Oral Squamous Carcinomas: Associations with SALL1 and SALL3 Methylation Status. DNA Cell Biol. 2019, 38, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Weinshilboum, R.M.; Raymond, F.A. Inheritance of low erythrocyte catechol-o-methyltransferase activity in man. Am. J. Hum. Genet. 1977, 29, 125–135. [Google Scholar]
- Cao, M.; Yin, D.; Qin, Y.; Liao, F.; Su, Y.; Xia, X.; Gao, J.; Zhu, Y.; Zhang, W.; Shu, Y.; et al. Screening of Novel Pharmacogenetic Candidates for Mercaptopurine-Induced Toxicity in Patients with Acute Lymphoblastic Leukemia. Front. Pharmacol. 2020, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Singh, G.; Rai, V. Evaluation of COMT Gene rs4680 Polymorphism as a Risk Factor for Endometrial Cancer. Indian J. Clin. Biochem. 2020, 35, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Peng, Q.; Qin, A.; Chen, Z.; Lin, L.; Deng, Y.; Xie, L.; Xu, J.; Li, H.; Li, T.; et al. Association of COMT Val158Met polymorphism and breast cancer risk: An updated meta-analysis. Diagn. Pathol. 2012, 7, 136. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Shiina, M.; Maekawa, S.; Kato, T.; Shahryari, V.; Kulkarni, P.; Dasgupta, P.; Yamamura, S.; Saini, S.; Tabatabai, Z.L.; et al. Suppressor effect of catechol-O-methyltransferase gene in prostate cancer. PLoS ONE 2021, 16, e0253877. [Google Scholar] [CrossRef]
