Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model
Abstract
1. Introduction
1.1. Expanding the Human Footprint
1.2. The Challenges of Spaceflight
1.3. Effects of Radiation on the GIT
1.4. Torpor
1.5. Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model
2. Materials and Methods
2.1. Zebrafish Husbandry
2.2. Locomotion Assay for Activity Score
2.3. Development of the Induced Torpor Model and Radiation Protocol
2.4. RNA Extraction and Sequencing
2.5. RNA-seq Data Processing and Differential Expression
2.6. Experimental Validation
2.7. Pathway Analysis
2.8. Network Analysis
3. Results
3.1. Temperature and Melatonin Reduce Zebrafish Activity
3.2. Transcriptomic Characterisation of Induced Torpor Model Reveals a Reduction in Metabolism
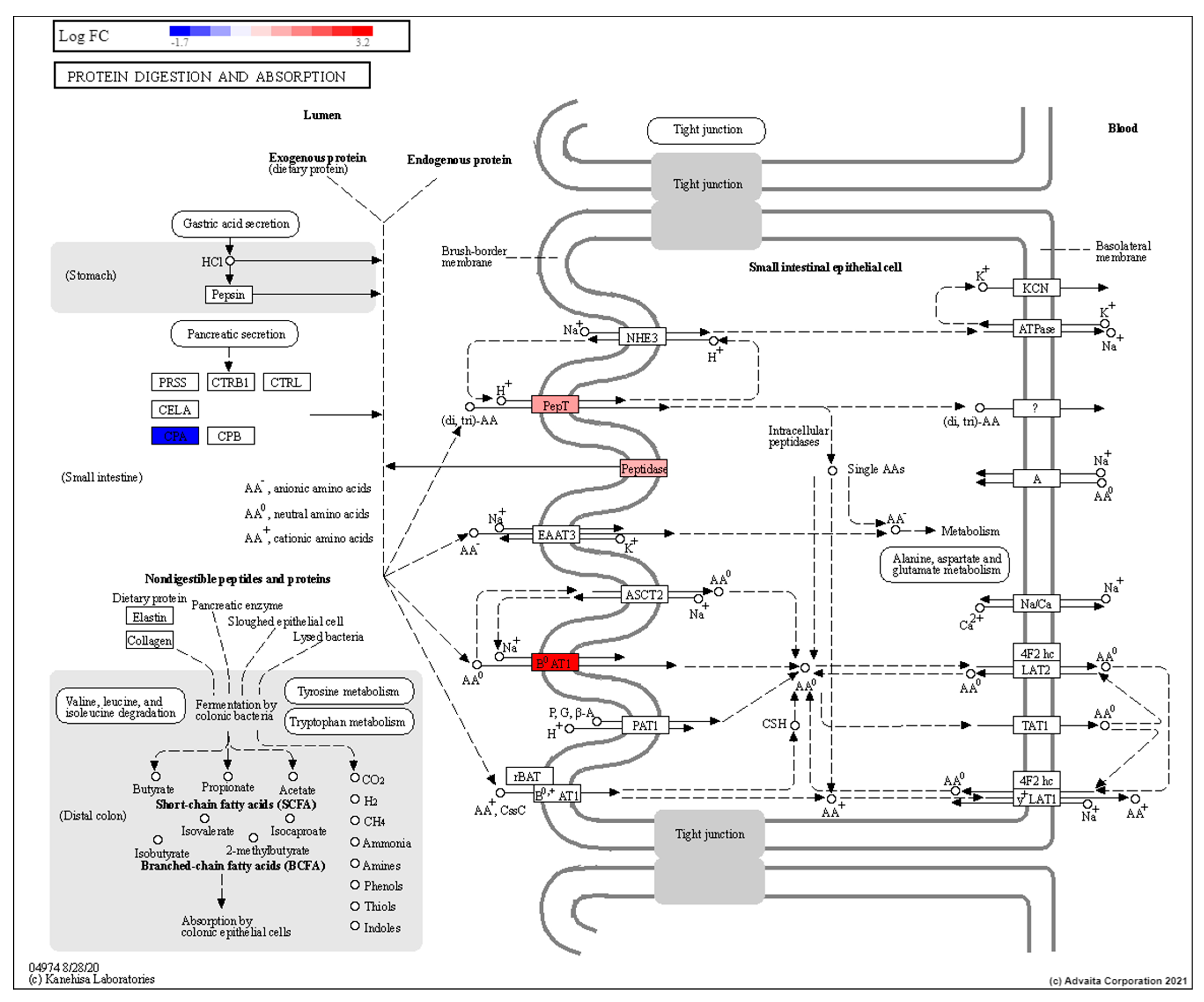
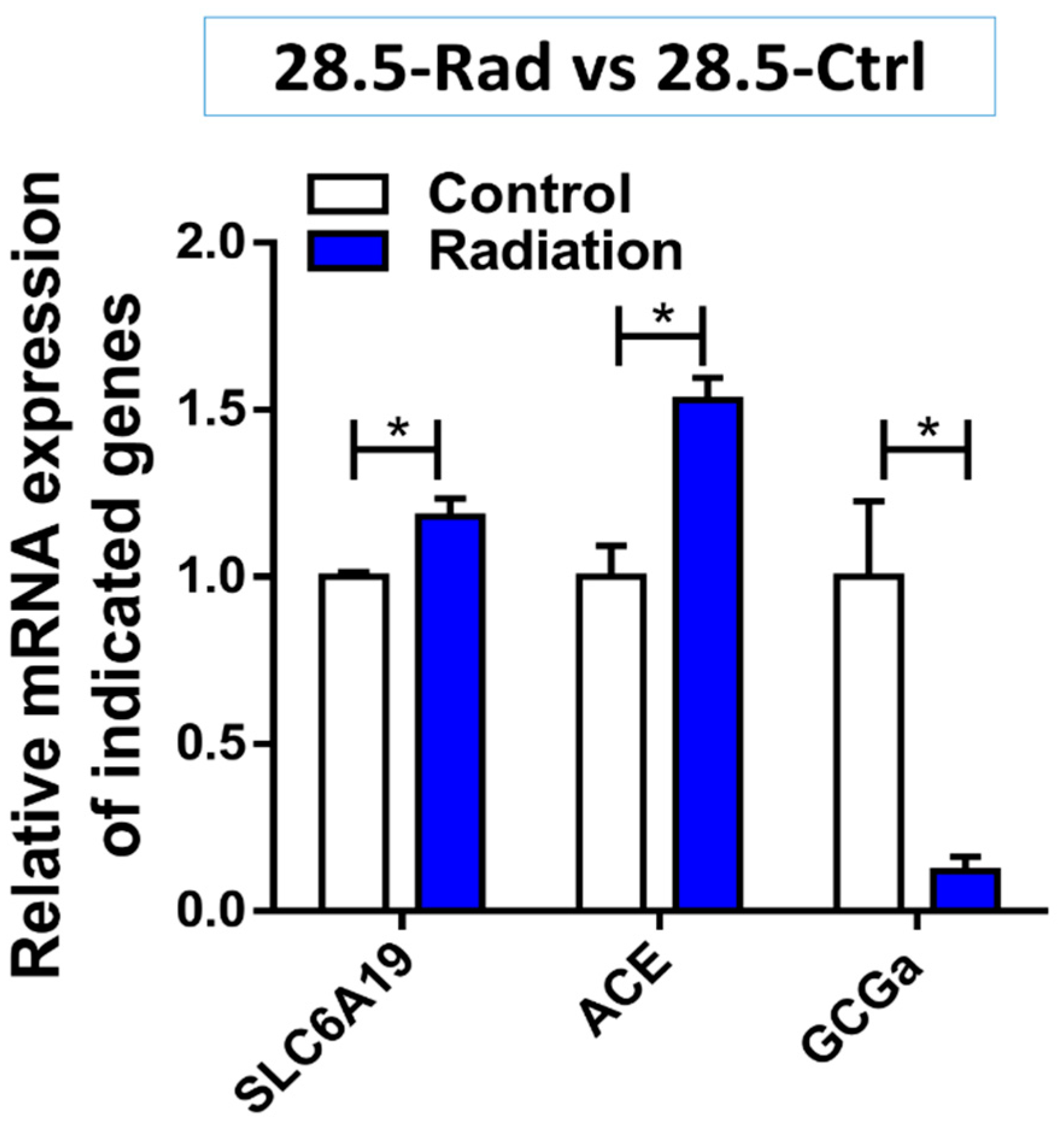
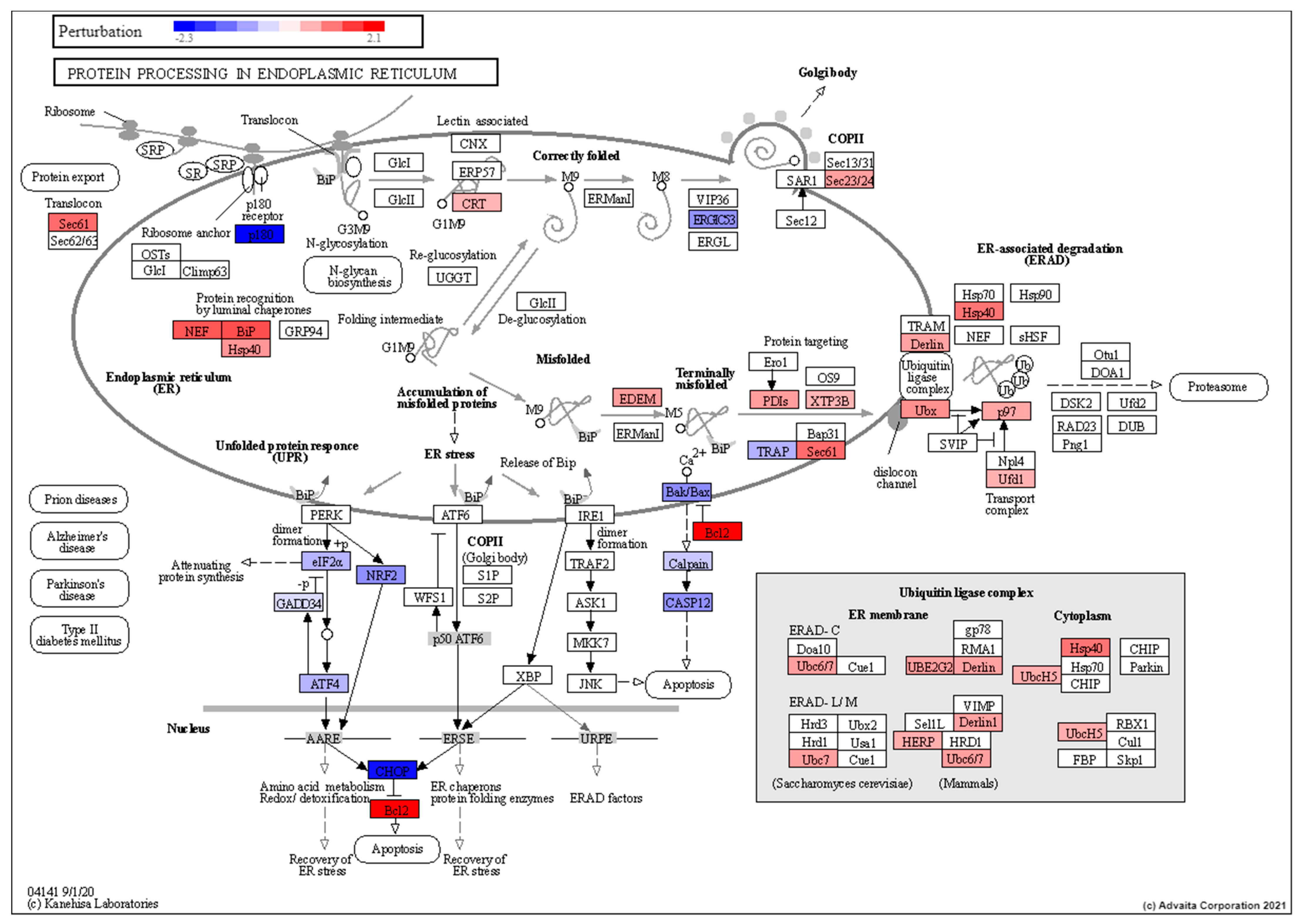
3.3. Low Dose Radiation Affects Metabolism and Absorption in the GIT
3.4. Pathway Analysis of Induced Torpor with Radiation Reveals Stress Response with Pro-survival Signals
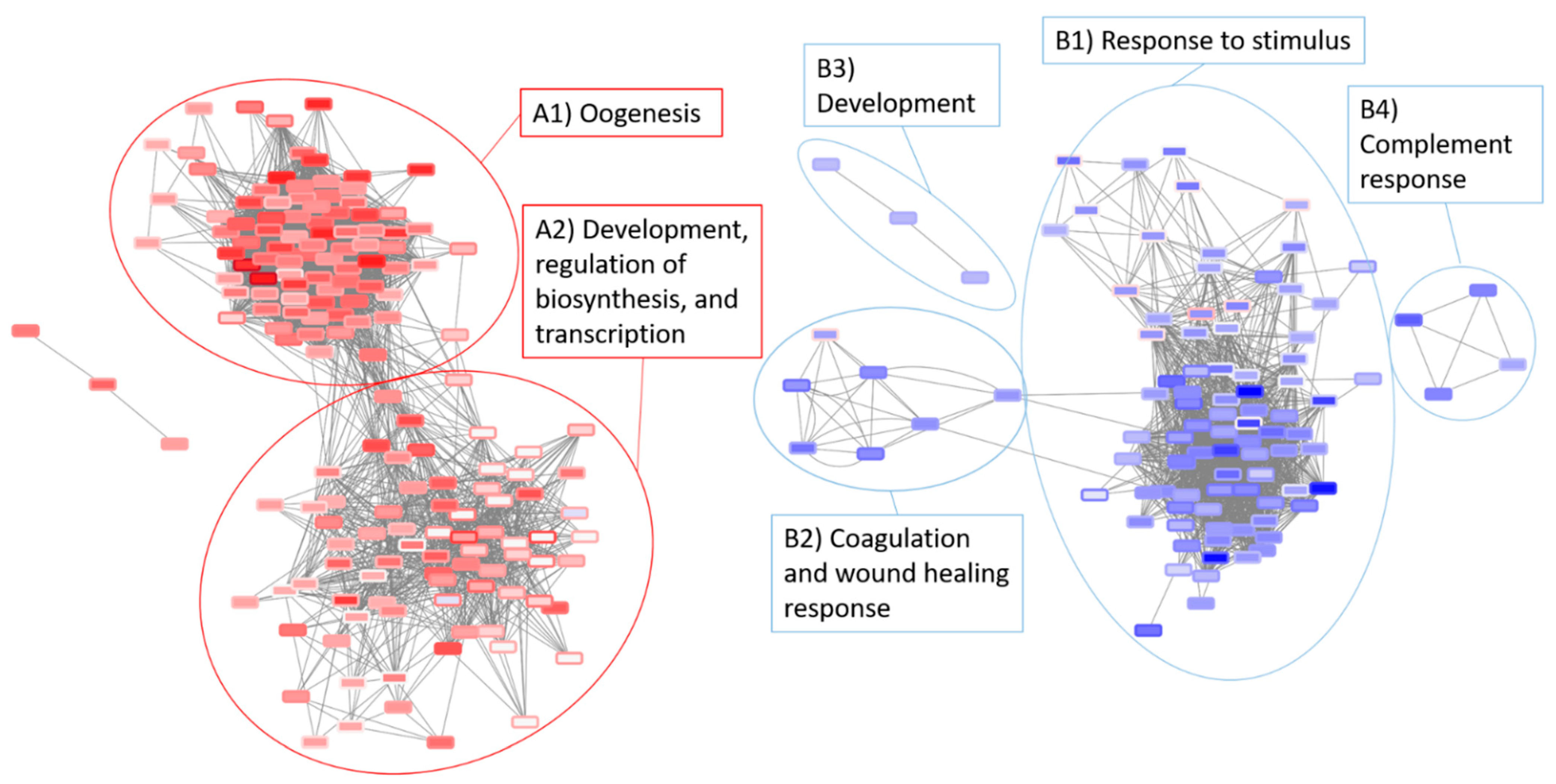
3.5. Network Analysis of the Radiation and Torpor + Radiation Groups Reveal Differential Regulation of Radio-Resistant Genes
4. Discussion
4.1. Induced Torpor Reduces Metabolism
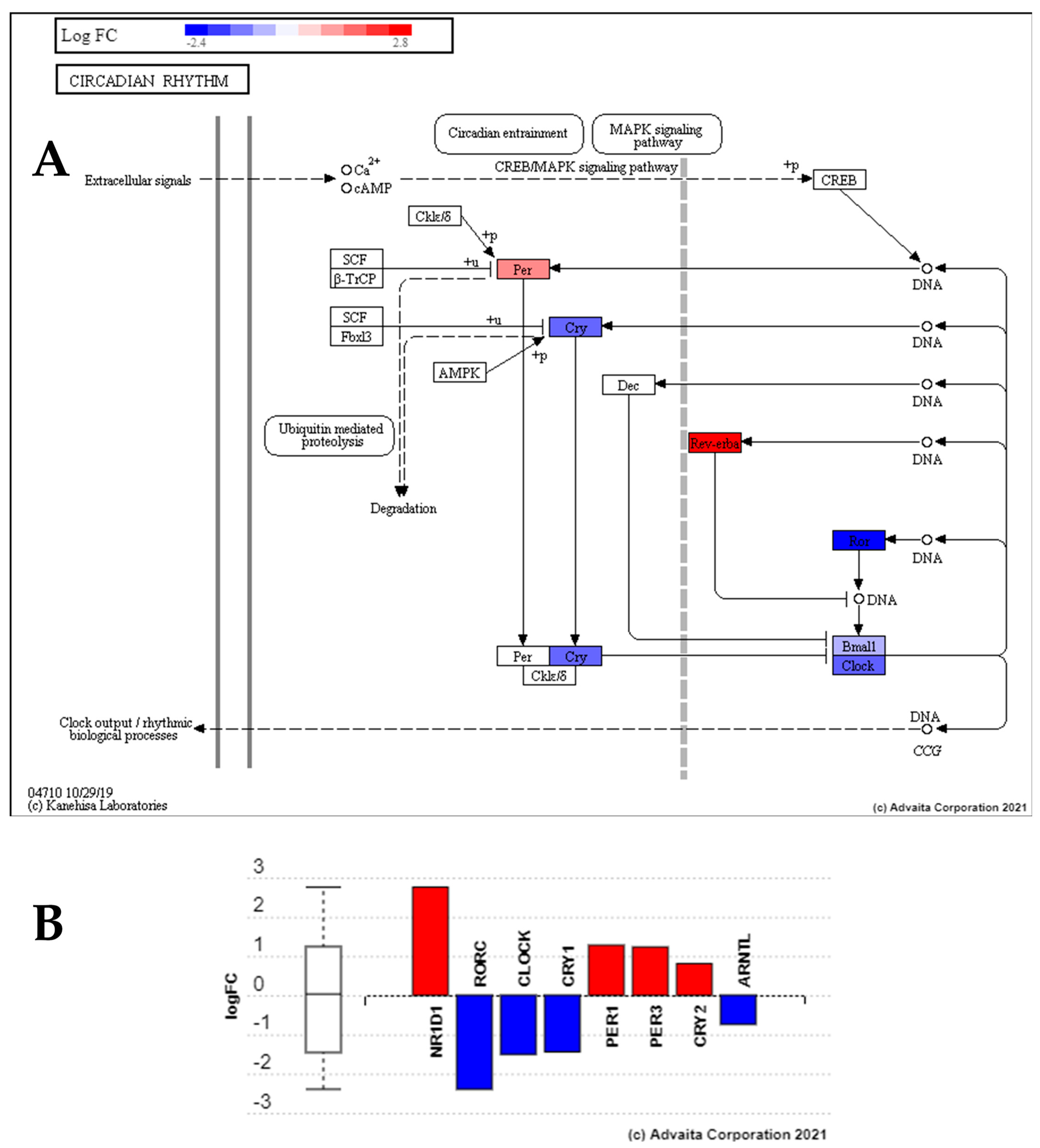
4.2. Low Dose Radiation Perturbed Key Circadian Rhythm Genes
4.3. Low Dose Radiation Induced a Glucocorticoid Stress Signalling Response
4.4. Low Dose Radiation May Affect Nutritional Status of Astronauts
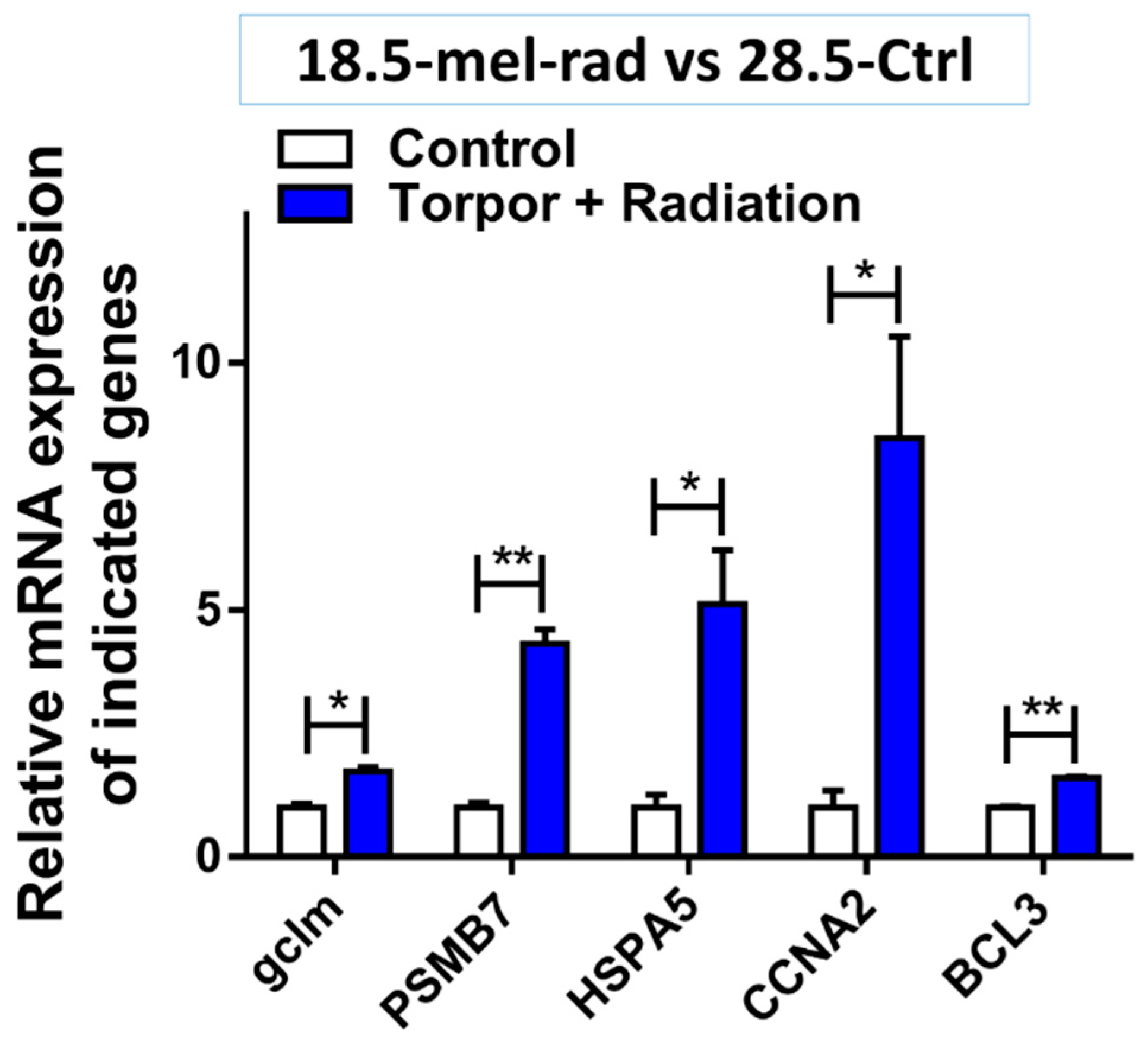
4.5. Torpor May Protect against Radiation through Increased Pro-Survival Signalling
4.6. Torpor May Reduce Radiation-Induced Oxidative Stress
4.7. Torpor May Lead to Removal of Radiation Damaged Proteins
4.8. Considerations for Inducing Torpor in Mammals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DE | Differentially expressed |
| FC | Fold change |
| FTI | Functional Target Identification |
| CR | Circadian Rhythm |
| GC | Glucocorticoid |
| GIT | Gastrointestinal Tract |
| IACUC | Institutional Animal Care and Use Committee |
| IMP | Integrative Multi-species Prediction |
| MUSC | Medical University of South Carolina |
| ORA | Over-Representation Analysis |
