Drosophila O-GlcNAcase Mutants Reveal an Expanded Glycoproteome and Novel Growth and Longevity Phenotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Stocks
2.2. Longevity Assay
2.3. Wing Size Measurement
2.4. Polytene Chromosome Staining and Imaging
2.5. Mass Spectrometry
3. Results
3.1. Adult O-GlcNAc Proteome Revealed Growth and Development Related Proteins
3.2. ogadel.1 Flies Have Larger Body Size Than Wild Type Flies
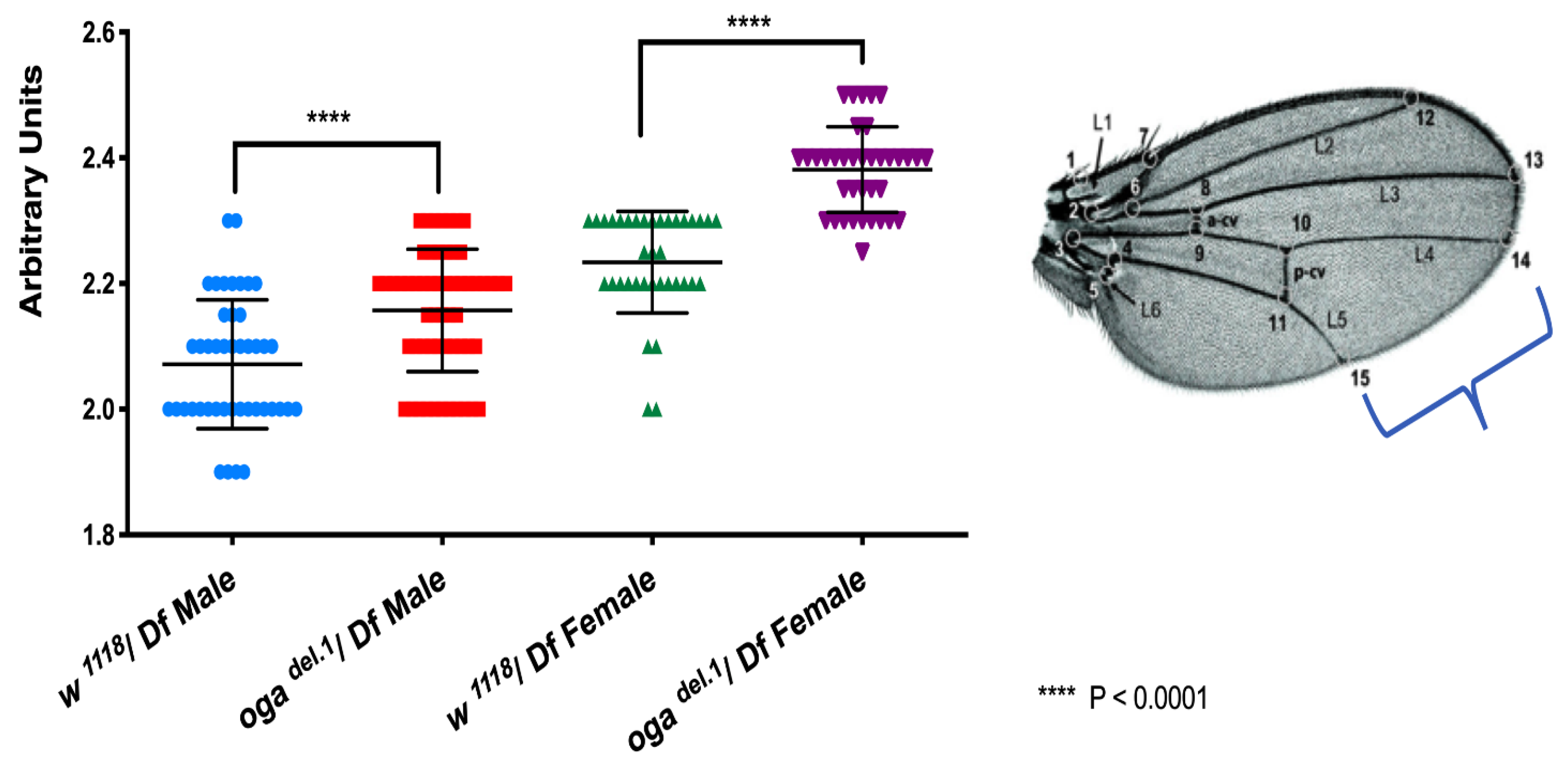
3.3. O-GlcNAc Levels Are Important for Imaginal Wing Disc
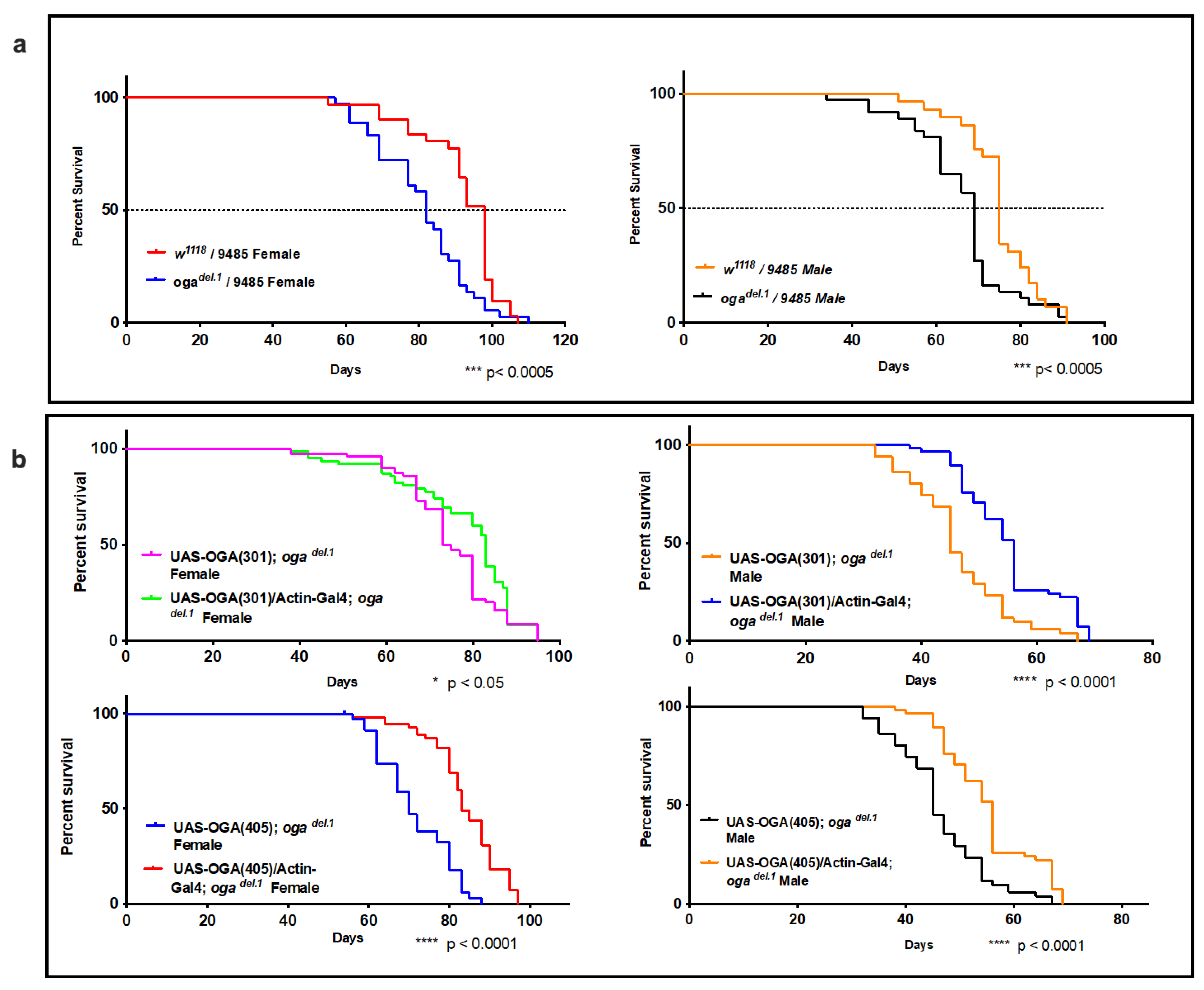
3.4. ogadel.1 Flies Display Shorter Life Span Compared to Wild Type Flies
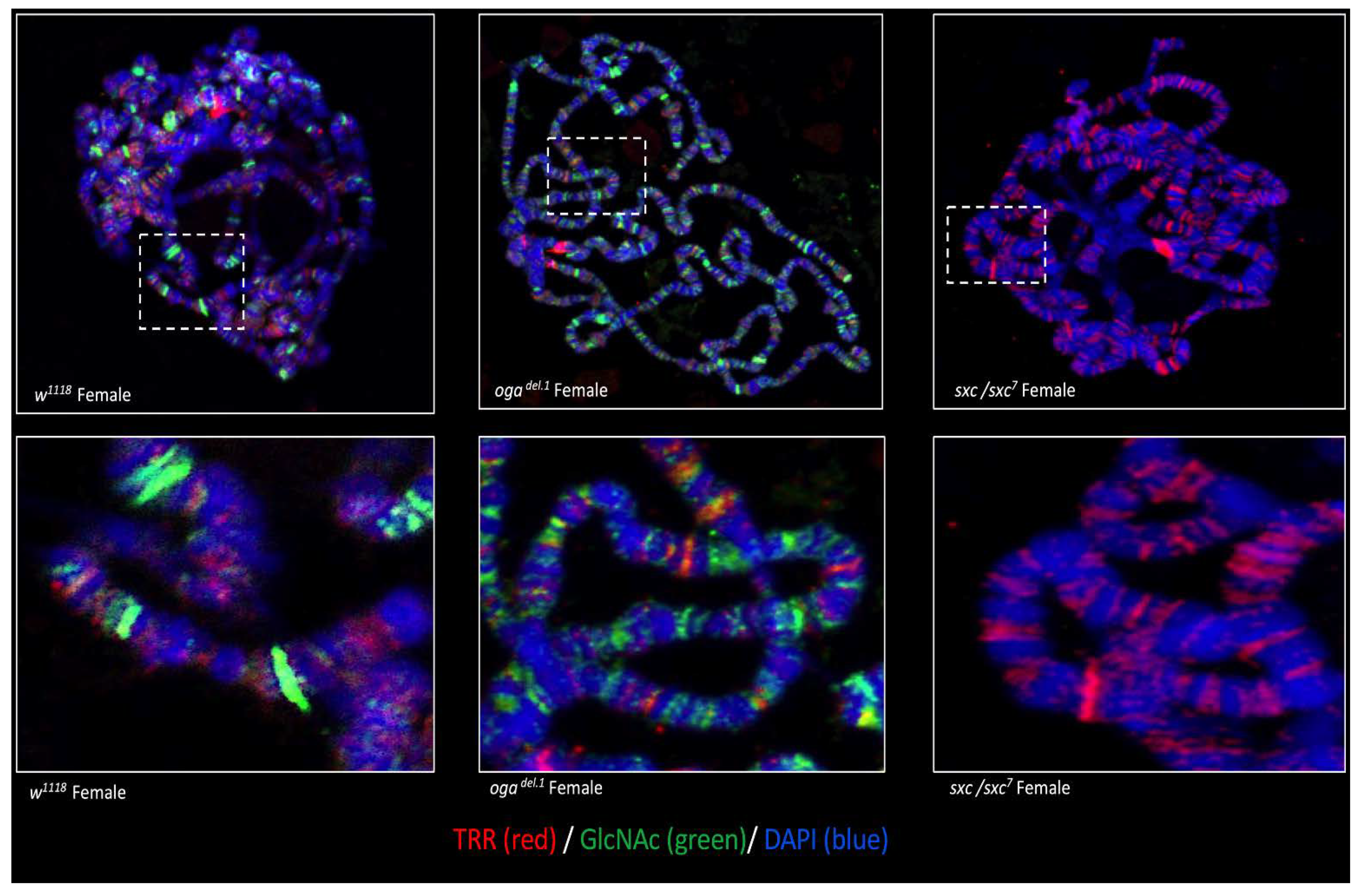
3.5. TRR Is Co-Stained with O-GlcNAc on Polytene Chromosomes
4. Discussion
4.1. Oga Mutant Flies Facilitates Glycoproteomics
4.2. Glycoproteomics Reveals Novel O-GlcNAc Modified Proteins Related to Growth and Longevity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell. Biol. 2015, 208, 869–880. [Google Scholar] [CrossRef]
- Jang, I.; Kim, H.B.; Seo, H.; Kim, J.Y.; Choi, H.; Yoo, J.S.; Kim, J.; Cho, J.W. O-GlcNAcylation of eIF2alpha regulates the phospho-eIF2alpha-mediated ER stress response. Biochim. Biophys. Acta 2015, 1853, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Liu, F.; Tao, T.; Zhang, D.; Liu, X.; Zhu, G.; Xu, Z.; Ni, R.; Shen, A. Modification of p27 with O-linked N-acetylglucosamine regulates cell proliferation in hepatocellular carcinoma. Mol. Carcinog. 2017, 56, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Udeshi, N.D.; Slawson, C.; Compton, P.D.; Sakabe, K.; Cheung, W.D.; Shabanowitz, J.; Hunt, D.F.; Hart, G.W. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010, 3, ra2. [Google Scholar] [CrossRef] [PubMed]
- Hanover, J.A.; Krause, M.W.; Love, D.C. Bittersweet memories: Linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 2012, 13, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell 1984, 37, 815–823. [Google Scholar] [CrossRef]
