Abstract
Breast cancer is one of the leading causes of cancer-related deaths in women worldwide, and its incidence is on the rise. A small fraction of cancer stem cells was identified within the tumour bulk, which are regarded as cancer-initiating cells, possess self-renewal and propagation potential, and a key driver for tumour heterogeneity and disease progression. Cancer heterogeneity reduces the overall efficacy of chemotherapy and contributes to treatment failure and relapse. The cell-surface and subcellular biomarkers related to breast cancer stem cell (BCSC) phenotypes are increasingly being recognised. These biomarkers are useful for the isolation of BCSCs and can serve as potential therapeutic targets and prognostic tools to monitor treatment responses. Recently, the role of noncoding microRNAs (miRNAs) has extensively been explored as novel biomarker molecules for breast cancer diagnosis and prognosis with high specificity and sensitivity. An in-depth understanding of the biological roles of miRNA in breast carcinogenesis provides insights into the pathways of cancer development and its utility for disease prognostication. This review gives an overview of stem cells, highlights the biomarkers expressed in BCSCs and describes their potential role as prognostic indicators.
1. Introduction
Breast cancer is the most commonly diagnosed cancer in women in most parts of the world, with an increasing trend observed. In the United States, the number of new cases of breast cancer is higher than the combined number of lung cancer in both sexes [1]. In Malaysia, it is also the most common cancer among women. The Malaysian National Cancer Registry report in 2019 showed a total of 18,206 new cases between 2012 and 2016, and this accounted for 34.1% of all cancers. The number of cases and the cumulative risk has gradually increased over a decade, from 3.4 (2007) to 3.8 (2016). There was also an increase in the percentage of cases presented at the late stage of the disease between the years 2012 and 2016 (47.9%), as compared to between 2007 and 2011 (43.2%) [2,3].
2. Types of Stem Cells
2.1. Adult and Embryonic Stem Cells
The pluripotent embryonic stem cells are derived from blastocysts of developing embryos and have the ability to self-renew and differentiate into every type of mature cell and tissue in the body [4,5]. In contrast, adult stem cells are isolated from various organs such as skin, liver, lung and mammary gland. Unlike embryonic stem cells, they have limited differentiation capability and, therefore, are multipotent or unipotent. The progenitor cells will gradually lose their self-renewal ability as they differentiate to produce mature cells [6], with a finite lifespan. In general, stem cells are rare in most of the adult tissues and are difficult to isolate [7].
2.2. Cancer Stem Cells (CSCs)
CSCs are a subpopulation of cells within malignant tumours. These cells possess stem cell characteristics such as self-renewal, differentiation and the ability to recapitulate the parental tumour when transplanted into a host. They contribute to the growth of tumours, development of chemotherapy resistance and metastasis [8]. In other words, they are the key player of tumour progression and aggressiveness [9]. They express embryonic stem cell factors like sex determining region Y-box 2 (SOX2), octamer binding transcription factor 4 (Oct4), Nanog and DNA (cytosine-5)-methyltransferase 1 (Dnmt1) [10,11,12]. The percentage of these cells varies between 0.02% and 25% depending on the tumour type [13].
CSCs have been identified in various types of tumours, including breast, brain, colon, leukaemia and lung [10]. Undifferentiated tumours have the highest percentage of CSCs. Conventional cancer treatments are not able to target CSCs, as they are highly resistant, resulting in tumour metastasis and recurrence [9].
2.3. Breast Cancer Stem Cells (BCSCs)
Al-Hajj et al. (2003) was the first to isolate a subpopulation of cancer cells from human breast tumours based on the expression of the CD44+, CD24−/low and epithelial specific antigen (ESA)+ [14], which was described as BCSCs. Interestingly, these cells were able to form new tumours in immunodeficient mice. Subsequently, Ginestier et al. (2007) reported that high aldehyde dehydrogenase 1 (ALDH-1) expression is also a characteristic feature for BCSCs, and it can be used to replace ESA as a biomarker [15]. The current accepted markers for BCSCs are CD44+, CD24−/low and ALDH-1+ [16]. The aim of this review is to explore the values of biomarkers of breast cancer stem cells as prognostic factors.
3. Prognostic Value of BCSC-Related Biomarkers
3.1. Oct4
Oct4, an embryonic stem cell marker, is a transcription factor encoded by the POU Class 5 Homeobox 1 (POU5F1) gene. In breast cancer, it was reported to be associated with ALDH-1 positivity, high Ki-67 proliferative index, high histological tumour grade and an independent prognostic factor. This association was found specifically in the hormone receptor positive breast cancer [17]. Bhatt et al. (2016) reported a possible association between Oct4 and tamoxifen resistance [18]. Oct4 overexpression was found to be a poor prognostic factor in various other tumours such as renal cell carcinoma [19], prostate carcinoma [20], bladder carcinoma [21] and cervical carcinoma [22].
3.2. Nanog
Nanog, an embryonic stem cell marker, is also a biomarker for CSCs. A study showed that its expression was higher in invasive breast cancer, and it was directly associated with tumour size, tumour grade, tumour stage and lymph node metastasis and a poor overall survival [23].
3.3. CD133
Croker et al. (2009) found a subpopulation of cells expressing CD133 in human invasive breast cancer cell lines [24], together with the other CSC markers like CD44/CD24 and ALDH-1. The number of CD133+ cells was between 1% and 2% in both luminal and human epidermal growth factor receptor 2 (HER2)+ cells. Interestingly, in basal-like cell lines, it was up to 80% [25,26]. This suggested that CD133 may be a marker for more aggressive types of breast cancer.
CD133 messenger RNA (mRNA) and protein expression were also found to be directly correlated with an increasing tumour grade, positive lymph node metastasis, negative progesterone receptor (PR) and oestrogen receptor (ER), positive HER2 gene status, advanced tumour, nodes and metastases (TNM) stage and a poor overall survival [27,28]. In addition, CD133 expression in both the cytoplasm and membrane was associated with a shorter survival. A high membrane expression of CD133 was seen in younger ages at the diagnosis of breast cancer [29]. Brugnoli et al. (2013) reported that, by silencing of CD133, it reduced the invasiveness of triple-negative breast cancer (TNBC) [30]. However, some studies described a negative correlation between CD133 expression and poor prognosis. They found an inverse relationship between CD133 levels with the clinical stage of TNBC tumours [31]. Cantile et al. (2013) suggested that a poor prognosis in TNBC is possibly due to a nuclear mislocalisation of CD133, which normally showed membrane positivity [32]. Collina et al. (2015) also reported no statistical association of CD133 expression with TNBC patient survivals [33]. Therefore, the value of CD133 as a prognostic factor is still questionable.