| PCA | Principal component analysis |
| RAS | Renin-Angiotensin System |
| ROS | Reactive oxygen species |
| TCA | Tricarboxylic acid |
References
- Dunbar, B. What is Artemis? Available online: https://www.nasa.gov/what-is-artemis (accessed on 25 September 2019).
- Cucinotta, F.A.; Kim, M.-H.Y.; Chappell, L.J.; Huff, J.L. How Safe Is Safe Enough? Radiation Risk for a Human Mission to Mars. PLoS ONE 2013, 8, e74988. [Google Scholar] [CrossRef] [PubMed]
- Cerri, M.; Tinganelli, W.; Negrini, M.; Helm, A.; Scifoni, E.; Tommasino, F.; Sioli, M.; Zoccoli, A.; Durante, M. Hibernation for space travel: Impact on radioprotection. Life Sci. Space Res. 2016, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cahill, T.; Hardiman, G. Nutritional challenges and countermeasures for space travel. Nutr. Bull. 2020, 45, 98–105. [Google Scholar] [CrossRef]
- Drysdale, A.; Ewert, M.; Hanford, A. Life support approaches for Mars missions. Adv. Space Res. 2003, 31, 51–61. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Kim, M.-H.Y.; Ren, L. Evaluating shielding effectiveness for reducing space radiation cancer risks. Radiat. Meas. 2006, 41, 1173–1185. [Google Scholar] [CrossRef]
- Teyssier, F.; Bay, J.O.; Dionet, C.; Verrelle, P. Cell cycle regulation after exposure to ionizing radiation. Bull Cancer 1999, 86, 345–357. [Google Scholar]
- Spitz, D.R.; Hauer-Jensen, M. Ionizing Radiation-Induced Responses: Where Free Radical Chemistry Meets Redox Biology and Medicine. Antioxid. Redox Signal. 2014, 20, 1407–1409. [Google Scholar] [CrossRef] [PubMed]
- Malkinson, F.D. Some principles of radiobiology: A selective review. J. Investig. Dermatol. 1981, 77, 32–38. [Google Scholar] [CrossRef]
- Castle, K.D.; Kirsch, D.G. Establishing the Impact of Vascular Damage on Tumor Response to High-Dose Radiation Therapy. Cancer Res. 2019, 79, 5685–5692. [Google Scholar] [CrossRef]
- Otterson, M.F. Effects of radiation upon gastrointestinal motility. World J. Gastroenterol. 2007, 13, 2684–2692. [Google Scholar] [CrossRef]
- Clément, G. Fundamentals of Space Medicine; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Shadad, A.K.; Sullivan, F.J.; Martin, J.D.; Egan, L.J. Gastrointestinal radiation injury: Symptoms, risk factors and mechanisms. World J. Gastroenterol. WJG 2013, 19, 185. [Google Scholar] [CrossRef]
