Abstract
Refine current agricultural practices considering environmental changes are crucial for finding tolerant rice varieties that can meet the demands of human consumption. To this end, stability analysis assesses a crop genotype’s ability to adapt to various conditions. Therefore, the objective of this study was to (1) examine the interaction between rice genotypes and environmental conditions; (2) evaluate the stability of twelve rice genotypes using various stability methods; (3) identify representative environments for multi-environment testing; and (4) determine superior genotypes for specific environments. The evaluated rice cultivars were Sakha 101, Sakha 104, Sakha 105, Sakha 106, Sakha 107, Sakha 108, Giza 177, Giza 178, Giza 179, Giza 182, Egyptian Yasmine, and Sakha super 300. The experiment followed a strip-plot design, with three replications. The findings revealed significant differences among the rice varieties across various environments for the majority of the assessed characteristics. The joint regression analysis of variance demonstrated highly significant differences among rice cultivars for all the studied traits in terms of genotype-by-environment interaction (G × E). The statistical significance of the interaction between genetic and environmental factors was evident for all variables demonstrating heritable variation among the rice cultivars, specifically Sakha 108, Sakha 104, Giza 177, and Giza 178, concerning grain yield per feddan. These rice cultivars exhibited stability parameters that were not significantly different from unity for the regression coefficient (bi) and from zero for the deviations from regression (S2di) for those traits. Overall, stability criteria are essential for ensuring reliable rice production, meeting human consumption, advancing genetic improvement, and promoting environmental sustainability in agriculture.
1. Introduction
Rice, a vital cereal that is cultivated as a staple food crop, faces challenges in meeting human consumption demands, which are expected to increase by an additional 112 million metric tons by the year 2035 [1]. To address this growing need, it is imperative to optimize existing farming methods, manage water resources effectively, and explore opportunities for developing new rice varieties [2].
Rice cultivation spans various agroecology and cropping systems, including rain-fed highland and lowland areas, irrigated fields, and mangrove ecosystems [3]. While upland rice constitutes a relatively minor portion (approximately 11%) of global rice output, it holds significant importance in local rice production in both arid and semiarid regions [4,5]. The challenges posed by limited water resources for lowland rice agriculture are exacerbated by the impacts of climate change. In response, upland rice emerges as a potentially sustainable solution to tackle food security concerns, as suggested by [3,5,6].
Before commercial release, plant breeders typically evaluate the performance of the new varieties across various environments to assess their stability and consistency [7]. Genotypes that demonstrate consistent performance across diverse environments are preferred, but the complexity of genotype–environment interactions poses challenges in genotype selection [8,9,10]. Therefore, it is crucial to employ statistical methods to evaluate genotype stability before commercial release [11,12]. To this end, the applied statistical methods for assessing genotype stability can be classified into parametric and nonparametric stability statistics [13,14]. Parametric stability statistics encompass various univariate and multivariate methods. Univariate methods comprise techniques such as Wricke’s equivalence (Wi2) [15], Shukla’s stability variance (S2_Shu.) [16], coefficient of variance (CV) [17], Environmental variance (S2_Env.) [18], Mean-variance component (q) [19], GE variance component (q’) [20], and regression coefficient (bi) [21], among others. On the other hand, multivariate approaches include the additive main effects and multiplicative interaction (AMMI) model [22] and the GGE biplot method [23], which can accurately predict genotype-by-environment interactions and identify optimal genotypes across diverse testing environments [24].
The objectives of this study are to (1) study the interaction of genotypes and the environment; (2) determine the stability of rice genotypes with various stability methods; (3) determine representative environments for multi-environment testing; (4) determine superior genotypes in specific environments; and (5) identify high-yielding rice genotypes with consistent performance across six distinct locations over two consecutive years (2020–2021) (three sowing dates during two seasons). This is achieved by evaluating the effectiveness of various univariate and multivariate stability criteria. The identification of high-yielding rice genotypes is essential for ensuring food security and enabling adaptation to climate change in rice-producing regions worldwide.