3.4. CD44
In tumours, the epithelial–mesenchymal transition (EMT) is an important step in disease progression. EMT is an embryonic program that is reactivated in tumour cells. It gives additional properties to the tumour cells, such as the ability to invade adjacent tissues and to disseminate under the influence of various cytokines produced by the surrounding stroma cells [34]. The BCSCs can exchange between the epithelial-mesenchymal phenotype and the mesenchymal-epithelial phenotype [35]. ALDH-1 and CD44 are the main markers associated with circulating CSCs, and their detection is able to identify higher metastatic potential [8,36,37].
CD44 is a transmembrane glycoprotein that binds to many extracellular matrix proteins. CD44 mRNA and protein overexpression were observed in the basal subtype of breast cancer. Patients with breast cancer expressing high levels of CD44 have significantly worse overall survival [38]. Although many studies have found that CD44 was associated with a poorer prognosis, the expression status of whether the association is related to a high level or loss of CD44 expression is still questionable. For example, a high CD44 expression was a poor prognostic factor for breast carcinoma [39], lymphoma [40] and thymoma [41], while, in other studies, a loss/reduced CD44 expression was correlated to a poorer prognosis, such as carcinoid tumour [42], gastrointestinal stroma tumour [43], laryngeal carcinoma [44] and lung carcinoma [45]. On the contrary, some studies found that a high level of CD44 was associated with a better prognosis in ovarian cancer and neuroblastoma [46,47].
3.5. ALDH-1
ALDH is a family of cytosolic enzymes that oxidises intracellular aldehydes and retinol during the differentiation of rudimentary stem cells [48]. ALDH-1 expression is increased in normal tissue and in many malignancies [49]. Studies on ALDH-1 as an independent prognostic marker in TNBC showed contradicting results. Ma et al. (2017) reported a significant correlation between ALDH-1 expression with tumour size, tumour stage and overall survival [50]. However, Panigoro et al. (2020) found that ALDH-1 can be used as a poor prognostic marker, but it is not an independent prognostic marker [51].
3.6. Autophagy-Related Genes (Autophagy Protein Light Chain 3 (LC3) and Beclin-1)
Chang et al. (2016) reported that the LC3−/CD44+/CD24− immunophenotype indicated a highly aggressive subgroup of TNBC, which was associated with poor prognosis [52]. LC3-negative TNBC has a significant negative association with overall survival [52]. Harmurcu et al. (2018) showed the knockdown of LC3 and Beclin-1 in MDA-MB-231 and BT-549 TNBC cells, resulting in the inhibition of various proto-oncogenic signalling pathways, such as cyclin D1, urokinase receptor (uPAR)/integrin-β1/ SRC proto-oncogene, nonreceptor tyrosine kinase (Src) and poly (ADP-ribose) polymerase 1 (PARP1) [53]. It suggested that LC3 and Beclin-1 are required for cell proliferation, survival, migration and invasion and may contribute to the tumour growth and progression of highly aggressive and metastatic TNBC tumours.
3.7. Zinc Finger E-box-Binding Homeobox 1 (ZEB1)
ZEB1 is an EMT inducer that downregulates E-cadherin and induces the epithelial to mesenchymal transition in breast and other carcinomas [54,55]. ZEB1 and the loss of E-cadherin were more commonly observed in metaplastic carcinomas than in other subtypes of breast cancer. ZEB1 expression was identified to be an independent poor prognostic factor for disease-free survival [54].
3.8. Other Potential Prognostic Markers
El Abbass et al. (2020) evaluated the BCSC marker expressions and the number of mammospheres in cultures of breast cancer tissues and correlated them with patients’ overall survival [56]. The study found that phosphatase and tensin homologue (PTEN), phosphoinositide 3-kinase (PI3K), protein kinase B (AKT), wingless-related integration site (Wnt) and β-catenin may play important roles in the development and progression of breast cancer, and they can be used as potential prognostic biomarkers. Lei et al. (2016) performed transcriptome sequencing on BCSCs, breast cancer cells, mammary cells and CD44+ mammary cells and found that carbonic anhydrase 12 (CA12) was a prognostic biomarker in HER2-positive breast cancer [57].
4. miRNAs in Stem Cells and Cancer Stem Cells
miRNAs consist of about 18–22 nucleotides and play an important role in virtually all biological processes, including stem cell maintenance, differentiation and development. Using a high-throughput sequencing platform, miRNAs were found in embryonic stem cells, neural stem cells and mesenchymal stem cells [58,59]. The miR-302 cluster comprises of a polycistronic cluster that houses five precursors of miRNAs, i.e., miR-302b, miR302c, miR-302a, miR-302d and miR-367, and is the most well-characterised human embryonic stem cell-specific miRNA [58,60]. Studies have shown that miRNAs are involved in the regulation of breast cancer stem cells [61], pancreatic cancer stem cells [62], colorectal cancer stem cells [63], liver cancer stem cells [64] and cervical cancer stem cells [65].
miRNAs that were found to be downregulated in BCSCs are let-7 family, miR-200, miR-30 family, miR-128, miR-34c and miR-16, while those miRNAs that were upregulated consisted of miR-181 and miR-495 [66]. As BCSCs play an important role in the pathogenesis of aggressive phenotypes, miRNAs that affect the maintenance of these cells could be valuable prognostic markers. Table 1 provides a summary of miRNAs that are associated with the regulation of BCSCs.

Table 1.
MicroRNAs associated with the regulation of breast cancer stem cells.