- Srere, H.K.; Wang, L.C.; Martin, S.L. Central role for differential gene expression in mammalian hibernation. Proc. Natl. Acad. Sci. USA 1992, 89, 7119–7123. [Google Scholar] [CrossRef]
- Heldmaier, G.; Ortmann, S.; Elvert, R. Natural hypometabolism during hibernation and daily torpor in mammals. Respir. Physiol. Neurobiol. 2004, 141, 317–329. [Google Scholar] [CrossRef]
- Barnes, B.M. Freeze avoidance in a mammal: Body temperatures below 0 degree C in an Arctic hibernator. Science 1989, 244, 1593–1595. [Google Scholar] [CrossRef] [PubMed]
- Barr, R.E.; Musacchia, X.J. Postirradiation Hibernation and Radiation Response of Ground Squirrels: Telemetry Surveillance. Radiat. Res. 1972, 51, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Hock, R.J. Seasonal variations in physiological functions of arctic ground squirrels and black bears. Bull. Museum Comp. Zool. Harvard. Coll. 1960, 124, 155–171. [Google Scholar]
- Geiser, F. Metabolic Rate and Body Temperature Reduction During Hibernation and Daily Torpor. Annu. Rev. Physiol. 2004, 66, 239–274. [Google Scholar] [CrossRef]
- Bartsiokas, A.; Arsuaga, J.-L. Hibernation in hominins from Atapuerca, Spain half a million years ago. L’Anthropologie 2020, 124, 102797. [Google Scholar] [CrossRef]
- Song, S.S.; Lyden, P.D. Overview of Therapeutic Hypothermia. Curr. Treat. Options Neurol. 2012, 14, 541–548. [Google Scholar] [CrossRef]
- Vergauwen, L.; Benoot, D.; Blust, R.; Knapen, D. Long-term warm or cold acclimation elicits a specific transcriptional response and affects energy metabolism in zebrafish. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 157, 149–157. [Google Scholar] [CrossRef]
- Chou, M.-Y.; Hsiao, C.-D.; Chen, S.-C.; Chen, I.-W.; Liu, S.-T.; Hwang, P.-P. Effects of hypothermia on gene expression in zebrafish gills: Upregulation in differentiation and function of ionocytes as compensatory responses. J. Exp. Biol. 2008, 211, 3077–3084. [Google Scholar] [CrossRef]
- Reed, B.; Jennings, M. Guidance on the Housing and Care of Zebrafish Danio Rerio. Available online: https://www.rspca.org.uk/webContent/staticImages/Downloads/HousingAndCareZebrafish.pdf (accessed on 14 April 2021).