2. Materials and Methods
This study, conducted at the rice research farm Sakha Agricultural station in Kafr Elsheikh, Egypt, aimed to assess 12 rice cultivars over the 2020 and 2021 seasons, which were subjected to three different sowing dates.
2.1. Rice Cultivars and Experimental Design
Twelve rice cultivars, namely Sakha 101, Sakha 104, Sakha 105, Sakha 106, Sakha 107, Sakha 108, Giza 177, Giza 178, Giza 179, Giza 182, Egyptian Yasmine, and Sakha super 300, were evaluated. The experiment was set using a split-plot design with three replications. Sowing dates were allocated in the main plots, and rice varieties were applied in the sub-plots. The plot size was 2 m × 5 m (10 m2).
The chemical and mechanical analyses of the soil and organic matter were conducted following the methods described by [25] at the Agricultural Research Center, Ministry of Agriculture, Egypt. The chemical and physical properties of the soil at the experimental site, measured at a depth of 0–30 cm, are shown in Table 1.

Table 1.
Mechanical and chemical soil analyses of the experimental site during the 2020 and 2021 seasons.
Weather data for the two seasons were collected from the Agriculture Research Center, Field Crops Research Institute, and Rice Research and Training Center in Sakha, Kafr El-Sheikh, Egypt (Table 2).

Table 2.
The monthly minimum and maximum air temperature (°C) and relative humidity (%) at Sakha Agricultural research station during the 2020 and 2021 rice growing seasons.
2.2. Field Preparation and Planting
The experiment was carried out in plots with seven rows, each 5 m in length, and a plant spacing of 20 m × 20 cm. Bed nursery and permanent field preparation followed the guidelines provided by the Rice Research and Training Center (RRTC). Phosphorus fertilizer in the form of mono-super phosphate (15%) at the rate of 36 kg P2O5/ha and potassium in the form of potassium sulfate (48% K2O) at the rate of 57 kg K2O/ha were applied during land preparation. Nitrogen in the form of urea (46.5% N) was applied in two splits, i.e., two-thirds as basal application incorporated into the soil immediately before flooding and one-third after 30 days from the first dose.
2.3. Transplantation and Spacing
Seedlings were transplanted from the nursery to the permanent field after 25 days.
Transplantation was performed into sub-sub-plots using plant spacing (20 × 20, cm) for distances between hills and rows. Seedlings were transplanted into hills at a density of 3–4 seedlings per hill with the following schedule: E1 = the first sowing date in the first season 20 April 2020; E2 = the second sowing date in the first season 10 May 2020; E3 = the third sowing date in the first season 30 May 2020; E4 = the first sowing date in the second season 20 April 2021; E5 = the second sowing date in the second season 10 May 2021; and E6 = the third sowing date in the second season 30 May 2021.
2.4. Studied Characteristics
This study proposed the evaluation of additional attributes, including the number of filled spikelets per panicle, seed set percentage, 1000-grain weight, and grain yield per plant. Overall, this study utilized a rigorous experimental design and methodology to evaluate the performance of different rice cultivars under varying sowing dates during the two seasons, providing valuable insights into rice production in the region.
2.5. Statistical Analyses
The stability and adaptability of rice genotypes were determined using parametric and nonparametric stability in addition to multivariate analysis. Parametric stability included linear regression coefficient (bi) and AMMI stability indexes, whereas nonparametric stability included Si. Multivariate analysis was performed using AMMI and GGE biplot.
Stability studies were conducted using the Eberhart and Russell model [26] with metan package in R software 2021, with particular attention to the G × E factor. A genotype with a regression coefficient of 1 (bi = 1) and a deviation not statistically significant from zero (S2di = 0) is deemed stable, exhibiting a unity response.