5. Prognostic Value of BCSC-Related MiRNAs
5.1. miR-1
Liu et al. (2015) analysed the miRNA expression of 45 breast cancer tissues and serum samples using RT-PCR [81]. They found that the miR-1 expression was significantly lower in basal-like tumours compared to luminal A, luminal B and HER2+ tumours. In cases with distant metastasis, the miR-1 level was higher compared to cases without metastasis. They concluded that miR-1 expression was underregulated in BCSCs and inversely related to the aggressiveness of tumours. Zhang et al. (2019) reported a down-regulation of miR-1 in BCSC and it triggered mitophagy of cancer stem cells by binding to the LRPPRC protein and targeting mitochondrial inner membrane organizing system 1 (MINOS1) and glycerol-3-phosphate dehydrogenase 2 (GPD2) mRNAs [82].
5.2. Let-7 miRNA
Let-7 is undetectable in embryonic stem cells, and it became upregulated upon differentiation [58]. In human BCSCs, let-7 suppressed self-renewal and differentiation by targeting a Harvey rat sarcoma viral oncogene homologue (H-RAS) and high-mobility group AT-hook 2 (HMGA2) [83]. Studies found that the downregulation of let-7 was observed in the CSCs, such as colon cancer and Wilms tumours [84,85].
5.3. miR-9 and miR-221
In a study of 206 patients, MiR-9 and miR-221 were found to be associated with stemness features, higher metastatic potential and EMT activation, as well as independently associated with a poorer overall survival and disease-free survival [67].
5.4. miR-24
miR-24 was reported to enhance the stemness property of breast cancer cells by expressing antiapoptotic protein BCL21-interacting protein BIM (BIML), and its overexpression promoted mammosphere formation in the T-47D, MCF-7 and MDA-MB-231 cell lines. The overexpression of miR-24 also induced the upregulation of Oct3/4 and Nanog stem cell-related genes. Other studies reported miR-24 induced BCSC resistance against cisplatin [69,92].
5.5. miR-27a
Tang et al. (2014) demonstrated that BCSCs treated with vascular endothelial growth factor (VEGF) increased miR-27a and suggested that the interaction between VEGF and miR-27a promoted angiogenesis and metastasis [70]. miR-27a was upregulated in breast cancer and was correlated with tumour size, lymph node metastasis and distant metastasis. They suggested that a high level of miR-27a is associated with overall poor survival [71].
5.6. miR-125a
A study showed that increasing the expression of miR-125a led to an increase in BCSC populations in MCF12A cells. In sphere-forming assays, the addition of miR-125a enhanced the sphere-forming ability by about 1.5-fold compared to mock-treated control cells. In contrast, miR-125a inhibition resulted in a decreased stem cells population and a decrease in sphere formation. They also found miR-125a overexpressing MCF12A spheres promoted SOX2 expression, a stem cell marker [72].
5.7. miR-128
Breast cancer stem cells and breast cancer cells downregulated miR-128-3p. A study showed miR-128-3p overexpression inhibited the proliferation, migration, invasion, self-renewal and tumorigenicity of BCSCs via the downregulation of serine/threonine-protein kinase Nek2 (NEK2), a gene involved in cell division. The study suggested that miR-128-3p inhibits the stem-like cell features of BCSCs [86].
5.8. miR-142
Isobe et al. (2014) reported that the ectopic expression of miR-142 in normal mouse mammary stem cells led to the formation of hyperproliferative mammary glands [73]. By knockdown miR-142, it suppressed the organoid formation by BCSCs and reduced the tumour growth initiated by BCSCs. miR-142 regulated the properties of BCSC by, in part, activating the Wnt-signalling pathway and miR-150. The study suggested that miR-142 is frequently upregulated in BCSCs.
5.9. miR-200
Shimono et al. (2009) isolated BCSCs directly from breast cancer samples and identified a subset of miRNAs that were differentially expressed between BCSCs and nontumourigenic cancer cells [75]. These miRNAs are the miR-200 family, let-7, miR-1 and miR-27. The miR-200 family comprises five members that are located in two clusters: the first cluster is on human chromosome 1 (miR−200a, miR−200b and miR−429), and the second cluster is on human chromosome 12 (miR−200c and miR−141) [76]. The miR-200 family modulates the self-renewal ability of CSCs by targeting the B-lymphoma Mo-MLV insertion region 1 homologue (BMI-1) and SUZ12 polycomb repressive complex 2 subunit (SUZ12) [77]. BMI-1 regulates the self-renewal and differentiation of several types of stem cells [78].
5.10. miR-205
Zhang et al. (2020) reported that miR-205 expression was reduced in CD44+/CD24−/low BCSCs compared with non-BCSCs [87]. The overexpression of miR-205 in the MB-231 cell line led to a reduced CD44+/CD24−/low population. They suggested that miR-205 could inhibit breast cancer malignancy by regulating RUNX2, and miR-205 is a tumour suppressor during breast cancer development.
5.11. miR-210
Tang et al. (2018) reported that the upregulation of miR-210 induced by a hypoxic microenvironment promoted breast cancer stem cell metastasis, proliferation and self-renewal by targeting E-cadherin [79]. They also found that miR-210 was upregulated in culture MCF-7 spheroid cells, which were high in BCSCs compared with MCF-7 parental cells. A high miR-210 expression was also detected in CD44+/CD24− MCF-7 cells and human CD44+/CD24− breast cancer cells.
5.12. miR-495
MiR-495 expression was found to be upregulated in BCSCs. The overexpression of miR-495 in breast cancer cells increased the colony-forming capacity in vitro and tumorigenesis in mice. The study also reported that miR-495 suppressed E-cadherin expression to promote cell invasion, while it inhibited the regulation in development and DNA damage responses 1 (REDD1) expression in a hypoxia environment to enhance cell proliferation. They concluded that miR-495 contributed to BCSC properties [80].
5.13. miR-590-5p
Zhou et al. (2017) evaluated the expression of miR-590-5p by real-time polymerase chain reaction (RT-PCR) on 49 breast cancer patients and compared them with nontumorous tissue and found that miR-590-5p expression was reduced in breast cancer tissues [88]. The overexpression of miR-590-5p significantly decreased the ALDH1-positive cell population. They also reported that miR-590-5p significantly downregulated SOX2 protein expression and vice versa. They concluded that miR-590-5p inhibited breast cancer cell stemness through targeting SOX2.