- Malek, R.L.; Sajadi, H.; Abraham, J.; Grundy, M.A.; Gerhard, G.S. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2004, 138, 363–373. [Google Scholar] [CrossRef]
- Zhdanova, I.V.; Wurtman, R.J.; Regan, M.M.; Taylor, J.A.; Shi, J.P.; Leclair, O.U. Melatonin treatment for age-related insomnia. J. Clin. Endocrinol. Metab. 2001, 86, 4727–4730. [Google Scholar] [CrossRef] [PubMed]
- Hacışevki, A.; Baba, B. An Overview of Melatonin as an Antioxidant Molecule: A Biochemical Approach. Melatonin Mol. Biol. Clin. Pharm. Approaches 2018, 2018, 59–85. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.R. Zebrafish Genome Overview. 2017. Available online: https://www.ncbi.nlm.nih.gov/grc/zebrafish (accessed on 14 April 2021).
- Noldus, L.P.J.J.; Spink, A.J.; Tegelenbosch, R.A.J. EthoVision: A versatile video tracking system for automation of behavioral experiments. Behav. Res. Methods Instrum. Comput. 2001, 33, 398–414. [Google Scholar] [CrossRef]
- Nishimura, Y.; Okabe, S.; Sasagawa, S.; Murakami, S.; Ashikawa, Y.; Yuge, M.; Kawaguchi, K.; Kawase, R.; Tanaka, T. Pharmacological profiling of zebrafish behavior using chemical and genetic classification of sleep-wake modifiers. Front. Pharmacol. 2015, 6, 257. [Google Scholar] [CrossRef] [PubMed]
- Cavallo, A.; Hassan, M. Stability of melatonin in aqueous solution. J. Pineal Res. 1995, 18, 90–92. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bibsonomy.org/bibtex/2b6052877491828ab53d3449be9b293b3/ozborn (accessed on 31 January 2021).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yates, A.; Akanni, W.; Amode, M.R.; Barrell, D.; Billis, K.; Carvalho-Silva, D.; Cummins, C.; Clapham, P.; Fitzgerald, S.; Gil, L.; et al. Ensembl 2016. Nucleic Acids Res. 2016, 44, D710–D716. [Google Scholar] [CrossRef] [PubMed]
- Huff, M.; Da Silveira, W.A.; Carnevali, O.; Renaud, L.; Hardiman, G. Systems Analysis of the Liver Transcriptome in Adult Male Zebrafish Exposed to the Plasticizer (2-Ethylhexyl) Phthalate (DEHP). Sci. Rep. 2018, 8, 2118. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.K.; Krishnan, A.; Yao, V.; Tadych, A.; Troyanskaya, O.G. IMP 2.0: A multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2015, 43, W128–W133. [Google Scholar] [CrossRef][Green Version]
- Overton, I.M.; Sims, A.H.; Owen, J.A.; Heale, B.S.E.; Ford, M.J.; Lubbock, A.L.R.; Pairo-Castineira, E.; Essafi, A. Functional Transcription Factor Target Networks Illuminate Control of Epithelial Remodelling. Cancers 2020, 12, 2823. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2013, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Satyanarayana, A.; Kaldis, P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009, 28, 2925–2939. [Google Scholar] [CrossRef]
- Ohno, H.; Takimoto, G.; McKeithan, T.W. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell 1990, 60, 991–997. [Google Scholar] [CrossRef]
- Edilova, M.I.; Abdul-Sater, A.A.; Watts, T.H. TRAF1 Signaling in Human Health and Disease. Front. Immunol. 2018, 9, 2969. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.M. Caspases: The executioners of apoptosis. Biochem. J. 1997, 326, 1–16. [Google Scholar] [CrossRef]
- Opferman, J.T.; Kothari, A. Anti-apoptotic BCL-2 family members in development. Cell Death Differ. 2018, 25, 37–45. [Google Scholar] [CrossRef]
- Zanini, I.M.; Soneson, C.; Lorenzi, L.E.; Azzalin, C.M. Human cactin interacts with DHX8 and SRRM2 to assure efficient pre-mRNA splicing and sister chromatid cohesion. J. Cell Sci. 2017, 130, 767–778. [Google Scholar] [CrossRef]
- Alexandrov, A.; Colognori, D.; Shu, M.-D.; Steitz, J.A. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc. Natl. Acad. Sci. USA 2012, 109, 21313–21318. [Google Scholar] [CrossRef]
- Tseng, C.-K.; Chung, C.-S.; Chen, H.-C.; Cheng, S.-C. A central role of Cwc25 in spliceosome dynamics during the catalytic phase of pre-mRNA splicing. RNA 2017, 23, 546–556. [Google Scholar] [CrossRef]
- Wang, J.; Lee, J.; Liem, D.; Ping, P. HSPA5 Gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene 2017, 618, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Okada, T.; Yoshida, H.; Kaufman, R.J.; Nagata, K.; Mori, K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 2006, 172, 383–393. [Google Scholar] [CrossRef]
- Liang, J.; Yin, C.; Doong, H.; Fang, S.; Peterhoff, C.; Nixon, R.A.; Monteiro, M.J. Characterization of erasin (UBXD2): A new ER protein that promotes ER-associated protein degradation. J. Cell Sci. 2006, 119, 4011–4024. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.; Elmer, K.R. Alternative splicing and gene expression play contrasting roles in the parallel phenotypic evolution of a salmonid fish. Mol. Ecol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cheng, C.-H.C.; Zhang, J.; Cao, L.; Chen, L.; Zhou, L.; Jin, Y.; Ye, H.; Deng, C.; Dai, Z.; et al. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc. Natl. Acad. Sci. USA 2008, 105, 12944–12949. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, A. Impact of Environmental and Genetic Regulation of Skeletal Muscle Metabolism on Metabolic Response in Women with Overweight or Obesity: Molecular and Cellular Analyses and Genetic Association Studies. Ph.D. Thesis, Université d’Ottawa/University of Ottawa, Ottawa, ON, Canada, 2018. [Google Scholar]
- Friedel, S.; Reichwald, K.; Scherag, A.; Brumm, H.; Wermter, A.-K.; Fries, H.-R.; Koberwitz, K.; Wabitsch, M.; Meitinger, T.; Platzer, M.; et al. Mutation screen and association studies in the Diacylglycerol O-acyltransferase homolog 2 gene (DGAT2), a positional candidate gene for early onset obesity on chromosome 11q13. BMC Genet. 2007, 8, 17. [Google Scholar] [CrossRef]