Mean comparisons among genotypes were conducted using the LSD test, employing a significance threshold of 5%. To calculate variance components, ANOVA was performed using a split-plot design. The GenStat statistical tool (12th edition) facilitated this analysis. A Bayesian network is a probabilistic directed acyclic graph, where nodes represent variables such as traits and edges represent causal or conditional dependencies among these variables. The aim of analysis algorithms in Bayesian networks is to infer the relations of conditional or causal dependence among network variables based on the Bayes theorem. So, Bayesian networks belong to the class of structural network models. Bayesian network was built using the package bnlearn in R software.
3. Results
Substantial variations were observed in the mean square values for the number of filled grains per panicle, seed set percentage, 1000-grain weight, and grain yield per panicle among the different Egyptian rice types and sowing dates (Table 3). Variations in the habitats and genotypes had a significant impact on all the tested traits. Moreover, clear interactions were observed between years and environments, as well as between genotypes and years, across all traits except for the 1000-grain weight, where the disparity was not statistically significant. Furthermore, the analysis revealed a substantial and significant mean square of interaction across genotypes, years, and environments for the number of filled grains per panicle and seed set percentage (Table 3). The results demonstrated that the investigated genotypes had varying responses to environmental variables, highlighting the significance of evaluating genotypes across multiple environments to identify the most well-suited genotypes for specific situations. The environmental mean squares were the primary contributor to the overall mean squares for all traits. Furthermore, the variations attributed to different environments were greater than the variations resulting from the interactions between genotypes and environments for all the observed traits. Hence, the primary factor influencing the variations in the performance of rice genotypes in these trials was the environment, rather than the differences arising from genotype-by-environment interaction.

Table 3.
Mean square of seed set (%), 1000-grain weight, and grain yield/plant of the 12 rice cultivars.
3.1. Environment Effect
The mean number of filled grains per panicle decreased by 11.6 grains when comparing the sowing dates condition to the third sowing date condition (Table 4). When comparing the second and third sowing dates to the first sowing date, the seed set percentage, 1000-grain weight (in grams), and grain yield per plant were lower. The results demonstrated that the diverse growing conditions led to a significant difference in average yields, ranging from 45.0 g/plant under favorable initial planting date conditions to 42.5 g/plant under the third stress condition (Table 5).

Table 4.
Means of the 12 rice genotypes over years and environments.

Table 5.
AMMI analysis for the 12 rice genotypes.
The AMMI 1 biplot (Figure 1a) represents the relationship between grain yield and IPCA 1. In this plot, rice genotypes that fall on the same vertical line indicate similarity in yield, while those on the same horizontal line indicate similarity in interaction pattern. According to the AMMI 1 biplot, the rice genotypes Sakha104, Sakha105, Sakha106, Sakha108, Giza177, Giza178, and SR300 exhibited a high level of stability in grain production, and they are considered extensively adapted lines (Figure 1a). The rice genotypes Sakha101, Sakha107, Giza179, Giza182, and Egyptian Yasmine exhibited relatively low stability in grain yield due to their considerable distance from the origin and their specific adaptability to particular environments. The genotype S101 exhibited superior performance in the E1 and E2 environments, while the genotypes G182 and EY were specifically adapted to environments E3 and E5, respectively. Furthermore, the S106, S108, and G178 genotypes were specifically identified as being well suited to environment E4. In the AMMI 2 biplot (Figure 1b), the scores of IPCA1 plotted against IPCA2 show the stability of the genotypes across different environments. Environments E1, E2, and E4 had relatively small radii and exhibited weak interacting forces, whereas environments E3, E5, E6, and E7 had long radii and exerted substantial interaction. Genotypes S104, S105, and G178 were located in close proximity to the origin, indicating their insensitivity to environmental interaction forces. Conversely, the remaining genotypes were situated further from the zero line, signifying their heightened responsiveness. The genotypes S106 and EY were found to be the most well suited for site E6, whereas the genotypes S108 and SR300 showed a close similarity in environment E4.