- Gambetta, M.C.; Muller, J. O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev. Cell 2014, 31, 629–639. [Google Scholar] [CrossRef]
- Gambetta, M.C.; Oktaba, K.; Müller, J. Essential Role of the Glycosyltransferase Sxc/Ogt in Polycomb Repression. Science 2009, 325, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Akan, I.; Love, D.C.; Harwood, K.R.; Bond, M.R.; Hanover, J.A. Drosophila O-GlcNAcase Deletion Globally Perturbs Chromatin O-GlcNAcylation. J. Biol. Chem. 2016, 291, 9906–9919. [Google Scholar] [CrossRef]
- Alfaro, J.F.; Gong, C.X.; Monroe, M.E.; Aldrich, J.T.; Clauss, T.R.; Purvine, S.O.; Wang, Z.; Camp, D.G.; Shabanowitz, J.; Stanley, P.; et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. USA 2012, 109, 7280–7285. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Cao, L.; Reece, E.A.; Zhao, Z. Impact of protein O-GlcNAcylation on neural tube malformation in diabetic embryopathy. Sci. Rep. 2017, 7, 11107. [Google Scholar] [CrossRef]
- Ma, J.; Banerjee, P.; Whelan, S.A.; Liu, T.; Wei, A.C.; Ramirez-Correa, G.; McComb, M.E.; Costello, C.E.; O’Rourke, B.; Murphy, A.; et al. Comparative Proteomics Reveals Dysregulated Mitochondrial O-GlcNAcylation in Diabetic Hearts. J. Proteome Res. 2016, 15, 2254–2264. [Google Scholar] [CrossRef]
- Trinidad, J.C.; Barkan, D.T.; Gulledge, B.F.; Thalhammer, A.; Sali, A.; Schoepfer, R.; Burlingame, A.L. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteom. 2012, 11, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Wulff-Fuentes, E.; Berendt, R.R.; Massman, L.; Danner, L.; Malard, F.; Vora, J.; Kahsay, R.; Olivier-Van Stichelen, S. The human O-GlcNAcome database and meta-analysis. Sci. Data 2021, 8, 25. [Google Scholar] [CrossRef]