- Darmoul, D.; Lacasa, M.; Baricault, L.; Marguet, D.; Sapin, C.; Krejbich-Trotot, P.; Barbat, A.; Trugnan, G. Dipeptidyl peptidase IV (CD 26) gene expression in enterocyte-like colon cancer cell lines HT-29 and Caco-2. Cloning of the complete human coding sequence and changes of dipeptidyl peptidase IV mRNA levels during cell differentiation. J. Biol. Chem. 1992, 267, 4824–4833. [Google Scholar] [CrossRef]
- Mazzola, J.L.; Sirover, M.A. Subcellular localization of human glyceraldehyde-3-phosphate dehydrogenase is independent of its glycolytic function. Biochim. Biophys. Acta (BBA) Gen. Subj. 2003, 1622, 50–56. [Google Scholar] [CrossRef]
- Ackrell, B.A. Cytopathies involving mitochondrial complex II. Mol. Asp. Med. 2002, 23, 369–384. [Google Scholar] [CrossRef]
- Chen, X.J.; Wang, X.; Kaufman, B.A.; Butow, R.A. Aconitase Couples Metabolic Regulation to Mitochondrial DNA Maintenance. Sci. 2005, 307, 714–717. [Google Scholar] [CrossRef]
- Fujii, T.; Khawaja, M.R.; Dinardo, C.D.; Atkins, J.T.; Janku, F. Targeting isocitrate dehydrogenase (IDH) in cancer. Discov. Med. 2016, 21, 373–380. [Google Scholar]
- Vogel, R.O.; Dieteren, C.E.J.; Heuvel, L.P.W.J.V.D.; Willems, P.H.G.M.; Smeitink, J.A.M.; Koopman, W.J.H.; Nijtmans, L.G.J. Identification of Mitochondrial Complex I Assembly Intermediates by Tracing Tagged NDUFS3 Demonstrates the Entry Point of Mitochondrial Subunits. J. Biol. Chem. 2007, 282, 7582–7590. [Google Scholar] [CrossRef]
- Solmaz, S.R.; Hunte, C. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J. Biol. Chem. 2008, 283, 17542–17549. [Google Scholar] [CrossRef]
- Coburn, C.T.; Hajri, T.; Ibrahimi, A.; Abumrad, N.A. Role of CD36 in Membrane Transport and Utilization of Long-Chain Fatty Acids by Different Tissues. J. Mol. Neurosci. 2001, 16, 117–122. [Google Scholar] [CrossRef]
- Turk, E.; Martín, M.G.; Wright, E.M. Structure of the human Na+/glucose cotransporter gene SGLT1. J. Biol. Chem. 1994, 269, 15204–15209. [Google Scholar] [CrossRef]
- Bröer, S. The role of the neutral amino acid transporter B0AT1 (SLC6A19) in Hartnup disorder and protein nutrition. IUBMB Life 2009, 61, 591–599. [Google Scholar] [CrossRef]
- Fredrick, K.; Ibba, M. Errors rectified in retrospect. Nat. Cell Biol. 2009, 457, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Andreani, T.S.; Itoh, T.Q.; Yildirim, E.; Hwangbo, D.-S.; Allada, R. Genetics of Circadian Rhythms. Sleep Med. Clin. 2015, 10, 413–421. [Google Scholar] [CrossRef]
- Bartek, J.; Lukas, J. Order from destruction. Science 2001, 294, 66–67. [Google Scholar] [CrossRef]
- Fittkau, M.; Grothey, A.; Gerlach, R.; Schmoll, H.-J. A low dose of ionizing radiation increases luminal release of intestinal peptidases in rats. J. Cancer Res. Clin. Oncol. 2001, 127, 96–100. [Google Scholar] [CrossRef]
- Lin, L.; Yee, S.W.; Kim, R.B.; Giacomini, K.M. SLC transporters as therapeutic targets: Emerging opportunities. Nat. Rev. Drug Discov. 2015, 14, 543–560. [Google Scholar] [CrossRef]
- Wenzel, U.; Meissner, B.; Döring, F.; Daniel, H. PEPT1-mediated uptake of dipeptides enhances the intestinal absorption of amino acids via transport system b(0,+). J Cell Physiol. 2001, 186, 251–259. [Google Scholar] [CrossRef]
- Garg, M.; Angus, P.W.; Burrell, L.M.; Herath, C.; Gibson, P.R.; Lubel, J.S. Review article: The pathophysiological roles of the renin–angiotensin system in the gastrointestinal tract. Aliment. Pharmacol. Ther. 2012, 35, 414–428. [Google Scholar] [CrossRef]
- Fawley, J.; Gourlay, D.M. Intestinal alkaline phosphatase: A summary of its role in clinical disease. J. Surg. Res. 2016, 202, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-J. Biochemical Characterization of Statin, a Protein Marker Specific for Nonproliferating Cells, and Identification of Its Associated Proteins in Cultured Human Diploid Fibroblasts; National Library of Canada: Ottawa, ON, Canada; McGill University: Montréal, QC, Canada, 1994. [Google Scholar]
- Tarling, E.J.; Vallim, T.Q.D.A.; Edwards, P.A. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 2013, 24, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.L.; Entenman, C. The Role of Bile Secretion in the Gastrointestinal Radiation Syndrome. Radiat. Res. 1959, 10, 67. [Google Scholar] [CrossRef]
- Matschinsky, F.M.; Wilson, D.F. The Central Role of Glucokinase in Glucose Homeostasis: A Perspective 50 Years After Demonstrating the Presence of the Enzyme in Islets of Langerhans. Front. Physiol. 2019, 10, 148. [Google Scholar] [CrossRef]
- Bouchard-Mercier, A.; Rudkowska, I.; Lemieux, S.; Couture, P.; Pérusse, L.; Vohl, M.-C. SREBF1 gene variations modulate insulin sensitivity in response to a fish oil supplementation. Lipids Heal. Dis. 2014, 13, 152. [Google Scholar] [CrossRef]
- Oeffinger, K.C.; Sklar, C.A. Abdominal radiation and diabetes: One more piece in the puzzle. Lancet Oncol. 2012, 13, 961–962. [Google Scholar] [CrossRef]
- Kawamura, Y.; Tanaka, Y.; Kawamori, R.; Maeda, S. Overexpression of Kruppel-Like Factor 7 Regulates Adipocytokine Gene Expressions in Human Adipocytes and Inhibits Glucose-Induced Insulin Secretion in Pancreatic β-Cell Line. Mol. Endocrinol. 2006, 20, 844–856. [Google Scholar] [CrossRef]
- Roche, M.; Neti, P.V.S.V.; Kemp, F.W.; Agrawal, A.; Attanasio, A.; Douard, V.; Muduli, A.; Azzam, E.I.; Norkus, E.; Brimacombe, M.; et al. Radiation-induced reductions in transporter mRNA levels parallel reductions in intestinal sugar transport. Am. J. Physiol. Integr. Comp. Physiol. 2010, 298, R173–R182. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Mary, C.; Scherrer, A.; Huck, L.; Lakkaraju, A.K.; Thomas, Y.; Johnson, A.E.; Strub, K. Residues in SRP9/14 essential for elongation arrest activity of the signal recognition particle define a positively charged functional domain on one side of the protein. RNA 2010, 16, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Calixto, C.P.G.; Tzioutziou, N.A.; James, A.B.; Hornyik, C.; Guo, W.; Zhang, R.; Nimmo, H.G.; Brown, J.W.S. Cold-Dependent Expression and Alternative Splicing of Arabidopsis Long Non-coding RNAs. Front. Plant Sci. 2019, 10, 235. [Google Scholar] [CrossRef]