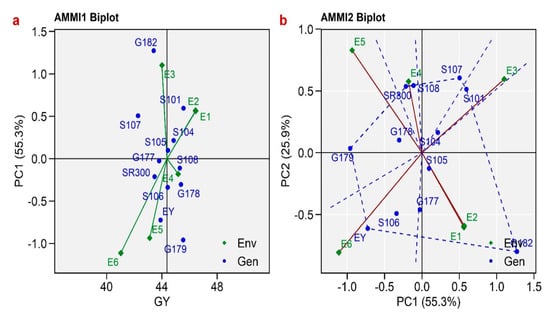
Figure 1.
(a) AMMI 1 (additive main effects and multiplicative interaction) biplot for grain yield of the 12 rice genotypes and 6 environments using genotypic and environmental scores; (b) AMMI 2 biplot for grain yield showing the interaction of IPCA1 (interaction principal component axes) against IPCA2 scores of the 12 rice genotypes in 6 environments.
3.2. Mean Performance
The average number of filled grains per panicle varied between 121.06 for Giza 179 and 168.0 grains for Giza 178, as shown in Table 6. The seed set percentages of the twelve rice genotypes exhibited substantial variations, as indicated by the data presented in Table 6. The seed set percentage was highest for Sakha 101 and lowest for E. Yasmin.

Table 6.
The average number of filled grains/panicle and seed sets for the twelve rice cultivars in different environments.
The mean 1000-grain weight was highest for Sakha 106, Sakha 105, and Sakha 108, with values of 29.1, 29.0, and 29.0, respectively. The genotype Giza 178 (22.1) had the lowest value among all genotypes (Table 7). The data presented clearly demonstrate that the genotypes had substantial variations in grain production per plant, which were statistically significant (Table 7). The Sakha 108, Giza 179, and Giza 178 varieties exhibited the highest average weights per plant, measuring 45.6, 45.5, and 45.4 g, respectively. The genotype Sakha 107 had the lowest yield, measuring 42.3 g per plant. The grain production per plant varied between 42.3 and 45.6 g per plant across the different genotypes. The analysis of variations was conducted on the combined data from six different environments, comprising three sowing dates and two seasons. Table 6 and Table 7 demonstrate notable variations in features across different contexts. The genotypic variation (s2g) was greater than the error variation (s2e) for the number of filled grains, seed set percentage, 1000-grain weight, and grain yield per plant. This indicates that environmental effects have little influence on these traits. Conversely, the s2g was significantly larger than s2 for all traits, indicating that genetic effects play a crucial role in the expression of these traits (Table 7).

Table 7.
Average 1000-grain weight and grain yield/plant for the twelve rice cultivars over environments.
The acquired results are consistent with those reported by [4]. The phenotypic (PCV) and genotypic (GCV) coefficients of variability were estimated and found to have modest variances for all traits evaluated across the six environments (Table 8). It is noteworthy that certain rice cultivars consistently exhibited similar characteristics across various conditions, including the number of panicles per plant, panicle weight, the number of filled grains per panicle, seed set percentage, 1000-grain weight, and grain yield per plant. However, certain types of rice varied in performance across different environments. For example, the rice cultivar Sakha 108 showed superiority in terms of the number of filled grains per panicle, 1000-grain weight, and grain weight per panicle in environment E1. On the other hand, Giza 178 performed better in terms of the number of filled grains per panicle, while Sakha 101 had a higher seed set percentage.

Table 8.
AMMI-based stability 12 indexes (low numbers indicate stability) and superiority index based on stability and yield (high numbers indicate superiority).