5.14. miR-628
A transfection study using miRNA mimics showed miR-628 suppressed the migration and invasiveness of BCSCs of MDA-MB-231 and MCF-7 cells by targeting the Son of sevenless homologue 1 (SOS1), which attenuated snails and vimentin, while enhancing E-cadherin activity. They also found that miR-628 was downregulated in bone metastatic breast cancer cells [89].
5.15. miR-638
Lin et al. (2020) investigated the relationship between miR-638 (a tumour suppressor gene) and E2F transcription factor 2 (E2F2) in BCSCs [90]. In breast cancer, miR-638 was downregulated, while E2F2 was elevated. The overexpression of miR-638 resulted in a reduction of CD24−/CD44+ cells and the stem cell markers like SOX2 and Oct4. In addition, miR-638 overexpression inhibited the abilities of BSCSs to self-renew, proliferate and invade. Overall, miR-638 overexpression caused a reduction in tumour growth.
5.16. miR-760
Han et al. (2016) evaluated the expression of miR-760 in the BT-549 cell line and MCF-7 cell line [91]. They found that the BT-549 cell line had a lower expression of miR-760 and a high level of Nanog expression. The BT-549 cell line had a greater number of CD44+/CD24−/low subpopulations. The overexpression of miR-760 in the BT-549 cell line resulted in reduced CD44+/CD24−/low cells and inhibited cell proliferation and migration by downregulating Nanog.
5.17. Other miRNAs
Several studies reported the association of miRNAs with disease progression. miR-33b targeted HMGA2, SALL4 and Twist1 and inhibited the stemness, migration and invasion of metastatic breast cancer cells [74]. miR-199a promoted BCSC propagation, tumour initiation and metastasis by the suppression of FOXP2 [93]. miR-20a enhanced the metastatic abilities of BCSCs by conferring the resistance of BCSCs to natural killer (NK) cell cytotoxicity [68].
6. Conclusions
In summary, these miRNAs exert their effects by regulating three main pathways: (1) by maintaining the stemness properties of BCSCs via upregulating Oct4; SOX2 and Nanog expressions (miR-24, miR-125a, miR-590-5p and miR-760); (2) by regulating the proliferation and self-renewal of BCSCs via the cell cycle and cell division genes like E2F2; HRAS; NEK2 and RUNX2 (let-7, miR-128, miR-205 and miR-638) and (3) by suppressing E-cadherin, a cell-cell adhesion molecule, to promote proliferation; invasion and metastasis (miR-210, miR-495 and miR-628) (Figure 1). As miRNAs can be isolated in the serum as membrane-bound vesicles, the identification of circulating BCSC-related miRNAs could be used as a form of prognostication of breast cancer.
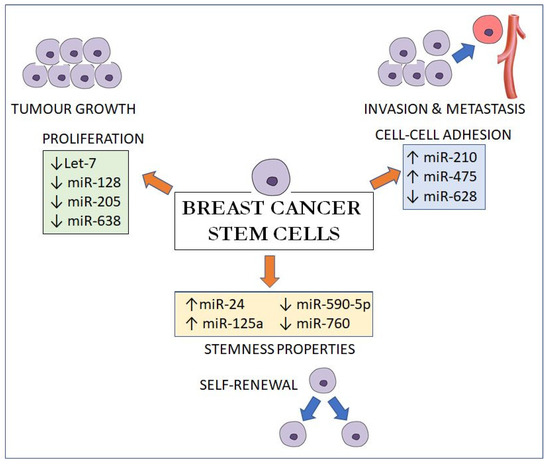
Figure 1.
MicroRNA effects on breast cancer stem cells on proliferation, cell-cell adhesion and stemness properties.
In the era of precision medicine, gene-targeting, immunomodulatory and cell-based therapies will be potential promising tools for breast cancer treatment. miRNA mimics and inhibitors could be used as therapeutic agents. Various studies have described the roles of miRNAs in the maintenance of BCSCs and the regulation of the stemness properties of these cells. The post-transcriptional regulation of miRNA is complex, as a single miRNA could target multiple mRNAs, while multiple miRNAs could target a single mRNA. Tumour growth and progression involve complex interactions between cancer cells, CSCs, inflammatory cells and fibroblasts within the tumour microenvironment. The development of miRNA-based therapy required intensive research to unravel the complex regulation of miRNAs in CSCs. Exploring the regulation of these miRNAs will further advance our knowledge of the roles of human BCSCs in tumour progression and their prognostic value in breast cancer.
Author Contributions
Writing—original draft preparation, G.C.T., Y.P.W., C.C.H.K. and W.K.C.; writing—review and editing, G.T.S. and Y.K.C.; project administration, C.C.H.K. and funding acquisition, G.T.S., Y.K.C. and G.C.T. The authors wish to acknowledge that the first two authors should be regarded as a joint first author. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Ministry of Higher Education of Malaysia through the Transdisciplinary Research Grant Scheme (TRGS/1/2014/UPM/02/6/3) and FF-2015-182.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviation
| AKT | Protein kinase B |
| ALDH1 | Aldehyde Dehydrogenase 1 |
| BMI-1 | B-lymphoma Mo-MLV insertion region 1 homolog |
| BIML | BCL21-interacting Protein BIM |
| BCSCs | Breast cancer stem cells |
| CSCs | Cancer stem cells |
| CA12 | Carbonic Anhydrase 12 |
| Dnmt1 | DNA (cytosine-5)-methyltransferase 1 |
| E2F2 | E2F Transcription Factor 2 |
| EMT | Epithelial–mesenchymal transition |
| ER | Oestrogen receptor |
| ESA | Epithelial specific antigen |
| FOXP2 | Forkhead box protein P2 |
| H-RAS | Harvey rat sarcoma viral oncogene homolog |
| HMGA2 | High-mobility group AT-hook 2 |
| HER2 | Human epidermal growth factor receptor 2 |
| MICA and MICB | MHC class I chain-related protein A and B |
| miRNAs | MicroRNAs |
| mRNA | Messenger RNA |
| NEK2 | Serine/threonine-protein kinase Nek2 |
| NK cells | Natural killer cells |
| Oct-4 | Octamer binding transcription factor 4 |
| PARP1 | Poly (ADP-ribose) polymerase 1 |
| PI3K | Phosphoinositide 3-kinase |
| POU5F1 | POU Class 5 Homeobox 1 |
| PR | Progesterone receptor |
| PTEN | Phosphatase and tensin homolog |
| REDD1 | Regulated in development and DNA damage responses 1 |
| RT-PCR | Real time polymerase chain reaction |
| RUNX2 | Runt-related transcription factor 2 |
| SALL4 | Sal-like protein 4 |
| SOS1 | Son of sevenless homolog 1 |
| SOX2 | SRY (sex determining region Y)-box 2 |
| Src | SRC Proto-Oncogene, Non-Receptor Tyrosine Kinase |
| SUZ12 | SUZ12 Polycomb Repressive Complex 2 Subunit |
| TNBC | Triple negative breast cancer |
| TNM stage | tumour (T), nodes (N), and metastases (M) stage |
| Twist1 | Twist-related protein 1 |
| uPAR | Urokinase receptor |
| VEGF | Vascular endothelial growth factor |
| Wnt | Wingless-related integration site |
| ZEB1 | Zinc Finger E-Box Binding Homeobox 1 |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA. Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Azizah, A.M.; Nor Saleha, I.T.; Noor Hashimah, A.; Asmah, Z.A.; Mastulu, W. Malaysian National Cancer Registry Report 2007–2011; National Cancer Institute, Ministry of Health: Putrajaya, Malaysia, 2016. Available online: https://www.crc.gov.my/wp-content/uploads/documents/report/MNCRRrepor2007-2011.pdf (accessed on 19 September 2020).