- Lodhi, I.J.; Semenkovich, C.F. Peroxisomes: A Nexus for Lipid Metabolism and Cellular Signaling. Cell Metab. 2014, 19, 380–392. [Google Scholar] [CrossRef]
- Fararjeh, A.S.; Chen, L.-C.; Ho, Y.-S.; Cheng, T.-C.; Liu, Y.-R.; Chang, H.-L.; Chang, H.-W.; Wu, C.-H.; Tu, S.-H. Proteasome 26S Subunit, non-ATPase 3 (PSMD3) Regulates Breast Cancer by Stabilizing HER2 from Degradation. Cancers 2019, 11, 527. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, R.S.; Sharma, D.; Checker, R.; Thoh, M.; Sandur, S.K. Spatio-temporal changes in glutathione and thioredoxin redox couples during ionizing radiation-induced oxidative stress regulate tumor radio-resistance. Free. Radic. Res. 2015, 49, 1218–1232. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Choudhary, D.; Upreti, M.; Rath, P.; Kale, R. Radiation induced oxidative stress: I. Studies in Ehrlich solid tumor in mice. Mol. Cell. Biochem. 2001, 223, 71–80. [Google Scholar] [CrossRef]
- Tang, D.; Loze, M.T.; Zeh, I.H.J.; Kang, R. The redox protein HMGB1 regulates cell death and survival in cancer treatment. Autophagy 2010, 6, 1181–1183. [Google Scholar] [CrossRef]
- Helson, L. Radiation-induced Demyelination and Remyelination in the Central Nervous System: A Literature Review. Anticancer. Res. 2018, 38, 4999–5002. [Google Scholar] [CrossRef]
- Ridley, A.; Hall, A. Signal transduction pathways regulating Rho-mediated stress fibre formation: Requirement for a tyrosine kinase. EMBO J. 1994, 13, 2600–2610. [Google Scholar] [CrossRef]
- Bourgier, C.; Haydont, V.; Milliat, F.; François, A.; Holler, V.; Lasser, P.; Bourhis, J.; Mathé, D.; Vozenin-Brotons, M.-C. Inhibition of Rho kinase modulates radiation induced fibrogenic phenotype in intestinal smooth muscle cells through alteration of the cytoskeleton and connective tissue growth factor expression. Gut 2005, 54, 336–343. [Google Scholar] [CrossRef]
- Whitnall, M.H.; Elliott, T.B.; Harding, R.A.; Inal, C.E.; Landauer, M.R.; Wilhelmsen, C.L.; McKinney, L.; Miner, V.L.; Jackson, W.E.; Loria, R.M.; et al. Androstenediol stimulates myelopoiesis and enhances resistance to infection in gamma-irradiated mice. Int. J. Immunopharmacol. 2000, 22, 1–14. [Google Scholar] [CrossRef]
- Kijima, Y.; Wantong, W.; Igarashi, Y.; Yoshitake, K.; Asakawa, S.; Suzuki, Y.; Watabe, S.; Kinoshita, S. Age-associated different transcriptome profiling in zebrafish and rat: Insight into diversity of vertebrate aging. bioRxiv 2018, 478438. [Google Scholar] [CrossRef]
- Tian, T.; Zhao, L.; Zhao, X.; Zhang, M.; Meng, A. A zebrafish gene trap line expresses GFP recapturing expression pattern of foxj1b. J. Genet. Genom. 2009, 36, 581–589. [Google Scholar] [CrossRef]
- Cai, C.; Sang, C.; Du, J.; Jia, H.; Tu, J.; Wan, Q.; Bao, B.; Xie, S.; Huang, Y.; Li, A.; et al. Knockout of tnni1b in zebrafish causes defects in atrioventricular valve development via the inhibition of the myocardial wnt signaling pathway. FASEB J. 2019, 33, 696–710. [Google Scholar] [CrossRef] [PubMed]
- Fukai, J.; Yokote, H.; Yamanaka, R.; Arao, T.; Nishio, K.; Itakura, T. EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Mol. Cancer Ther. 2008, 7, 2768–2778. [Google Scholar] [CrossRef] [PubMed]
- Danesin, C.; Darche-Gabinaud, R.; Escalas, N.; Bouguetoch, V.; Cochard, P.; Al Oustah, A.; Ohayon, D.; Glise, B.; Soula, C. Sulf2a controls Shh-dependent neural fate specification in the developing spinal cord. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Peng, Y.; Zhang, W.; Lv, N.; Tang, J.; Chen, C.; Zhang, C.; Gao, S.; Chen, H.; Zhi, G.; et al. Myosin Light Chain Kinase Is Central to Smooth Muscle Contraction and Required for Gastrointestinal Motility in Mice. Gastroenterol. 2008, 135, 610–620.e2. [Google Scholar] [CrossRef]
- Connell, L.E.; Helfman, D.M. Myosin light chain kinase plays a role in the regulation of epithelial cell survival. J. Cell Sci. 2006, 119, 2269–2281. [Google Scholar] [CrossRef]
- Schwartz, M. Rho signalling at a glance. J. Cell Sci. 2004, 117, 5457–5458. [Google Scholar] [CrossRef]
- Liu, R.-Z.; Denovan-Wright, E.M.; Degrave, A.; Thisse, C.; Thisse, B.; Wright, J.M. Differential expression of duplicated genes for brain-type fatty acid-binding proteins (fabp7a and fabp7b) during early development of the CNS in zebrafish (Danio rerio). Gene Expr. Patterns 2004, 4, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Quan, F.B.; Desban, L.; Mirat, O.; Kermarquer, M.; Roussel, J.; Koëth, F.; Marnas, H.; Djenoune, L.; Lejeune, F.-X.; Tostivint, H.; et al. Somatostatin 1.1 contributes to the innate exploration of zebrafish larva. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kossack, M.E.; High, S.K.; Hopton, R.E.; Yan, Y.-L.; Postlethwait, J.H.; Draper, B.W. Female Sex Development and Reproductive Duct Formation Depend on Wnt4a in Zebrafish. Genetics 2019, 211, 219–233. [Google Scholar] [CrossRef]
- Navajas Acedo, J.; Voas, M.G.; Alexander, R.; Woolley, T.; Unruh, J.R.; Li, H.; Moens, C.; Piotrowski, T. PCP and Wnt pathway components act in parallel during zebrafish mechanosensory hair cell orientation. Nat. Commun. 2019, 10, 3993. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.N.; Cheng, C.N.; Adeeb, B.; Addiego, A.; Wesselman, H.M.; Chambers, B.E.; Chambers, J.M.; Wingert, R.A. Iroquois transcription factor irx2a is required for multiciliated and transporter cell fate decisions during zebrafish pronephros development. Sci. Rep. 2019, 9, 6454. [Google Scholar] [CrossRef]
- Gavriouchkina, D.; Williams, R.M.; Lukoseviciute, M.; Hochgreb-Hägele, T.; Senanayake, U.; Chong-Morrison, V.; Thongjuea, S.; Repapi, E.; Mead, A.; Sauka-Spengler, T. From pioneer to repressor: Bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. bioRxiv 2017, 213611. [Google Scholar] [CrossRef]