3.3. Stability Parameters
The predictability of genotypes for the number of filled grains per panicle varied from 0.04 for Sakha 106 to 1.37 for Sakha super 300 (Table 9). The examined genotypes were classified into three categories based on the regression coefficient (bi) values. The initial group consisted of the most genetically stable genotypes, specifically Sakha 107, which exhibited the coefficient of regression bi values equal to 1. The second group consisted of genotypes that were better suited to unfavorable environments, namely Sakha 104, Sakha 106, and Giza 178 (with the lowest bi values). On the other hand, genotypes Sakha 101, Sakha 105, Sakha 108, Giza 177, Giza 179, Giza 182, E. Yasmin, and Sakha super 300 were sensitive to input and adapted to high-potential environments. Regarding the grain yield, the rice cultivars Sakha 106, Sakha 107, Sakha 108, and Giza 178 had a bi value close to 1 (Table 9), suggesting that these cultivars were well suited to varied environmental conditions. Genotypes Giza 177, E. Yasmin, and Sakha super 300 exhibited bi values below 1, showing their suitability for unfavorable conditions. Sakha 101, Sakha 104, Sakha 105, and Giza 182 displayed a bi value greater than 1.

Table 9.
Mean performance and phenotypic stability measurements of the number of filled grains/panicle and seed set (%) for the twelve genotypes in three environments.
3.4. Environmental Indices
An environment index is a quantitative metric that assesses the appropriateness of a specific environment in a given area. By analyzing these indicators, we may determine the optimal conditions required to fully use the genetic potential. If an environment has a positive index value, it can be inferred that it is a favorable environment for genotypes. The indices of E1 and E4 were 2.08 and 2.07, respectively. The value for E4 was 0.8, while the values for E3, E5, and E6 were −0.37, −1.25, and −3.34, correspondingly.
Figure 2 displays the results of a GGE biplot analysis, which is widely regarded as the most efficient method for summarizing both genotypes and the interaction between genotypes and the environment. This analysis helps select the most suitable genotype for each environment and evaluate the stability of the tested genotypes. Figure 2a demonstrates that genotypes located closer to the origin exhibit greater stability compared to those located further away from the origin. Furthermore, the environments were grouped into distinct clusters, namely (E1, E2, E3, and E4) and (E5 and E6). Furthermore, the genotype characterized by a long-dashed line has a higher GY (grain yield) compared to the genotype with a short-dashed line. The genotypes positioned on the vertices of the polygon in Figure 2b are the most responsive to different settings, meaning that they show greater variability in their performance. On the other hand, the genotypes inside the polygon are more stable across environments, indicating that they are less influenced by changes in environmental conditions. The lines of equality partition the graph (Figure 2b) into five sectors. A total of six habitats were preserved, divided into two sectors, and further categorized into two mega-environments.
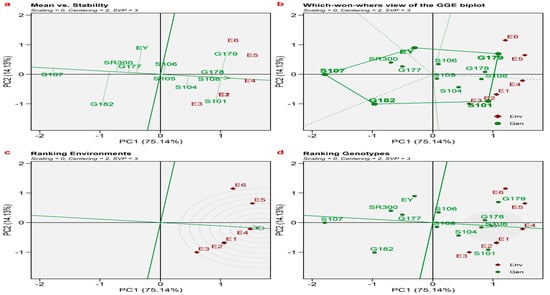
Figure 2.
(a) Means versus stability; (b) polygon view of GGE biplot (which-won-where model); (c) ranking environment; (d) ranking genotypes.
In the first mega-environment, which included E1, E2, E3, and E4, the top-performing varieties were Sakha 101, Sakha 104, Sakha 105, and Sakha 108. Conversely, in the second mega-environment encompassing E5 and E6, the leading varieties were Sakha 106, Giza 178, and Giza 179. Figure 2c,d depict the respective rankings of environments and genotypes.