- Azizah, A.M.; Hashimah, B.; Nirmal, K.; Siti Zubaidah, A.R.; Puteri, N.A.; Nabihah, A.; Sukumaran, R.; Balqis, B.; Nadia, S.M.R.; Sharifah, S.S.S.; et al. National Cancer Registry Report 2012–2016; National Cancer Registry Department, National Cancer Institute: Putrajaya, Malaysia, 2019; Available online: https://drive.google.com/file/d/1BuPWrb05N2Jez6sEP8VM5r6JtJtlPN5W/view (accessed on 19 September 2020).
- Donovan, P.J.; Gearhart, J. The end of the beginning for pluripotent stem cells. Nature 2001, 414, 92–97. [Google Scholar] [CrossRef]
- Hall, P.A.; Watt, F.M. Stem cells: The generation and maintenance of cellular diversity. Development 1989, 106, 619–633. [Google Scholar] [PubMed]
- Spillane, J.B.; Henderson, M.A. Cancer stem cells: A review. ANZ. J. Surg. 2007, 77, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Scioli, M.G.; Storti, G.; D’Amico, F.; Gentile, P.; Fabbri, G.; Cervelli, V.; Orlandi, A. The role of breast cancer stem cells as a prognostic marker and a target to improve the efficacy of breast cancer therapy. Cancers 2019, 11, 1021. [Google Scholar] [CrossRef]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef]
- Desai, A.; Yan, Y.; Gerson, S.L. Concise reviews: Cancer stem cell targeted therapies: Toward clinical success. Stem Cells Transl. Med. 2019, 8, 75–81. [Google Scholar] [CrossRef]
- Dragu, D.L.; Necula, L.G.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Therapies targeting cancer stem cells: Current trends and future challenges. World J. Stem Cells 2015, 7, 1185–1201. [Google Scholar]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Toledo-Guzmán, M.E.; Bigoni-Ordóñez, G.D.; Ibáñez Hernández, M.; Ortiz-Sánchez, E. Cancer stem cell impact on clinical oncology. World J. Stem Cells. 2018, 10, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitão, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J Clin. Pathol. 2011, 64, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Gwak, J.M.; Kim, M.; Kim, H.J.; Jang, M.H.; Park, S.Y. Expression of embryonal stem cell transcription factors in breast cancer: Oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget 2017, 8, 36305–36318. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Stender, J.D.; Joshi, S.; Wu, G.; Katzenellenbogen, B.S. OCT-4: A novel estrogen receptor-α collaborator that promotes tamoxifen resistance in breast cancer cells. Oncogene 2016, 35, 5722–5734. [Google Scholar] [CrossRef]
- Rasti, A.; Mehrazma, M.; Madjd, Z.; Abolhasani, M.; Zanjani, L.S.; Asgari, M. Co-expression of cancer stem cell markers OCT4 and NANOG predicts poor prognosis in renal cell carcinomas. Sci. Rep. 2018, 8, 11739. [Google Scholar] [CrossRef]
- Kosaka, T.; Mikami, S.; Yoshimine, S.; Miyazaki, Y.; Daimon, T.; Kikuchi, E.; Miyajima, A.; Oya, M. The prognostic significance of OCT4 expression in patients with prostate cancer. Hum. Pathol. 2016, 51, 1–8. [Google Scholar] [CrossRef]
- Huang, P.; Chen, J.; Wang, L.; Na, Y.; Kaku, H.; Ueki, H.; Sasaki, K.; Yamaguchi, K.; Zhang, K.; Saika, T.; et al. Implications of transcriptional factor, OCT-4, in human bladder malignancy and tumor recurrence. Med. Oncol. 2012, 29, 829–834. [Google Scholar] [CrossRef]
- Kim, B.W.; Cho, H.; Choi, C.H.; Ylaya, K.; Chung, J.Y.; Kim, J.H.; Hewitt, S.M. Clinical significance of OCT4 and SOX2 protein expression in cervical cancer. BMC Cancer 2015, 15, 1015. [Google Scholar] [CrossRef]
- Zhai, Y.; Han, Y.; Han, Z. Aberrant expression of WWOX and its association with cancer stem cell biomarker expression. Int. J. Clin. Exp. Pathol. 2020, 13, 1176–1184. [Google Scholar] [PubMed]
- Croker, A.K.; Goodale, D.; Chu, J.; Postenka, C.; Hedley, B.D.; Hess, D.A.; Allan, A.L. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J. Cell Mol. Med. 2009, 13(8B), 2236–2252. [Google Scholar] [CrossRef] [PubMed]
- Borgna, S.; Armellin, M.; di Gennaro, A.; Maestro, R.; Santarosa, M. Mesenchymal traits are selected along with stem features in breast cancer cells grown as mammospheres. Cell Cycle 2012, 11, 4242–4251. [Google Scholar] [CrossRef] [PubMed]
- Brugnoli, F.; Grassilli, S.; Al-Qassab, Y.; Capitani, S.; Bertagnolo, V. CD133 in breast cancer cells: More than a stem cell marker. J. Oncol. 2019, 2019, 7512632. [Google Scholar] [CrossRef] [PubMed]
- Currie, M.J.; Beardsley, B.E.; Harris, G.C.; Gunningham, S.P.; Dachs, G.U.; Dijkstra, B.; Morrin, H.R.; Wells, J.E.; Robinson, B.A. Immunohistochemical analysis of cancer stem cell markers in invasive breast carcinoma and associated ductal carcinoma in situ: Relationships with markers of tumor hypoxia and microvascularity. Hum. Pathol. 2013, 44, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Xia, P. CD133 mRNA may be a suitable prognostic marker for human breast cancer. Stem Cell Investig. 2017, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Joseph, C.; Arshad, M.; Kurozomi, S.; Althobiti, M.; Miligy, I.M.; Al-Izzi, S.; Toss, M.S.; Goh, F.Q.; Johnston, S.J.; Martin, S.G.; et al. Overexpression of the cancer stem cell marker CD133 confers a poor prognosis in invasive breast cancer. Breast Cancer Res. Treat 2019, 174, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Brugnoli, F.; Grassilli, S.; Piazzi, M.; Palomba, M.; Nika, E.; Bavelloni, A.; Capitani, S.; Bertagnolo, V. In triple negative breast tumor cells, PLC-β2 promotes the conversion of CD133high to CD133low phenotype and reduces the CD133-related invasiveness. Mol. Cancer 2013, 12, 165. [Google Scholar] [CrossRef]