- Liu, T.W.; Myschyshyn, M.; Sinclair, D.A.; Cecioni, S.; Beja, K.; Honda, B.M.; Morin, R.D.; Vocadlo, D.J. Genome-wide chemical mapping of O-GlcNAcylated proteins in Drosophila melanogaster. Nat. Chem. Biol. 2017, 13, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Selvan, N.; Williamson, R.; Mariappa, D.; Campbell, D.G.; Gourlay, R.; Ferenbach, A.T.; Aristotelous, T.; Hopkins-Navratilova, I.; Trost, M.; van Aalten, D.M. A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat. Chem. Biol. 2017, 13, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Akan, I.; Olivier-Van Stichelen, S.; Bond, M.R.; Hanover, J.A. Nutrient-driven O-GlcNAc in proteostasis and neurodegeneration. J. Neurochem. 2018, 144, 7–34. [Google Scholar] [CrossRef]
- Gong, C.X.; Liu, F.; Iqbal, K. O-GlcNAcylation: A regulator of tau pathology and neurodegeneration. Alzheimer’s Dement 2016, 12, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.-C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nat. Cell Biol. 2007, 448, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Kaasik, K.; Kivimäe, S.; Allen, J.J.; Chalkley, R.J.; Huang, Y.; Baer, K.; Kissel, H.; Burlingame, A.L.; Shokat, K.M.; Ptáček, L.J.; et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013, 17, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Golic, K.G.; Hawley, R.S. Drosophila: A Laboratory Handbook; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2005. [Google Scholar]
- Palladino, M.J.; Hadley, T.J.; Ganetzky, B. Temperature-sensitive paralytic mutants are enriched for those causing neurodegeneration in Drosophila. Genetics 2002, 161, 1197–1208. [Google Scholar] [CrossRef]
- Rulifson, E.J.; Kim, S.K.; Nusse, R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 2002, 296, 1118–1120. [Google Scholar] [CrossRef]
- Lavrov, S.; Déjardin, J.; Cavalli, G. Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol. Biol. 2004, 247, 289–303. [Google Scholar]