The Bayesian network in Figure 3 depicts the impact of treatments on traits, as well as the influence of traits on other traits, using the Bayesian information criterion (BIC). A greater negative BIC value implies a more pronounced effect. The data showed that genotypes had a significant and direct impact on plant height (PH), days to heading (DTH), grain weight (PW), thousand-kernel weight (TKW), the total grain number per plant (TGNP), and the filled grain number per plant (FGNP). Additionally, the influence of genotypes on FGNP was mediated indirectly through its direct effect on TGNP. Conversely, the environment had a significant direct impact on PL (primary productivity) and PNPP (primary net primary productivity), as well as an indirect impact on PL through PNPP. Additionally, the environment had an indirect impact on FGNP (final gross net primary productivity) and PW (plant water) through PNPP, as well as an indirect impact on GY (growth yield) through PL. PW was influenced by both genotypes and PNPP, but GY was significantly impacted by HI. To summarize, genetics exerted a greater influence on traits compared to the environment.
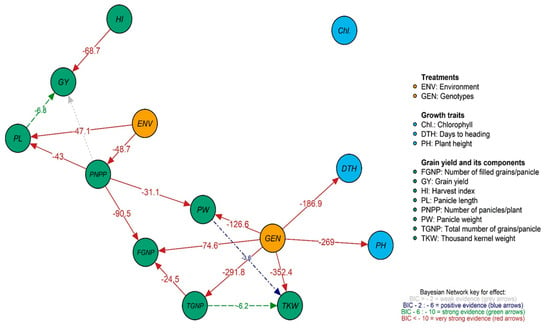
Figure 3.
Bayesian network.
3.5. Stability and Superiority Indexes
The total of twelve stability indexes, along with one superiority index, is displayed (Table 9). Based on the stability indexes, Sakha 105 exhibited the highest level of stability among the genotypes, whereas Giza 182 displayed the lowest level of stability. The stability indices primarily consider the stability of genotypes in different conditions while disregarding the yield potential of the genotype. In contrast, the superiority index takes both the stability and prospective yield of the genotype into consideration. According to the superiority index, Giza 178 exhibited the highest level of superiority among the genotypes, whilst Giza 182 displayed the lowest level of superiority.
4. Discussion
Multi-environmental trials are crucial in plant breeding projects since they assess crop yield and adaptability across several settings. To enhance the accuracy of genotype selection, one can assess the genotype-by-environment interaction using mega-environment analysis, genotype evaluation, and multi-environment stability analysis [27]. Genetic diversity can be achieved by breeding inter-subspecific cultivars or accessions having distinct genetic lineages [28,29,30]. Environmental variability can be obtained by increasing the magnitudes of location factors such as geographic distance, variances in climatic regions (temperate versus tropical), and differences in elevation [31,32].
Our breeding operations primarily prioritize the preservation of the commercial worth of cultivars. Crop enhancements are primarily achieved by the interbreeding of superior cultivars within restricted genetic pools, resulting in limited genetic diversity among the cultivars [33,34,35]. Furthermore, the test environments were located in geographically proximate agriculturally advantageous flat regions. The stability of a cultivar/genotype pertains to the degree of variation in the phenotypic manifestation of crop performance across diverse environmental conditions. The aim of this study was to assess the idea of “dynamic” stability, which refers to the performance of a stable genotype in different environments. Consequently, not all genotypes exhibited the same response to the environment [36,37].
The results of our study indicate that the genotypes we tested in various locations over two consecutive years are extremely susceptible to climatic zones and environmental factors [38]. The variations seen may be attributed to the disparities in terrain and climatic circumstances among various sites and years where the experiments were carried out. Achieving effective breeding outcomes for yield and related traits in rice necessitates precise measurement and analysis of genotype, environment, years, and their interactions within the breeding protocol [39]. The current discovery of noteworthy sources of variation has been previously documented in rice and other cereal crops [10,27]. Significant effects of both linear and nonlinear components of G × E interaction were seen for all examined traits, as evidenced by highly significant mean squares resulting from G × E (linear) interaction and pooled deviation. The reported results are consistent with those obtained by [40]. The grain production of different rice varieties exhibited significant variations in response to changes in climatic variables, such as planting dates and years [41].