- Zhao, P.; Lu, Y.; Jiang, X.; Li, X. Clinicopathological significance and prognostic value of CD133 expression in triple-negative breast carcinoma. Cancer Sci. 2011, 102, 1107–1111. [Google Scholar] [CrossRef]
- Cantile, M.; Collina, F.; D’Aiuto, M.; Rinaldo, M.; Pirozzi, G.; Borsellino, C.; Franco, R.; Botti, G.; Di Bonito, M. Nuclear localization of cancer stem cell marker CD133 in triple-negative breast cancer: A case report. Tumori 2013, 99, e245–e250. [Google Scholar] [CrossRef]
- Collina, F.; Di Bonito, M.; Li Bergolis, V.; De Laurentiis, M.; Vitagliano, C.; Cerrone, M.; Nuzzo, F.; Cantile, M.; Botti, G. Prognostic value of cancer stem cells markers in triple-negative breast cancer. Biomed Res. Int. 2015, 2015, 158682. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. EMT: When epithelial cells decide to become mesenchymal-like cells. J. Clin. Invest. 2009, 119, 1417–1419. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2013, 2, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Ginestier, C.; Bertucci, F.; Cabaud, O.; Wicinski, J.; Finetti, P.; Josselin, E.; Adelaide, J.; Nguyen, T.T.; Monville, F.; et al. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res. 2013, 73, 7290–7300. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Liu, Y.; Wu, H.; Liu, Q.; Wu, G.S.; Wu, K. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. Onco. Targets Ther. 2016, 9, 431–444. [Google Scholar]
- Watanabe, O.; Kinoshita, J.; Shimizu, T.; Imamura, H.; Hirano, A.; Okabe, T.; Aiba, M.; Ogawa, K. Expression of a CD44 variant and VEGF-C and the implications for lymphatic metastasis and long-term prognosis of human breast cancer. J. Exp. Clin Cancer Res. 2005, 24, 75–82. [Google Scholar]
- Niitsu, N.; Iijima, K. High serum soluble CD44 is correlated with a poor outcome of aggressive non-Hodgkin’s lymphoma. Leuk. Res. 2002, 26, 241–248. [Google Scholar] [CrossRef]
- Lee, S.C.; Harn, H.J.; Lin, T.S.; Yeh, K.T.; Liu, Y.C.; Tsai, C.S.; Cheng, Y.L. Prognostic significance of CD44v5 expression in human thymic epithelial neoplasms. Ann. Thorac. Surg. 2003, 76, 213–218. [Google Scholar] [CrossRef]
- Sun, X.; Gong, Y.; Talamonti, M.S.; Rao, M.S. Expression of cell adhesion molecules, CD44s and E-cadherin, and microvessel density in carcinoid tumors. Mod. Pathol. 2002, 15, 1333–1338. [Google Scholar] [CrossRef][Green Version]
- Montgomery, E.; Abraham, S.C.; Fisher, C.; Deasel, M.R.; Amr, S.S.; Sheikh, S.S.; House, M.; Lilliemoe, K.; Choti, M.; Brock, M.; et al. CD44 loss in gastric stromal tumors as a prognostic marker. Am. J. Surg. Pathol. 2004, 28, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Esteban, F.; Bravo, J.J.; González-Moles, M.A.; Bravo, M.; Ruiz-Avila, I.; Gil-Montoya, J.A. Adhesion molecule CD44 as a prognostic factor in laryngeal cancer. Anticancer Res. 2005, 25, 1115–1121. [Google Scholar]
- Pirinen, R.; Hirvikoski, P.; Böhm, J.; Moisio, K.; Viren, M.; Johansson, R.; Hollmen, S.; Kosma, V.M. Reduced expression of CD44v3 variant isoform is associated with unfavorable outcome in non-small cell lung carcinoma. Hum. Pathol. 2000, 31, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Combaret, V.; Gross, N.; Lasset, C.; Frappaz, D.; Peruisseau, G.; Philip, T.; Beck, D.; Favrot, M.C. Clinical relevance of CD44 cell-surface expression and N-myc gene amplification in a multicentric analysis of 121 pediatric neuroblastomas. J. Clin. Oncol. 1996, 14, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Sillanpää, S.; Anttila, M.A.; Voutilainen, K.; Tammi, R.H.; Tammi, M.I.; Saarikoski, S.V.; Kosma, V.M. CD44 expression indicates favorable prognosis in epithelial ovarian cancer. Clin. Cancer Res. 2003, 9, 5318–5324. [Google Scholar] [PubMed]
- Balicki, D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell 2007, 1, 485–487. [Google Scholar] [CrossRef]
- Shima, H.; Yamada, A.; Ishikawa, T.; Endo, I. Are breast cancer stem cells the key to resolving clinical issues in breast cancer therapy? Gland Surg. 2017, 6, 82–88. [Google Scholar] [CrossRef]
- Ma, F.; Li, H.; Li, Y.; Ding, X.; Wang, H.; Fan, Y.; Lin, C.; Qian, H.; Xu, B. Aldehyde dehydrogenase 1 (ALDH1) expression is an independent prognostic factor in triple negative breast cancer (TNBC). Med. 2017, 96, e6561. [Google Scholar] [CrossRef]