- Mohan, M.; Herz, H.-M.; Smith, E.R.; Zhang, Y.; Jackson, J.; Washburn, M.P.; Florens, L.; Eissenberg, J.C.; Shilatifard, A. The COMPASS Family of H3K4 Methylases in Drosophila. Mol. Cell. Biol. 2011, 31, 4310–4318. [Google Scholar] [CrossRef]
- Boersema, P.J.; Raijmakers, R.; Lemeer, S.; Mohammed, S.; Heck, A.J.R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009, 4, 484–494. [Google Scholar] [CrossRef]
- Halim, A.; Larsen, I.S.B.; Neubert, P.; Joshi, H.J.; Petersen, B.L.; Vakhrushev, S.Y.; Strahl, S.; Clausen, H. Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast. Proc. Natl. Acad. Sci. USA 2015, 112, 15648–15653. [Google Scholar] [CrossRef] [PubMed]
- Halim, A.; Westerlind, U.; Pett, C.; Schorlemer, M.; Rüetschi, U.; Brinkmalm, G.; Sihlbom, C.; Lengqvist, J.; Larson, G.; Nilsson, J. Assignment of Saccharide Identities through Analysis of Oxonium Ion Fragmentation Profiles in LC–MS/MS of Glycopeptides. J. Proteome Res. 2014, 13, 6024–6032. [Google Scholar] [CrossRef] [PubMed]
- Larsen, I.S.B.; Narimatsu, Y.; Joshi, H.J.; Yang, Z.; Harrison, O.J.; Brasch, J.; Shapiro, L.; Honig, B.; Vakhrushev, S.Y.; Clausen, H.; et al. Mammalian O-mannosylation of cadherins and plexins is independent of protein O-mannosyltransferases 1 and 2. J. Biol. Chem. 2017, 292, 11586–11598. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Dorighi, K.M.; Tamkun, J.W. Drosophila Kismet Regulates Histone H3 Lysine 27 Methylation and Early Elongation by RNA Polymerase II. PLoS Genet. 2008, 4, e1000217. [Google Scholar] [CrossRef] [PubMed]
- Hallson, G.; Hollebakken, R.E.; Li, T.; Syrzycka, M.; Kim, I.; Cotsworth, S.; Fitzpatrick, K.A.; Sinclair, D.A.R.; Honda, B.M. dSet1 Is the Main H3K4 Di- and Tri-Methyltransferase Throughout Drosophila Development. Genetics 2012, 190, 91–100. [Google Scholar] [CrossRef]