Hence, the cultivar that exhibits consistent and predictable crop output is suitable for implementing risk management strategies to mitigate yield losses in challenging lowland environments [42]. A statistically significant difference has been observed across cultivars and settings in a linear manner, as opposed to a nonlinear manner, in which the deviations are aggregated. This finding highlights the significance of the interplay between genotype and environment in determining grain yield performance and its components across several locations [28,43]. Therefore, the phenotypic stability assessment took into account both the linear (bi) and nonlinear (S2di) components of G × E interactions [23,44]. Moreover, it has been documented that a high average value of the linear regression coefficient, equivalent to the nonlinear coefficient, is advised for achieving a diverse range. Furthermore, it has been suggested that nonlinear regression can be employed to assess stability, whereas linear regression can be utilized to evaluate the impact of different environmental circumstances on varietal response [45]. Hence, it is crucial to take into account the average and the deviation from the regression of each cultivar when assessing the stability and linear regression for evaluating the responsiveness of the variety. Stresses can limit the growth resources available to plants, resulting in a reduction in the size of plant organs such as leaves, tillers, and spikes [41].
The stability regression coefficient (bi) and deviation from regression (S2di) for the genotypes under study are shown in Table 7. A stable genotype refers to a genotype that exhibits a consistently high level of performance, with a regression coefficient (bi) equal to 1 and a deviation from regression of 0. This indicates that the system is well suited for favorable situations, but its performance will be subpar in unfavorable environments. Therefore, these rice cultivars possess significant value for conducting investigations in harsh environments, as the identified types have been suggested for their suitability in unfavorable environmental conditions, as indicated in Table 9 [2].
In the context of selecting and developing superior rice cultivars, genetic studies on yield and yield quality serve as crucial benchmarks for both farmers and industry professionals. As highlighted in previous research, evaluations incorporating various stability statistics offer a more comprehensive understanding of genotype performance across different environments. For instance, a study on sweet potato genotypes in West Java, Indonesia, demonstrated the importance of genotype-by-environment interactions (GEIs) in identifying high-yield and stable genotypes. This research utilized a randomized block design across three environments and found significant effects of genotypes, environments, and GEIs on yield and quality traits. Specifically, GEIs contributed substantially to the variability observed in yield, sweetness, moisture content, tuber diameter, and tuber length. We identified five genotypes (G4, G6, G7, G31, and G32) as both high-yielding and stable across environments. Furthermore, genotype-by-yield ×trait (GYT) analysis revealed seven superior genotypes based on yield and quality, namely G7, G15, G4, G20, G6, G31, and G14. Notably, G4, G6, G7, and G31 were recognized for their high and stable yields combined with favorable quality traits. These findings underscore the value of integrating multiple stability measures and GYT analysis to identify and release superior varieties [46].
The results of pooled ANOVA for stability according to Eberhart and Russell [29], as well as the AMMI model, revealed that the variance of genotypes was significant for rice grain yield, indicating that the performance of rice genotypes was different; the genotypes also had differential responses to the changes in years and locations (environments).
In this study, the genotype Sakha 105 had regression coefficients (bi) of 1.11 and was observed to be stable. According to the Eberhart and Russell [29] model, a slope of >1.0 with high mean yield and nonsignificant squared deviation (S2di) is suitable for a favorable environment.
The AMMI model defines the genotype–environment interaction. The AMMI model revealed that a major part of the variation in rice grain yield is explained by the environment, which indicates environmental diversity. The AMMI1 biplot analysis showed that the variation was due to the main effect of rice grain yield and the interaction effect. Genotypes with IPCA1 scores close to zero were characterized as having low interaction effects and consequently considered stable.