- Panigoro, S.S.; Kurnia, D.; Kurnia, A.; Haryono, S.J.; Albar, Z.A. ALDH1 cancer stem cell marker as a prognostic factor in triple-negative breast cancer. Int. J. Surg. Oncol. 2020, 2020, 7863243. [Google Scholar] [CrossRef]
- Chang, S.J.; Ou-Yang, F.; Tu, H.P.; Lin, C.H.; Huang, S.H.; Kostoro, J.; Hou, M.F.; Chai, C.Y.; Kwan, A.L. Decreased expression of autophagy protein LC3 and stemness (CD44+/CD24-/low) indicate poor prognosis in triple-negative breast cancer. Hum. Pathol. 2016, 48, 48–55. [Google Scholar] [CrossRef]
- Hamurcu, Z.; Delibaşı, N.; Geçene, S.; Sener, E.F.; Dönmez-Altuntaş, H.; Özkul, Y.; Canatan, H.; Ozpolat, B. Targeting LC3 and Beclin-1 autophagy genes suppresses proliferation, survival, migration and invasion by inhibition of Cyclin-D1 and uPAR/Integrin β1/ Src signaling in triple negative breast cancer cells. J. Cancer Res. Clin. Oncol. 2018, 144, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.H.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Park, S.Y. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 2015, 46, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Eger, A.; Aigner, K.; Sonderegger, S.; Dampier, B.; Oehler, S.; Schreiber, M.; Berx, G.; Cano, A.; Beug, H.; Foisner, R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005, 24, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- El Abbass, K.A.; Abdellateif, M.S.; Gawish, A.M.; Zekri, A.N.; Malash, I.; Bahnassy, A.A. The role of breast cancer stem cells and some related molecular biomarkers in metastatic and nonmetastatic breast cancer. Clin. Breast Cancer 2020, S1526-820930756-6. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Zhang, X.Y.; Zhou, J.P.; Mu, G.N.; Li, Y.W.; Zhang, Y.X.; Pang, D. Transcriptome sequencing of HER2-positive breast cancer stem cells identifies potential prognostic marker. Tumour Biol. 2016, 37, 14757–14764. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.C.; Chan, E.; Molnar, A.; Sarkar, R.; Alexieva, D.; Isa, I.M.; Robinson, S.; Zhang, S.; Ellis, P.; Langford, C.F.; et al. 5’ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014, 42, 9424–9435. [Google Scholar] [CrossRef]
- Tan, G.C.; Dibb, N. IsomiRs have functional importance. Malays. J. Pathol. 2015, 37, 73–81. [Google Scholar]
- Tan, G.C.; Wong, Y.P.; Cui, W.; Dibb, N. Construction of a doxycycline inducible lentivirus that expresses stem cell-specific miR-302 cluster. Malays. J. Pathol. 2020, 42, 91–97. [Google Scholar]
- Shimono, Y.; Mukohyama, J.; Nakamura, S.; Minami, H. MicroRNA regulation of human breast cancer stem cells. J. Clin. Med. 2015, 5, 2. [Google Scholar] [CrossRef]
- Xu, Y.F.; Hannafon, B.N.; Ding, W.Q. microRNA regulation of human pancreatic cancer stem cells. Stem Cell Investig. 2017, 4, 5. [Google Scholar] [CrossRef]
- Mukohyama, J.; Isobe, T.; Hu, Q.; Hayashi, T.; Watanabe, T.; Maeda, M.; Yanagi, H.; Qian, X.; Yamashita, K.; Minami, H.; et al. miR-221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res. 2019, 79, 5151–5158. [Google Scholar] [CrossRef] [PubMed]
- Hao, P.; Yue, F.; Xian, X.; Ren, Q.; Cui, H.; Wang, Y. Inhibiting effect of MicroRNA-3619-5p/PSMD10 axis on liver cancer cell growth in vivo and in vitro. Life Sci. 2020, 254, 117632. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.O.N.; Deng, W.; Ye, T.M.; Ngan, H.Y.S.; Tsao, S.W.; Cheung, A.N.Y.; Ziru, N.; Yuen, D.C.K.; Pang, R.T.K.; Yeung, W.S.B. MicroRNA-135a-induced formation of CD133+ subpopulation with cancer stem cell properties in cervical cancer. Carcinogenesis 2020, bgaa025. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbacher, D.; Balic, M.; Pichler, M. The role of microRNAs in breast cancer stem cells. Int. J. Mol. Sci. 2013, 14, 14712–14723. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.W.; Yu, J.C.; Hsieh, Y.H.; Liao, W.L.; Shieh, J.C.; Yao, C.C.; Lee, H.J.; Chen, P.M.; Wu, P.E.; Shen, C.Y. Increased cellular levels of microRNA-9 and microRNA-221 correlate with cancer stemness and predict poor outcome in human breast cancer. Cell Physiol. Biochem. 2018, 48, 2205–2218. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Q.; Wang, Z.; Jiang, J.; Yu, S.C.; Ping, Y.F.; Yang, J.; Xu, S.L.; Ye, X.Z.; Xu, C.; et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014, 74, 5746–5757. [Google Scholar] [CrossRef]
- Roscigno, G.; Puoti, I.; Giordano, I.; Donnarumma, E.; Russo, V.; Affinito, A.; Adamo, A.; Quintavalle, C.; Todaro, M.; Vivanco, M.D.; et al. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget 2017, 8, 19507–19521. [Google Scholar] [CrossRef]
- Tang, W.; Yu, F.; Yao, H.; Cui, X.; Jiao, Y.; Lin, L.; Chen, J.; Yin, D.; Song, E.; Liu, Q. miR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene 2014, 33, 2629–2638. [Google Scholar] [CrossRef]
- Tang, W.; Zhu, J.; Su, S.; Wu, W.; Liu, Q.; Su, F.; Yu, F. MiR-27 as a prognostic marker for breast cancer progression and patient survival. PLoS ONE 2012, 7, e51702. [Google Scholar] [CrossRef]