- Dorighi, K.M.; Tamkun, J.W. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development 2013, 140, 4182–4192. [Google Scholar] [CrossRef]
- Parker, J.B.; Yin, H.; Vinckevicius, A.; Chakravarti, D. Host Cell Factor-1 Recruitment to E2F-Bound and Cell-Cycle-Control Genes Is Mediated by THAP11 and ZNF143. Cell Rep. 2014, 9, 967–982. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Saiakhova, A.R.; Howard, B.; Monteith, K.E.; Scacheri, P.C.; Cosgrove, M.S.; Harte, P.J. Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development 2014, 141, 1129–1139. [Google Scholar] [CrossRef]
- Wang, P.; Lazarus, B.D.; Forsythe, M.E.; Love, D.C.; Krause, M.W.; Hanover, J.A. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 2012, 109, 17669–17674. [Google Scholar] [CrossRef] [PubMed]
- Fergestad, T.; Ganetzky, B.; Palladino, M.J. Neuropathology in Drosophila Membrane Excitability Mutants. Genetics 2006, 172, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Mazucanti, C.H.; Cabral-Costa, J.V.; Vasconcelos, A.R.; Andreotti, D.Z.; Scavone, C.; Kawamoto, E.M. Longevity Pathways (mTOR, SIRT, Insulin/IGF-1) as Key Modulatory Targets on Aging and Neurodegeneration. Curr. Top. Med. Chem. 2015, 15, 2116–2138. [Google Scholar] [CrossRef]
- Palladino, M.J.; Bower, J.E.; Kreber, R.; Ganetzky, B. Neural Dysfunction and Neurodegeneration inDrosophila Na+/K+ ATPase Alpha Subunit Mutants. J. Neurosci. 2003, 23, 1276–1286. [Google Scholar] [CrossRef] [PubMed]
- Yuzwa, S.A.; Shan, X.; Jones, B.A.; Zhao, G.; Woodward, M.L.; Li, X.; Zhu, Y.; McEachern, E.J.; Silverman, M.A.; Watson, N.V.; et al. Pharmacological inhibition of O-GlcNAcase (OGA) prevents cognitive decline and amyloid plaque formation in bigenic tau/APP mutant mice. Mol. Neurodegener. 2014, 9, 42. [Google Scholar] [CrossRef]
- Yuzwa, S.A.; Shan, X.; Macauley, M.S.; Clark, T.; Skorobogatko, Y.; Vosseller, K.; Vocadlo, D.J. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 2012, 8, 393–399. [Google Scholar] [CrossRef]