- Jin, X.; Zhou, H.; Song, J.; Cui, H.; Luo, Y.; Jiang, H. P3H4 Overexpression Serves as a Prognostic Factor in Lung Adenocarcinoma. Comput. Math. Methods Med. 2021, 2021, 9971353. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Pang, K.; Pang, H.; Zhang, J.; Zhang, Z.; He, H.; Zhou, R.; Shi, Z.; Han, C. Knockdown of P3H4 inhibits proliferation and invasion of bladder cancer. Aging 2020, 12, 2156–2168. [Google Scholar] [CrossRef]
- Wan, B.; Zeng, Q.; Tang, X.Z.; Tang, Y.X. P3H4 affects renal carcinoma through up-regulating miR-1/133a. Eur. Rev. Med. Pharm. Sci. 2018, 22, 5180–5186. [Google Scholar] [CrossRef]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar]
- Wang, Q.; Guo, X.; Li, L.; Gao, Z.; Su, X.; Ji, M.; Liu, J. N6-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis. 2020, 11, 911. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Gao, S.; Liu, W.; Wong, C.C.; Wu, J.; Wu, J.; Liu, D.; Gou, H.; Kang, W.; Zhai, J.; et al. RNA N(6)-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m(6)A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology 2021, 160, 1284–1300. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pan, C.; Wang, X.; Xu, D.; Ma, Y.; Hu, J.; Chen, P.; Xiang, Z.; Rao, Q.; Han, X. Silencing of METTL3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics 2021, 11, 7640–7657. [Google Scholar] [CrossRef]
- Xia, T.; Wu, X.; Cao, M.; Zhang, P.; Shi, G.; Zhang, J.; Lu, Z.; Wu, P.; Cai, B.; Miao, Y.; et al. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol. Res. Pract. 2019, 215, 152666. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Lv, X.; Liu, D.; Guo, H.; Yao, G.; Wang, L.; Liang, X.; Yang, Y. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021, 28, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Yang, C.; Zhang, S.; Cheng, M.; Guo, S.; Zhu, Y.; Ma, J.; Liang, Y.; Wang, L.; Zheng, S.; et al. METTL3-mediated m(6)A mRNA modification promotes esophageal cancer initiation and progression via Notch signaling pathway. Mol. Ther. Nucleic Acids 2021, 26, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, J.Z.; Yang, X.; Yu, H.; Zhou, R.; Lu, H.C.; Yuan, W.B.; Lu, J.C.; Zhou, Z.J.; Lu, Q.; et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 2019, 18, 110. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N(6)-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Chang, L.; Liu, C.; Chen, X.; Zhu, X. The study of METTL3 and METTL14 expressions in childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Mol. Genet. Genom. Med. 2019, 7, e00933. [Google Scholar] [CrossRef] [PubMed]
- Melstrom, L.; Chen, J. RNA N6-methyladenosine modification in solid tumors: New therapeutic frontiers. Cancer Gene Ther. 2020, 27, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, H.; Esaki, Y.; Kelly, F.; Yan, W. Cullin3 is a KLHL10-interacting protein preferentially expressed during late spermiogenesis. Biol. Reprod. 2006, 74, 102–108. [Google Scholar] [CrossRef]
- Miyamoto, T.; Minase, G.; Okabe, K.; Ueda, H.; Sengoku, K. Male infertility and its genetic causes. J. Obstet. Gynaecol. Res. 2015, 41, 1501–1505. [Google Scholar] [CrossRef]
- Liang, C.; Zhao, Y.; Chen, C.; Huang, S.; Deng, T.; Zeng, X.; Tan, J.; Zha, X.; Chen, S.; Li, Y. Higher TOX Genes Expression Is Associated with Poor Overall Survival for Patients With Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 740642. [Google Scholar] [CrossRef]