- Nandy, S.B.; Arumugam, A.; Subramani, R.; Pedroza, D.; Hernandez, K.; Saltzstein, E.; Lakshmanaswamy, R. MicroRNA-125a influences breast cancer stem cells by targeting leukemia inhibitory factor receptor which regulates the Hippo signaling pathway. Oncotarget 2015, 6, 17366–17378. [Google Scholar] [CrossRef]
- Isobe, T.; Hisamori, S.; Hogan, D.J.; Zabala, M.; Hendrickson, D.G.; Dalerba, P.; Cai, S.; Scheeren, F.; Kuo, A.H.; Sikandar, S.S.; et al. miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. eLife 2014, 3, e01977. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, A.Y.; Fan, C.; Zheng, H.; Li, Y.; Zhang, C.; Wu, S.; Yu, D.; Huang, Z.; Liu, F.; et al. MicroRNA-33b Inhibits Breast Cancer Metastasis by Targeting HMGA2, SALL4 and Twist1. Sci Rep. 2015, 5, 9995. [Google Scholar] [CrossRef] [PubMed]
- Shimono, Y.; Ugalde, M.Z.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Down-regulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodal, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Lindahl-Allen, M.; Polytarchou, C.; Hirsch, H.A.; Tsichlis, P.N.; Struhl, K. Loss of miR-200 inhibition of SUZ12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef]
- Molofsky, A.V.; Pardal, R.; Iwashita, T.; Park, I.K.; Clarke, M.F.; Morrison, S.J. BMI-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 2003, 425, 962–967. [Google Scholar] [CrossRef]
- Tang, T.; Yang, Z.; Zhu, Q.; Wu, Y.; Sun, K.; Alahdal, M.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. Up-regulation of miR-210 induced by a hypoxic microenvironment promotes breast cancer stem cells metastasis, proliferation, and self-renewal by targeting E-cadherin. FASEB J. 2018, fj201801013R. [Google Scholar] [CrossRef]
- Hwang-Verslues, W.W.; Chang, P.H.; Wei, P.C.; Yang, C.Y.; Huang, C.K.; Kuo, W.H.; Shew, J.Y.; Chang, K.J.; Lee, Y.H.P.; Lee, W.H. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene 2011, 30, 2463–2474. [Google Scholar] [CrossRef]
- Liu, T.; Hu, K.; Zhao, Z.; Chen, G.; Ou, X.; Zhang, H.; Zhang, X.; Wei, X.; Wang, D.; Cui, M.; et al. MicroRNA-1 down-regulates proliferation and migration of breast cancer stem cells by inhibiting the Wnt/β-catenin pathway. Oncotarget 2015, 6, 41638–41649. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, C.; Zhang, X. Mitochondrial Damage Mediated by miR-1 Overexpression in Cancer Stem Cells. Mol. Ther. Nucleic Acids 2019, 18, 938–953. [Google Scholar] [CrossRef]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Pode-Shakked, N.; Shukrun, R.; Mark-Danieli, M.; Tsvetkov, P.; Bahar, S.; Pri-Chen, S.; Goldstein, R.S.; Rom-Gross, E.; Mor, Y.; Fridman, E.; et al. The isolation and characterization of renal cancer initiating cells from human Wilms’ tumour xenografts unveils new therapeutic targets. EMBO Mol. Med. 2013, 5, 18–37. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.E. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle. 2009, 8, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, N.; Liu, L.; Dong, H.; Liu, X. microRNA-128-3p overexpression inhibits breast cancer stem cell characteristics through suppression of Wnt signalling pathway by down-regulating NEK2. J. Cell Mol. Med. 2020, 24, 7353–7369. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, L.; Xu, H.; Wang, G.; Fan, C.; Zheng, Q.; Li, F. miR-205/RunX2 axis negatively regulates CD44+/CD24− breast cancer stem cell activity. Am. J. Cancer Res. 2020, 10, 1871–1887. [Google Scholar]
- Zhou, L.; Zhao, L.C.; Jiang, N.; Wang, X.L.; Zhou, X.N.; Luo, X.L.; Ren, J. MicroRNA miR-590-5p inhibits breast cancer cell stemness and metastasis by targeting SOX2SOX2. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 87–94. [Google Scholar]
- Lin, C.; Gao, B.; Yan, X.; Lei, Z.; Chen, K.; Li, Y.; Zeng, Q.; Chen, Z.; Li, H. MicroRNA 628 suppresses migration and invasion of breast cancer stem cells through targeting SOS1. Onco. Targets Ther. 2018, 11, 5419–5428. [Google Scholar] [CrossRef]
- Lin, Q.Y.; Wang, J.Q.; Wu, L.L.; Zheng, W.E.; Chen, P.R. miR-638 represses the stem cell characteristics of breast cancer cells by targeting E2F2. Breast Cancer. 2020, 27, 147–158. [Google Scholar] [CrossRef]
- Han, M.L.; Wang, F.; Gu, Y.T.; Pei, X.H.; Guo, G.C.; Li, L.; Duan, X.; Zhu, M.Z.; Wang, Y.M. MicroR-760 suppresses cancer stem cell subpopulation and breast cancer cell proliferation and metastasis: By down-regulating NANOG. Biomed. Pharmacother. 2016, 80, 304–310. [Google Scholar] [CrossRef]
- Das, P.K.; Siddika, M.A.; Asha, S.Y.; Aktar, S.; Rakib, M.A.; Khanam, J.A.; Pillai, S.; Islam, F. MicroRNAs, a promising target for breast cancer stem cells. Mol. Diagn. Ther. 2020, 24, 69–83. [Google Scholar] [CrossRef]
- Cuiffo, B.G.; Campagne, A.; Bell, G.W.; Lembo, A.; Orso, F.; Lien, C.E.; Bhasin, M.K.; Raimo, M.; Hanson, S.E.; Marusyk, A.; et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell. 2014, 15, 762–774. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).