- Mangone, M.; Myers, M.P.; Herr, W. Role of the HCF-1 Basic Region in Sustaining Cell Proliferation. PLoS ONE 2010, 5, e9020. [Google Scholar] [CrossRef]
- McDonel, P.; Demmers, J.; Tan, D.W.; Watt, F.; Hendrich, B.D. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev. Biol. 2012, 363, 62–73. [Google Scholar] [CrossRef]
- Shi, F.-T.; Kim, H.; Lu, W.; He, Q.; Liu, D.; Goodell, M.A.; Wan, M.; Songyang, Z. Ten-Eleven Translocation 1 (Tet1) Is Regulated by O-Linked N-Acetylglucosamine Transferase (Ogt) for Target Gene Repression in Mouse Embryonic Stem Cells. J. Biol. Chem. 2013, 288, 20776–20784. [Google Scholar] [CrossRef]
- Xin, T.; Xuan, T.; Tan, J.; Li, M.; Zhao, G.; Li, M. The Drosophila putative histone acetyltransferase Enok maintains female germline stem cells through regulating Bruno and the niche. Dev. Biol. 2013, 384, 1–12. [Google Scholar] [CrossRef][Green Version]
- Yan, D.; Neumüller, R.A.; Buckner, M.; Ayers, K.; Li, H.; Hu, Y.; Yang-Zhou, D.; Pan, L.; Wang, X.; Kelley, C.; et al. A Regulatory Network of Drosophila Germline Stem Cell Self-Renewal. Dev. Cell 2014, 28, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Housley, M.P.; Rodgers, J.T.; Udeshi, N.D.; Kelly, T.J.; Shabanowitz, J.; Hunt, D.F.; Puigserver, P.; Hart, G.W. O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 2008, 283, 16283–16292. [Google Scholar] [CrossRef] [PubMed]
- Housley, M.P.; Udeshi, N.D.; Rodgers, J.T.; Shabanowitz, J.; Puigserver, P.; Hunt, D.F.; Hart, G.W. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J. Biol. Chem. 2009, 284, 5148–5157. [Google Scholar] [CrossRef] [PubMed]
- Mouchiroud, L.; Houtkooper, R.H.; Moullan, N.; Katsyuba, E.; Ryu, D.; Cantó, C.; Mottis, A.; Jo, Y.-S.; Viswanathan, M.; Schoonjans, K.; et al. The NAD+/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 2013, 154, 430–441. [Google Scholar] [CrossRef]