- Tessema, M.; Yingling, C.M.; Grimes, M.J.; Thomas, C.L.; Liu, Y.; Leng, S.; Joste, N.; Belinsky, S.A. Differential epigenetic regulation of TOX subfamily high mobility group box genes in lung and breast cancers. PLoS ONE 2012, 7, e34850. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Hao, J.; Dong, B.; Li, Y.; Zhu, X.; Ding, J.; Ren, S.; Zhao, H.; Wu, S.; et al. Reduced cytosolic carboxypeptidase 6 (CCP6) level leads to accumulation of serum polyglutamylated DNAJC7 protein: A potential biomarker for renal cell carcinoma early detection. Oncotarget 2016, 7, 22385–22396. [Google Scholar] [CrossRef]
- Wallmeier, J.; Shiratori, H.; Dougherty, G.W.; Edelbusch, C.; Hjeij, R.; Loges, N.T.; Menchen, T.; Olbrich, H.; Pennekamp, P.; Raidt, J.; et al. TTC25 Deficiency Results in Defects of the Outer Dynein Arm Docking Machinery and Primary Ciliary Dyskinesia with Left-Right Body Asymmetry Randomization. Am. J. Hum. Genet. 2016, 99, 460–469. [Google Scholar] [CrossRef]
- Pacholewska, A.; Marti, E.; Leeb, T.; Jagannathan, V.; Gerber, V. LPS-induced modules of co-expressed genes in equine peripheral blood mononuclear cells. BMC Genom. 2017, 18, 34. [Google Scholar] [CrossRef]
- Barrow, T.M.; Wong Doo, N.; Milne, R.L.; Giles, G.G.; Willmore, E.; Strathdee, G.; Byun, H.M. Analysis of retrotransposon subfamily DNA methylation reveals novel early epigenetic changes in chronic lymphocytic leukemia. Haematologica 2021, 106, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kim, T.N.; Kim, S.; Baek, S.H.; Kim, J.H.; Lee, S.R.; Kim, J.R. Overexpression of mitochondrial thioredoxin reductase and peroxiredoxin III in hepatocellular carcinomas. Anticancer Res. 2002, 22, 3331–3335. [Google Scholar] [PubMed]
- Bu, L.; Li, W.; Ming, Z.; Shi, J.; Fang, P.; Yang, S. Inhibition of TrxR2 suppressed NSCLC cell proliferation, metabolism and induced cell apoptosis through decreasing antioxidant activity. Life Sci. 2017, 178, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Karyadi, D.M.; Geybels, M.S.; Karlins, E.; Decker, B.; McIntosh, L.; Hutchinson, A.; Kolb, S.; McDonnell, S.K.; Hicks, B.; Middha, S.; et al. Whole exome sequencing in 75 high-risk families with validation and replication in independent case-control studies identifies TANGO2, OR5H14, and CHAD as new prostate cancer susceptibility genes. Oncotarget 2017, 8, 1495–1507. [Google Scholar] [CrossRef] [PubMed]
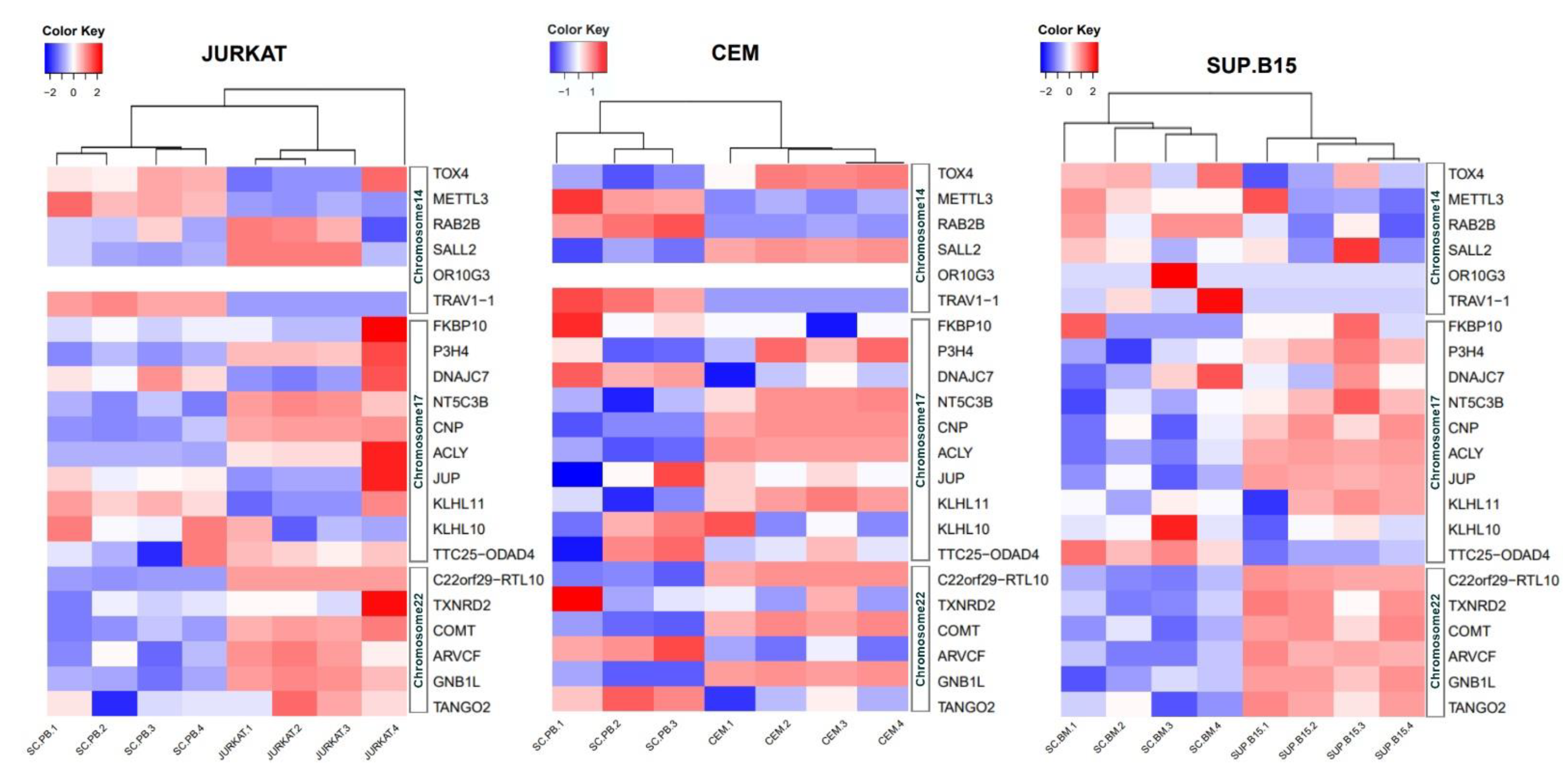
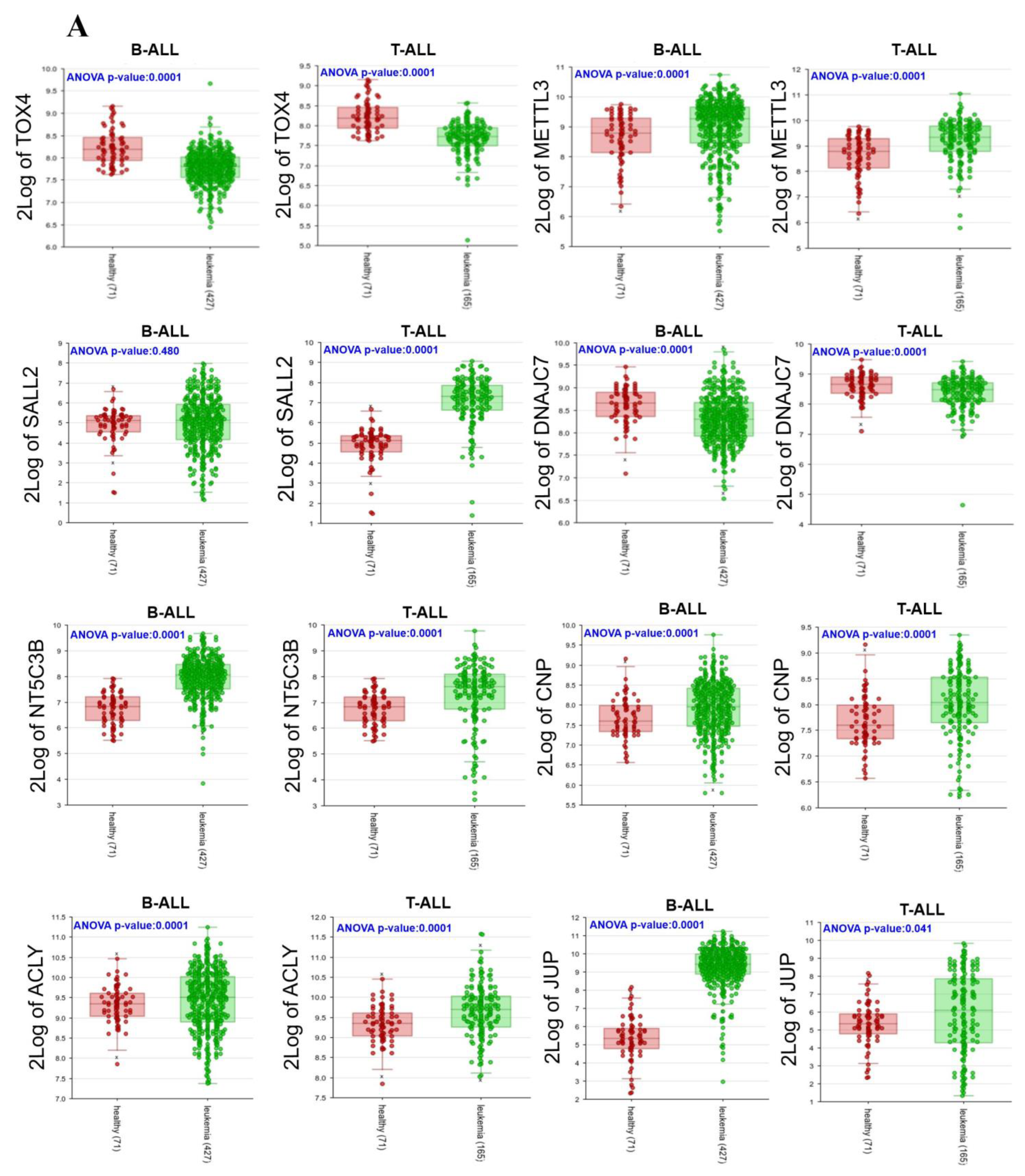
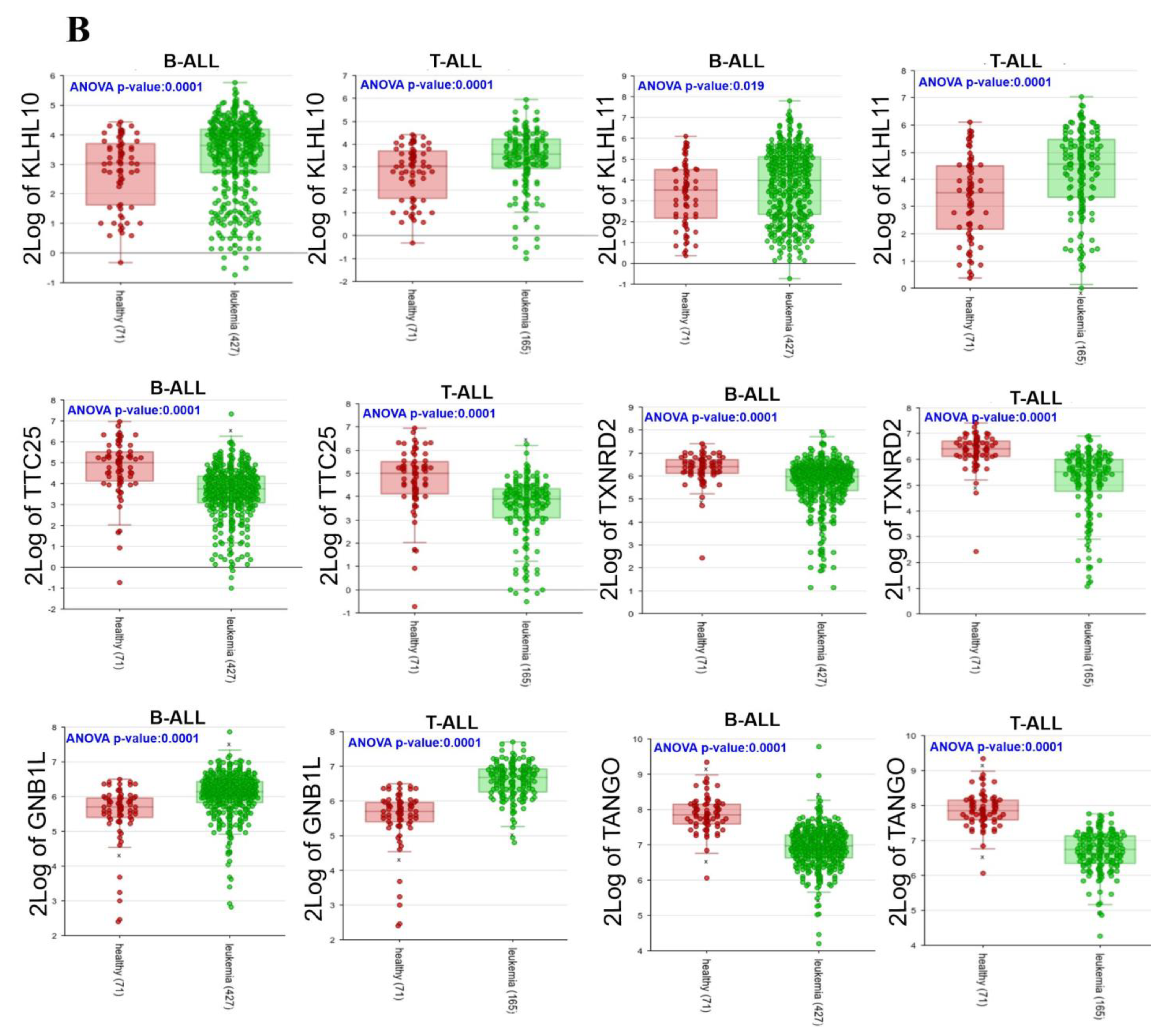
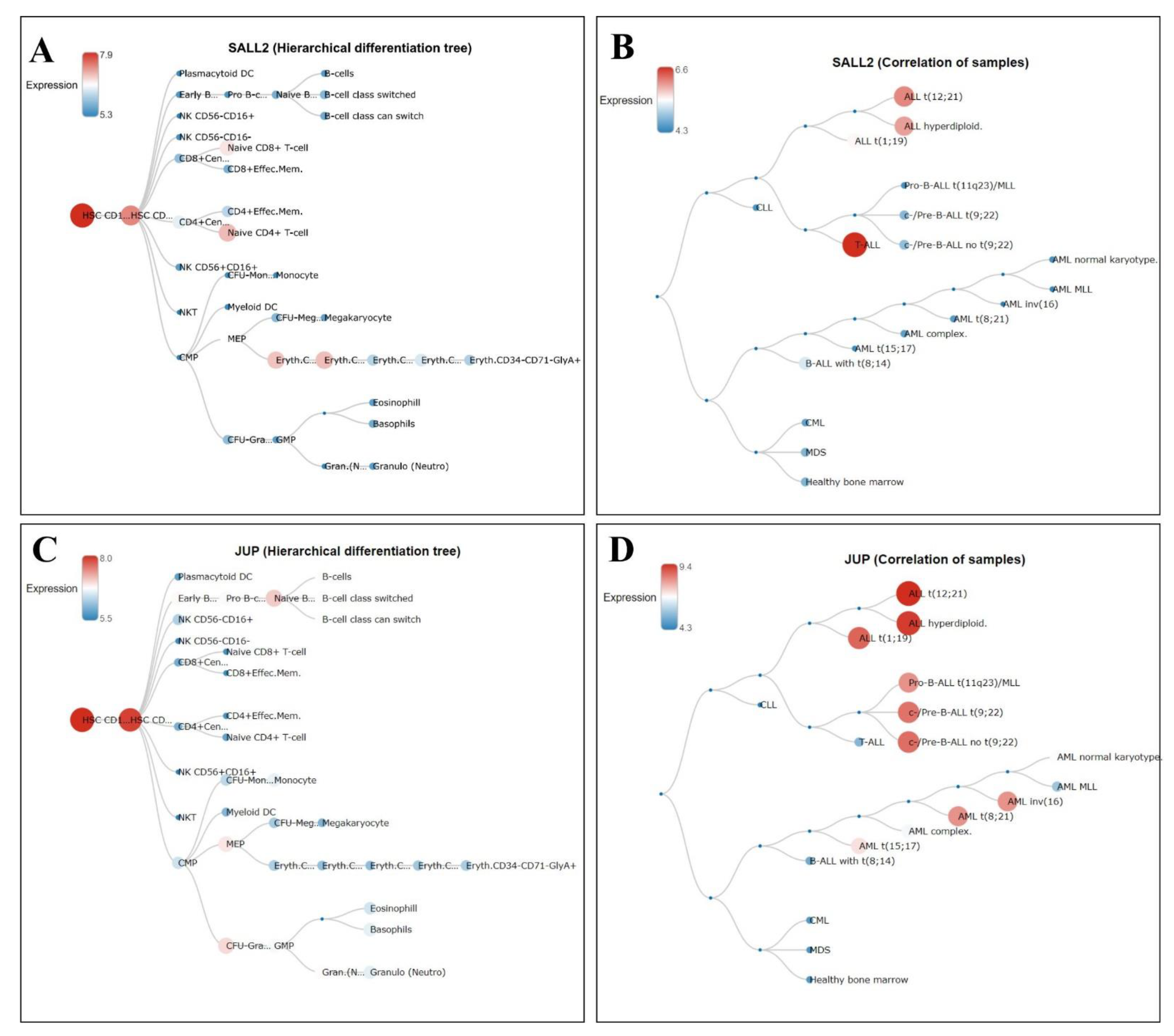
| Gene | Forward | Reverse | Size amplicon |
|---|---|---|---|
| JUP | AGCAGCCCTACACGGATG | GATGTTCTCCACCGACGAGTA | 161 bp |
| RPS18 | CGATGGGCGGCGGAAAA | CAGTCGCTCCAGGTCTTCACGG | 283 bp |
| RPLP0 | CCTCATATCCGGGGGAATGTG | GCAGCAGCTGGCACCTTATTG | 100 pb |
| Sample | Sex | Age | Type of Leukemia | Chromosome with Gains |
|---|---|---|---|---|
| JURKAT | M | 14 | T-ALL | 1,3,5,6,11,13,15,17,22 |
| CEM | F | 4 | T-ALL | 5,8,11,12,13,14,15,16,17,18,19,20,21,X |