- Desai, K.; Spikings, E.; Zhang, T. Effect of chilling on sox2, sox3 and sox19a gene expression in zebrafish (Danio rerio) embryos. Cryobiol. 2011, 63, 96–103. [Google Scholar] [CrossRef]
- Grajevskaja, V.; Camerota, D.; Bellipanni, G.; Balciuniene, J.; Balciunas, D. Analysis of a conditional gene trap reveals that tbx5a is required for heart regeneration in zebrafish. PLoS ONE 2018, 13, e0197293. [Google Scholar] [CrossRef]
- Liu, J.; An, H.; Yuan, W.; Feng, Q.; Chen, L.; Ma, J. Prognostic Relevance and Function of MSX2 in Colorectal Cancer. J. Diabetes Res. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Nechiporuk, A.; Linbo, T.; Poss, K.D.; Raible, D.W. Specification of epibranchial placodes in zebrafish. Development 2006, 134, 611–623. [Google Scholar] [CrossRef]
- Lecaudey, V.; Anselme, I.; Dildrop, R.; Ruther, U.; Schneider-Maunoury, S. Expression of the zebrafish Iroquois genes during early nervous system formation and patterning. J. Comp. Neurol. 2005, 492, 289–302. [Google Scholar] [CrossRef]
- Viktorin, G.; Chiuchitu, C.; Rissler, M.; Varga, Z.M.; Westerfield, M. Emx3 is required for the differentiation of dorsal telencephalic neurons. Dev. Dyn. 2009, 238, 1984–1998. [Google Scholar] [CrossRef] [PubMed]
- Hilinski, W.C.; Bostrom, J.R.; England, S.J.; Juárez-Morales, J.L.; de Jager, S.; Armant, O.; Legradi, J.; Strähle, U.; Link, B.A.; Lewis, K.E. Lmx1b is required for the glutamatergic fates of a subset of spinal cord neurons. Neural. Dev. 2016, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Cheng, C.-H.; Chen, G.-D.; Hung, C.-C.; Yang, C.-H.; Hwang, S.-P.L.; Kawakami, K.; Wu, B.-K.; Huang, C.-J. Recapitulation of zebrafish sncga expression pattern and labeling the habenular complex in transgenic zebrafish using green fluorescent protein reporter gene. Dev. Dyn. 2009, 238, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Blundon, J.A.; Rong, Y.; Zakharenko, S.S.; Morgan, J.I. Impaired Locomotor Learning and Altered Cerebellar Synaptic Plasticity in pep-19/pcp4-Null Mice. Mol. Cell. Biol. 2011, 31, 2838–2844. [Google Scholar] [CrossRef] [PubMed]
- Mione, M.; Lele, Z.; Kwong, C.T.; Concha, M.L.; Clarke, J.D. Expression of pcp4a in subpopulations of CNS neurons in zebrafish. J. Comp. Neurol. 2006, 495, 769–787. [Google Scholar] [CrossRef]
- Kandemir, Y.B.; Sarikcioglu, L. Melatonin and its therapeutic actions on peripheral nerve regeneration. Folia Morphol. 2015, 74, 283–289. [Google Scholar] [CrossRef]
- Leung, J.W.-H.; Cheung, K.-K.; Ngai, S.P.-C.; Tsang, H.W.-H.; Lau, B.W.-M. Protective Effects of Melatonin on Neurogenesis Impairment in Neurological Disorders and Its Relevant Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 5645. [Google Scholar] [CrossRef]
- Liu, R.K.; Walford, R.L. The effect of lowered body temperature on lifespan and immune and non-immune processes. Gerontology 1972, 18, 363–388. [Google Scholar] [CrossRef]
- Wang, Q.; Tan, X.; Jiao, S.; You, F.; Zhang, P.-J. Analyzing Cold Tolerance Mechanism in Transgenic Zebrafish (Danio rerio). PLoS ONE 2014, 9, e102492. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.W.; Halver, J. Fish Nutrition; Elsevier: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Guo, J.-H.; Qu, W.-M.; Chen, S.-G.; Chen, X.-P.; Lv, K.; Huang, Z.-L.; Wu, Y.-L. Keeping the right time in space: Importance of circadian clock and sleep for physiology and performance of astronauts. Mil. Med Res. 2014, 1, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Xiao, H.; Luo, D.; Zhang, X.; Zhao, S.; Zheng, Q.; Li, Y.; Zhao, Y.; Dong, J.; Li, H.; et al. Circadian Rhythm Shapes the Gut Microbiota Affecting Host Radiosensitivity. Int. J. Mol. Sci. 2016, 17, 1786. [Google Scholar] [CrossRef] [PubMed]
- Konturek, P.C.; Brzozowski, T.; Konturek, S.J. Gut clock: Implication of circadian rhythms in the gastrointestinal tract. J. Physiol. Pharmacol. 2011, 62, 139–150. [Google Scholar]
- Ijiri, K.; Potten, C.S. Circadian Rhythms in the Incidence of Apoptotic Cells and Number of Clonogenic Cells in Intestinal Crypts after Radiation Using Normal and Reversed Light Conditions. Int. J. Radiat. Biol. 1988, 53, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, S.; Souffriau, J.; Libert, C. A General Introduction to Glucocorticoid Biology. Front. Immunol. 2019, 10, 1545. [Google Scholar] [CrossRef]
- Kumar, S.; Suman, S.; Fornace, A.J.; Datta, K. Space radiation triggers persistent stress response, increases senescent signaling, and decreases cell migration in mouse intestine. Proc. Natl. Acad. Sci. USA 2018, 115, E9832–E9841. [Google Scholar] [CrossRef]
- Moghadam-Kia, S.; Werth, V.P. Prevention and treatment of systemic glucocorticoid side effects. Int. J. Dermatol. 2010, 49, 239–248. [Google Scholar] [CrossRef]
- Black, R.J.; Hill, C.L.; Lester, S.; Dixon, W.G. The Association between Systemic Glucocorticoid Use and the Risk of Cataract and Glaucoma in Patients with Rheumatoid Arthritis: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0166468. [Google Scholar] [CrossRef] [PubMed]
- Chylack, L.T.; Peterson, L.E.; Feiveson, A.H.; Wear, M.L.; Manuel, F.K.; Tung, W.H.; Hardy, D.S.; Marak, L.J.; Cucinotta, F.A. NASA Study of Cataract in Astronauts (NASCA). Report 1: Cross-Sectional Study of the Relationship of Exposure to Space Radiation and Risk of Lens Opacity. Radiat. Res. 2009, 172, 10–20. [Google Scholar] [CrossRef]
- Zwart, S.; Gibson, C.; Smith, S. Space Flight Ophthalmic Changes, Diet, and Vitamin Metabolism. In Handbook of Nutrition, Diet and the Eye; Academic Press: New York, NY, USA, 2014; pp. 393–399. [Google Scholar] [CrossRef]
- Lang, T.; Van Loon, J.J.; Bloomfield, S.; Vico, L.; Chopard, A.; Rittweger, J.; Kyparos, A.; Blottner, D.; Vuori, I.; Gerzer, R.; et al. Towards human exploration of space: The THESEUS review series on muscle and bone research priorities. npj Microgravity 2017, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dickmeis, T. Glucocorticoids and the circadian clock. J. Endocrinol. 2008, 200, 3–22. [Google Scholar] [CrossRef] [PubMed]