- Robida-Stubbs, S.; Glover-Cutter, K.; Lamming, D.W.; Mizunuma, M.; Narasimhan, S.D.; Neumann-Haefelin, E.; Sabatini, D.M.; Blackwell, T.K. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012, 15, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Love, D.C.; Ghosh, S.; Mondoux, M.A.; Fukushige, T.; Wang, P.; Wilson, M.A.; Iser, W.B.; Wolkow, C.A.; Krause, M.W.; Hanover, J.A. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 7413–7418. [Google Scholar] [CrossRef] [PubMed]
- Barnes, V.L.; Bhat, A.; Unnikrishnan, A.; Heydari, A.R.; Arking, R.; Pile, L.A. SIN3 is critical for stress resistance and modulates adult lifespan. Aging 2014, 6, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ebata, A.; Dong, Y.; Rizki, G.; Iwata, T.; Lee, S.S. Caenorhabditis elegans HCF-1 Functions in Longevity Maintenance as a DAF-16 Regulator. PLoS Biol. 2008, 6, e233. [Google Scholar] [CrossRef] [PubMed]
- Rizki, G.; Iwata, T.N.; Li, J.; Riedel, C.G.; Picard, C.L.; Jan, M.; Murphy, C.T.; Lee, S.S. The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO. PLoS Genet. 2011, 7, e1002235. [Google Scholar] [CrossRef] [PubMed]
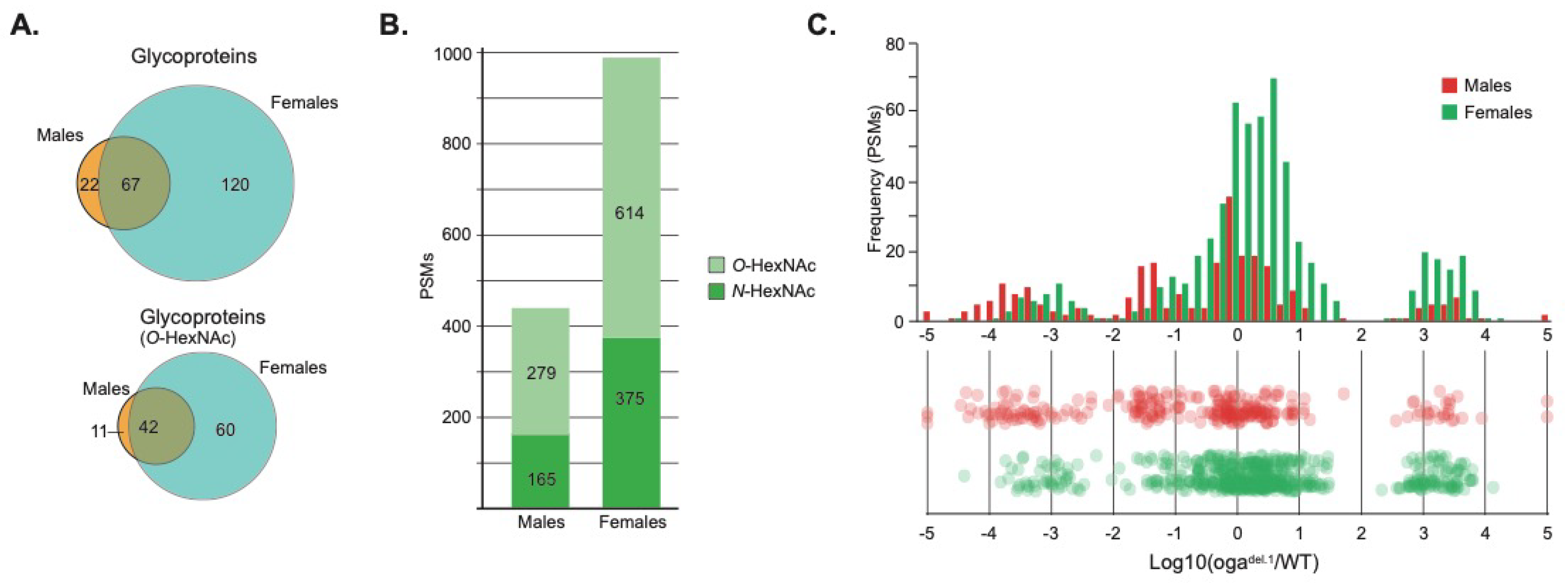

| GO Term | p-Value |
|---|---|
| positive regulation of gene expression [GO:0010628] | 1.06 × 10−4 |
| biological regulation [GO:0065007] | 1.68 × 10−4 |
| positive regulation of cellular process [GO:0048522] | 1.95 × 10−4 |
| regulation of cellular process [GO:0050794] | 3.71 × 10−4 |
| regulation of biological process [GO:0050789] | 3.94 × 10−4 |
| positive regulation of biological process [GO:0048518] | 8.27 × 10−4 |
| positive regulation of macromolecule biosynthetic process [GO:0010557] | 0.001605 |
| positive regulation of macromolecule metabolic process [GO:0010604] | 0.001687 |
| growth [GO:0040007] | 0.003118 |