| M5 | F | 49 | Pre-B-ALL | 3,5,7,9,10,11,12,13,14,15,16,17,18,19,20,22 |
| M12 | M | 77 | T-ALL-T | 1,14,17 |
| M13 | M | 65 | T-ALL | 7,14,17 |
| M15 | M | 17 | Pre-B-ALL | 5,14,15,19,21,22 |
| M19 | M | 45 | Pre-B-ALL | 14,17 |
| M28 | M | 16 | B-LLA | 4,14,17,Y |
| M29 | F | 50 | B-LLA | 1,6,8,10,14,15,17,18,19,22,X |
| M32 | F | 31 | T-LLA | 14,17 |
| M34 | M | 20 | B-LLA | 1,12,14,15,16,17,18,19,22,X |
| M35 | M | 16 | B-LLA | 7,9,10,12,14,15,17,19,22,Y |
| Sample ID | Chr | Position of Gain Pb | Size (Pb) |
|---|---|---|---|
| JURKAT | 17 | 41,757,408–42,009,906 | 252,498 |
| 22 | 14,507,171–30,395,708 | 15,888,537 | |
| CEM | 17 | 36,734,811–46,367,425 | 9,632,614 |
| M5 | 22 | 19,825,156–20,037,430 | 212,274 |
| M12 | 14 | 21,444,377–21,624,239 | 179,862 |
| M13 | 14 | 21,441,167–21,723,152 | 281,985 |
| 17 | 41,755,952–42,135,245 | 379,293 | |
| M15 | 22 | 19,718,309–20,037,430 | 319,121 |
| M19 | 17 | 35,353,354–42,945,517 | 7,592,163 |
| M28 | 14 | 21,441,167–22,013,159 | 571,992 |
| 17 | 40,964,001–43,704,741 | 2,740,740 | |
| M29 | 17 | 41,307,415–43,247,247 | 1,939,832 |
| 22 | 19,720,528–20,037,430 | 316,902 | |
| M32 | 14 | 21,424,744–21,961,837 | 537,093 |
| 17 | 40,741,630–43,408,899 | 2,667,269 | |
| M34 | 14 | 21,444,229–22,055,727 | 611,498 |
| 22 | 19,797,537–20,037,970 | 240,433 | |
| M35 | 14 | 21,441,167–22,046,172 | 605,005 |
| 17 | 41,526,491–42,128,942 | 602,451 | |