- So, A.Y.-L.; Bernal, T.U.; Pillsbury, M.L.; Yamamoto, K.R.; Feldman, B.J. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl. Acad. Sci. USA 2009, 106, 17582–17587. [Google Scholar] [CrossRef] [PubMed]
- Ota, S.; Hut, R.; Riede, S.; Crosby, P.; Suchecki, D.; Meerlo, P. Social stress and glucocorticoids alter PERIOD2 rhythmicity in the liver, but not in the suprachiasmatic nucleus. Horm. Behav. 2020, 120, 104683. [Google Scholar] [CrossRef]
- Peluso, I.; Campolongo, P.; Valeri, P.; Romanelli, L.; Palmery, M. INTESTINAL MOTILITY DISORDER INDUCED BY FREE RADICALS: A NEW MODEL MIMICKING OXIDATIVE STRESS IN GUT. Pharmacol. Res. 2002, 46, 533–538. [Google Scholar] [CrossRef]
- Tseng, Y.-C.; Hwang, P.-P. Some insights into energy metabolism for osmoregulation in fish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2008, 148, 419–429. [Google Scholar] [CrossRef]
- Bradford, J.; Merrel, B.; Schaffer, M.; Talk, D. Advancing Torpor Inducing Transfer Habitats for Human Stasis to Mars; SpaceWorks: Atlanta, GA, USA, 2018. [Google Scholar]
- Imamura, S.; Ojima, N.; Yamashita, M. Cold-inducible expression of the cell division cycle gene CDC48 and its promotion of cell proliferation during cold acclimation in zebrafish cells1. FEBS Lett. 2003, 549, 14–20. [Google Scholar] [CrossRef]
- Conger, A.D. The Effect of Oxygen on the Radiosensitivity of Mammalian Cells. Radiol. 1956, 66, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Monobe, M.; Hino, M.; Sumi, M.; Uzawa, A.; Hirayama, R.; Ando, K.; Kojima, S. Protective effects of melatonin on gamma-ray induced intestinal damage. Int. J. Radiat Biol. 2005, 81, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Pajonk, F.; McBride, W.H. Ionizing radiation affects 26s proteasome function and associated molecular responses, even at low doses. Radiother. Oncol. 2001, 59, 203–212. [Google Scholar] [CrossRef]
- Fader, S.C.; Yu, Z.; Spotila, J.R. Seasonal variation in heat shock proteins (hsp 70) in stream fish under natural conditions. J. Therm. Biol. 1994, 19, 335–341. [Google Scholar] [CrossRef]
- Todgham, A.E.; Hoaglund, E.A.; Hofmann, G.E. Is cold the new hot? Elevated ubiquitin-conjugated protein levels in tissues of Antarctic fish as evidence for cold-denaturation of proteins in vivo. J. Comp. Physiol. B 2007, 177, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Somero, G.N.; Hochachka, P.W. Biochemical adaptation to the environment. Am. Zool. 1971, 11, 159–167. [Google Scholar] [CrossRef]
- Hochachka, P.W.; Lewis, J.K. Enzyme variants in thermal acclimation. Trout liver citrate synthases. J. Biol. Chem. 1970, 245, 6567–6573. [Google Scholar] [CrossRef]
- Healy, T.M.; Schulte, P.M. Patterns of alternative splicing in response to cold acclimation in fish. J. Exp. Biol. 2019, 222, jeb193516. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.M.; Sunagawa, G.A.; Soya, S.; Abe, M.; Sakurai, K.; Ishikawa, K.; Yanagisawa, M.; Hama, H.; Hasegawa, E.; Miyawaki, A.; et al. A discrete neuronal circuit induces a hibernation-like state in rodents. Nat. Cell Biol. 2020, 583, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Hrvatin, S.; Sun, S.; Wilcox, O.F.; Yao, H.; Lavin-Peter, A.J.; Cicconet, M.; Assad, E.G.; Palmer, M.E.; Aronson, S.; Banks, A.S.; et al. Neurons that regulate mouse torpor. Nat. Cell Biol. 2020, 583, 115–121. [Google Scholar] [CrossRef]
- Ju, H.; So, H.; Ha, K.; Park, K.; Lee, J.-W.; Chung, C.-M.; Choi, I. Sustained torpidity following multi-dose administration of 3-iodothyronamine in mice. J. Cell. Physiol. 2010, 226, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Glossmann, H.H.; Lutz, O.M.D. Torpor: The Rise and Fall of 3-Monoiodothyronamine from Brain to Gut—From Gut to Brain? Front. Endocrinol. 2017, 8, 118. [Google Scholar] [CrossRef]
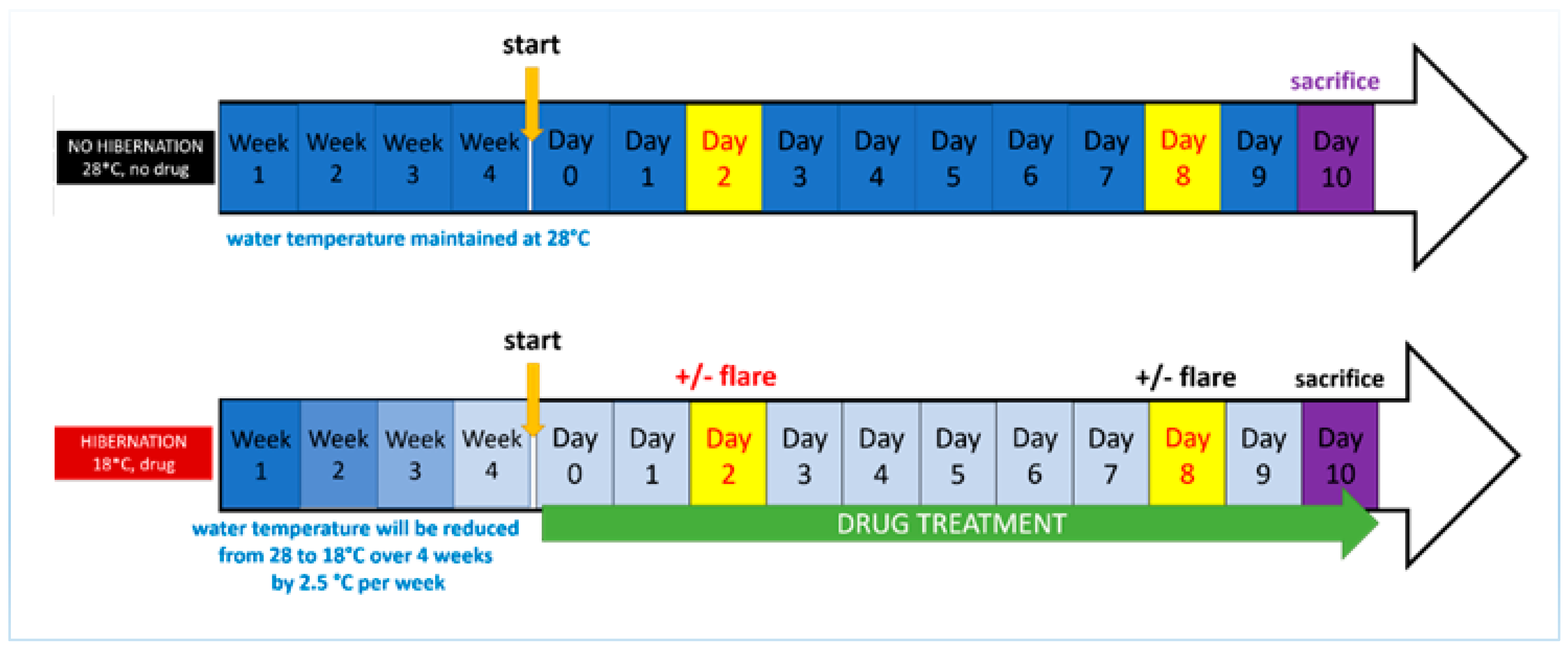
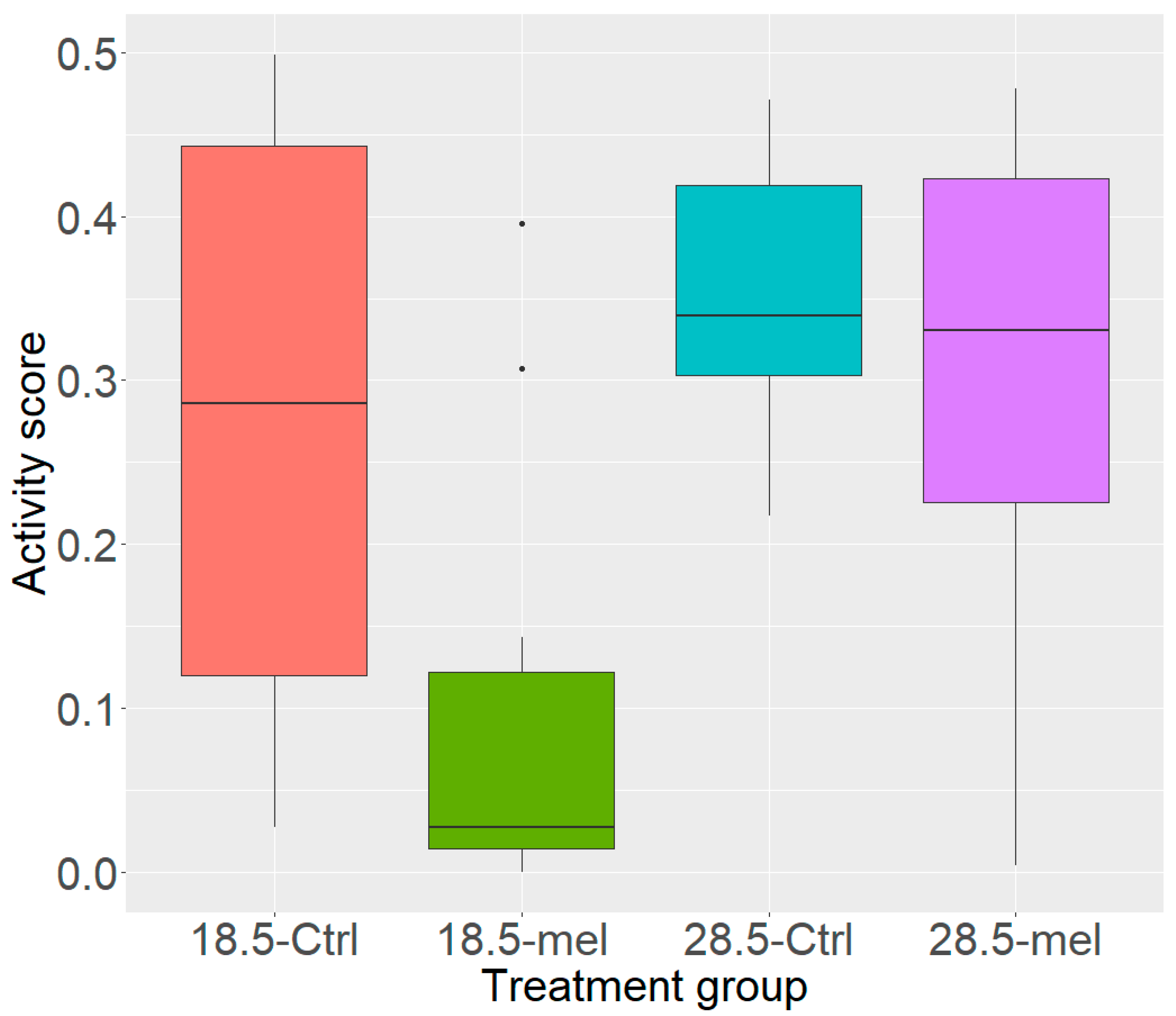
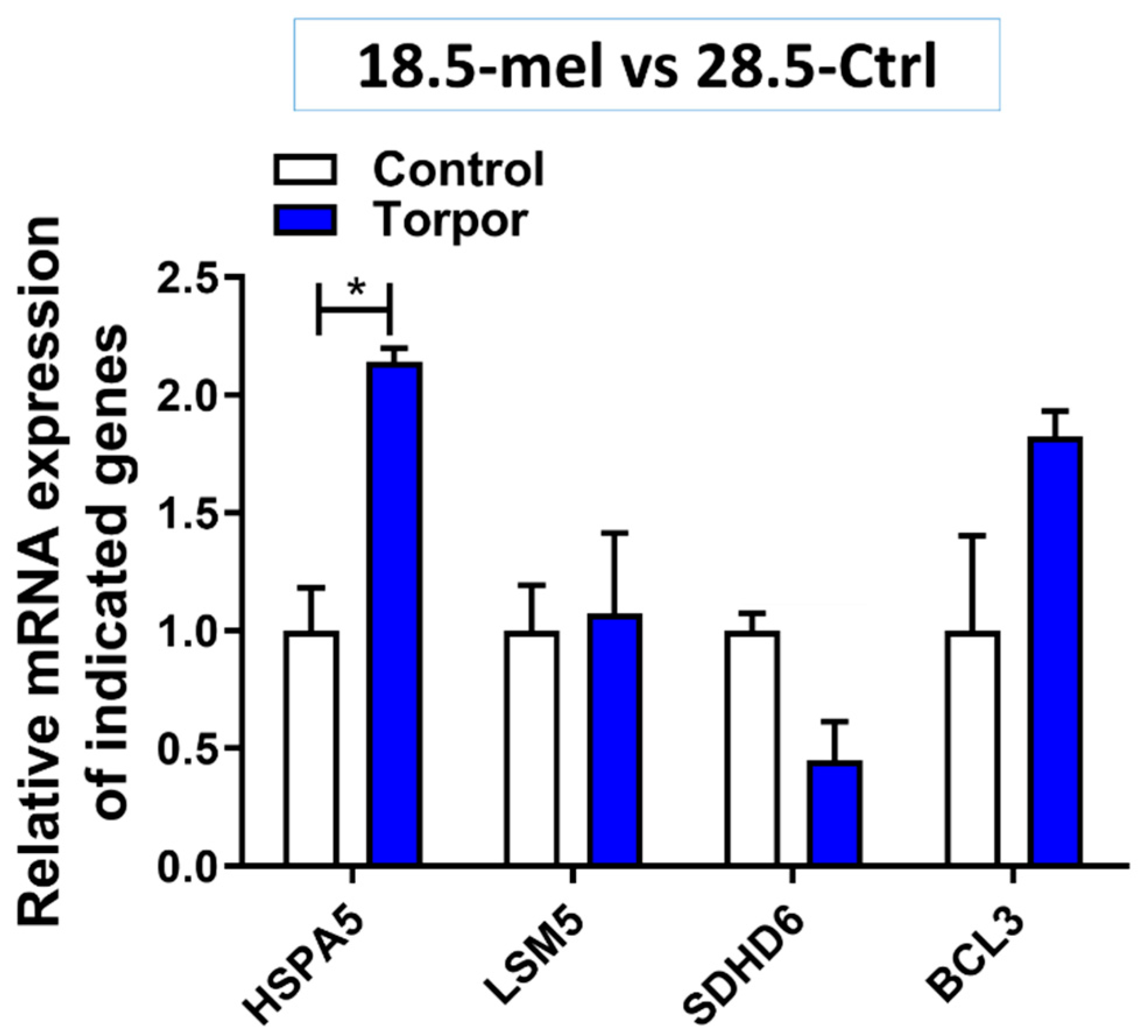
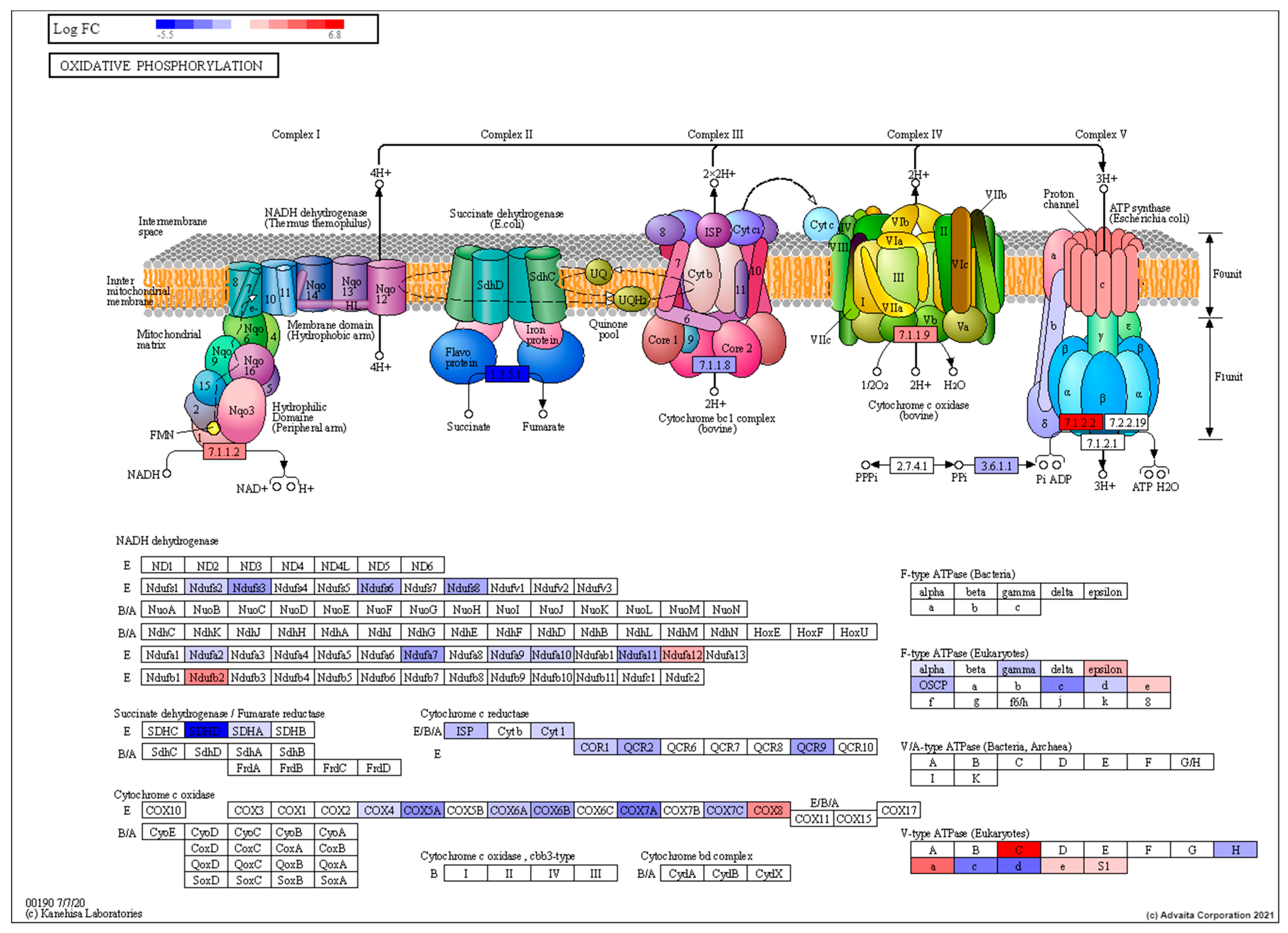
| Group | Key | Sample (N) | Radiation (cGy) | Water Temperature (°C) | Melatonin (µM) |
|---|---|---|---|---|---|
| Control | Ctrl | 6 | 0 | 28.5 | 0 |
| Melatonin | 28.5-mel | 6 | 0 | 28.5 | 24 |
| Temperature | 18.5-Ctrl | 6 | 0 | 18.5 | 0 |
| Torpor | 18.5-mel | 6 | 0 | 18.5 | 24 |
| Radiation | 28.5-rad | 6 | 32.64 | 28.5 | 0 |
| Torpor + radiation | 18.5-mel-rad | 6 | 32.64 | 18.5 | 24 |
| radNET_Only | OPP_13 | torpor_NET_Only |
|---|---|---|
| rho mpz slc6a1b hoxb1b | mpx hmgb3a fabp7a myh7 mylk ins epha4b sst1.1 foxj1b tnni1b sulf2a actn2b sncga | msx2b lmx1bb wnt11f2 myl7 tbx5a smyd1b pomca sox19a wnt4 gbgt1l4 emx3 pcp4a irx2a foxi1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cahill, T.; da Silveira, W.A.; Renaud, L.; Williamson, T.; Wang, H.; Chung, D.; Overton, I.; Chan, S.S.L.; Hardiman, G. Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model. Cells 2021, 10, 906. https://doi.org/10.3390/cells10040906
Cahill T, da Silveira WA, Renaud L, Williamson T, Wang H, Chung D, Overton I, Chan SSL, Hardiman G. Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model. Cells. 2021; 10(4):906. https://doi.org/10.3390/cells10040906
Chicago/Turabian StyleCahill, Thomas, Willian Abraham da Silveira, Ludivine Renaud, Tucker Williamson, Hao Wang, Dongjun Chung, Ian Overton, Sherine S. L. Chan, and Gary Hardiman. 2021. "Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model" Cells 10, no. 4: 906. https://doi.org/10.3390/cells10040906
APA StyleCahill, T., da Silveira, W. A., Renaud, L., Williamson, T., Wang, H., Chung, D., Overton, I., Chan, S. S. L., & Hardiman, G. (2021). Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model. Cells, 10(4), 906. https://doi.org/10.3390/cells10040906