| regulation of gene expression [GO:0010468] | 0.003782 |
| positive regulation of cellular biosynthetic process [GO:0031328] | 0.00389 |
| positive regulation of biosynthetic process [GO:0009891] | 0.003967 |
| positive regulation of metabolic process [GO:0009893] | 0.005195 |
| cellular component organization [GO:0016043] | 0.007664 |
| regulation of cellular macromolecule biosynthetic process [GO:2000112] | 0.022871 |
| cellular component organization or biogenesis [GO:0071840] | 0.023153 |
| regulation of macromolecule biosynthetic process [GO:0010556] | 0.02408 |
| anatomical structure development [GO:0048856] | 0.02648 |
| positive regulation of nitrogen compound metabolic process [GO:0051173] | 0.027266 |
| developmental process [GO:0032502] | 0.029356 |
| neuron development [GO:0048666] | 0.032766 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akan, I.; Halim, A.; Vakhrushev, S.Y.; Clausen, H.; Hanover, J.A. Drosophila O-GlcNAcase Mutants Reveal an Expanded Glycoproteome and Novel Growth and Longevity Phenotypes. Cells 2021, 10, 1026. https://doi.org/10.3390/cells10051026
Akan I, Halim A, Vakhrushev SY, Clausen H, Hanover JA. Drosophila O-GlcNAcase Mutants Reveal an Expanded Glycoproteome and Novel Growth and Longevity Phenotypes. Cells. 2021; 10(5):1026. https://doi.org/10.3390/cells10051026
Chicago/Turabian StyleAkan, Ilhan, Adnan Halim, Sergey Y. Vakhrushev, Henrik Clausen, and John A. Hanover. 2021. "Drosophila O-GlcNAcase Mutants Reveal an Expanded Glycoproteome and Novel Growth and Longevity Phenotypes" Cells 10, no. 5: 1026. https://doi.org/10.3390/cells10051026
APA StyleAkan, I., Halim, A., Vakhrushev, S. Y., Clausen, H., & Hanover, J. A. (2021). Drosophila O-GlcNAcase Mutants Reveal an Expanded Glycoproteome and Novel Growth and Longevity Phenotypes. Cells, 10(5), 1026. https://doi.org/10.3390/cells10051026