| 22 | 15,567,273–26,691,944 | 11,124,671 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapata-García, J.A.; Riveros-Magaña, A.R.; Ortiz-Lazareno, P.C.; Hernández-Flores, G.; Jave-Suárez, L.F.; Aguilar-Lemarroy, A. Comparative Genomic Hybridization and Transcriptome Sequencing Reveal Genes with Gain in Acute Lymphoblastic Leukemia: JUP Expression Emerges as a Survival-Related Gene. Diagnostics 2022, 12, 2788. https://doi.org/10.3390/diagnostics12112788
Zapata-García JA, Riveros-Magaña AR, Ortiz-Lazareno PC, Hernández-Flores G, Jave-Suárez LF, Aguilar-Lemarroy A. Comparative Genomic Hybridization and Transcriptome Sequencing Reveal Genes with Gain in Acute Lymphoblastic Leukemia: JUP Expression Emerges as a Survival-Related Gene. Diagnostics. 2022; 12(11):2788. https://doi.org/10.3390/diagnostics12112788
Chicago/Turabian StyleZapata-García, Jessica Alejandra, Alma Rocío Riveros-Magaña, Pablo Cesar Ortiz-Lazareno, Georgina Hernández-Flores, Luis Felipe Jave-Suárez, and Adriana Aguilar-Lemarroy. 2022. "Comparative Genomic Hybridization and Transcriptome Sequencing Reveal Genes with Gain in Acute Lymphoblastic Leukemia: JUP Expression Emerges as a Survival-Related Gene" Diagnostics 12, no. 11: 2788. https://doi.org/10.3390/diagnostics12112788
APA StyleZapata-García, J. A., Riveros-Magaña, A. R., Ortiz-Lazareno, P. C., Hernández-Flores, G., Jave-Suárez, L. F., & Aguilar-Lemarroy, A. (2022). Comparative Genomic Hybridization and Transcriptome Sequencing Reveal Genes with Gain in Acute Lymphoblastic Leukemia: JUP Expression Emerges as a Survival-Related Gene. Diagnostics, 12(11), 2788. https://doi.org/10.3390/diagnostics12112788