5. Conclusions
This study highlights that environmental factors are the primary drivers of variations in rice yield and related traits among Egyptian rice genotypes, with genotype–environment interactions playing a secondary role. Certain genotypes, like Sakha 104 and Giza 178, demonstrated stable performance across different environments, while others were better suited to specific conditions. Notably, Sakha 106, Sakha 107, Sakha 108, and Giza 178 showed bi values close to 1, indicating they are well adapted across diverse environments. In contrast, Giza 177, E. Yasmin, and Sakha Super 300, with bi values below 1, were more suitable for poor environments, while Sakha 101, Sakha 104, Sakha 105, and Giza 182, with bi values above 1, were better suited to favorable environments but may underperform in less favorable conditions. These findings underscore the importance of multi-environmental trials for selecting stable, high-yielding rice cultivars. The results provide a foundation for targeted breeding programs aimed at optimizing rice productivity by focusing on genotypes that consistently perform well across various growing conditions, considering both stability and adaptability.
Author Contributions
Conceptualization, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; methodology, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; software, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; validation, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; formal analysis, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; investigation, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; resources, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; data curation, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; writing—original draft preparation, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; writing—review and editing, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; visualization, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; supervision, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; project administration, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T.; funding acquisition, M.S.A.E.-A., M.I.A.-Y., F.A.S., M.S., M.A.G., O.M.I., M.Y.K., W.H.A.-Q., M.A.A.-M. and A.M.E.-T. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the researchers supporting Project Number (RSP2024R293), King Saud University, Riyadh, Saud Arabia.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Habib, M.A.; Azam, M.G.; Haque, A.; Hassan, L.; Khatun, M.S.; Nayak, S.; Abdullah, H.M.; Ullah, R.; Ali, E.A.; Hossain, N.; et al. Climate-smart rice (Oryza sativa L.) genotypes identification using stability analysis, multi-trait selection index, and genotype-environment interaction at different irrigation regimes with adaptation to universal warming. Sci. Rep. 2024, 14, 13836. [Google Scholar] [CrossRef] [PubMed]
- El-Aty, M.S.A.; Katta, Y.S.; Abd, A.E.M.B.E.; Mahmoud, S.; Ibrahim, O.M.; Wali, A.M.; El-Shehawi, A.M.; Elseehy, M.M.; El-Saadony, M.T.; El-Tahan, A.M. Assessment of grain quality traits in rice under normal and water deficit condition. Saudi J. Biol. Sci. 2022. [Google Scholar] [CrossRef]
- Al Azzawi, T.N.I.; Khan, M.; Hussain, A.; Shahid, M.; Imran, Q.M.; Mun, B.-G.; Lee, S.-U.; Yun, B.-W. Evaluation of Iraqi Rice Cultivars for Their Tolerance to Drought Stress. Agronomy 2020, 10, 1782. [Google Scholar] [CrossRef]
- Abd-El-Aty, M.S.; Abo-Youssef, M.I.; Bahgt, M.M.; Ibrahim, O.M.; Faltakh, H.; Nouri, H.; Korany, S.M.; Alsherif, E.A.; AbdElgawad, H.; El-Tahan, A.M. Mode of gene action and heterosis for physiological, biochemical, and agronomic traits in some diverse rice genotypes under normal and drought conditions. Front. Plant Sci. 2023, 14, 1108977. [Google Scholar] [CrossRef] [PubMed]
- Akanksha, A.; Jaiswal, H.K. Combining ability studies for yield and quality parameters in basmati rice (Oryza sativa L.) genotypes using diallel approach. Electron. J. Plant Breed. 2019, 10, 9–17. [Google Scholar] [CrossRef]
- Aslam, M.M.; Rashid, M.A.R.; Siddiqui, M.A.; Khan, M.T.; Farhat, F.; Yasmeen, S.; Khan, I.A.; Raja, S.; Rasool, F.; Sial, M.A.; et al. Recent Insights into Signaling Responses to Cope Drought Stress in Rice. Rice Sci. 2022, 29, 105–117. [Google Scholar] [CrossRef]
- Aswidinnoor, H.; Listiyanto, R.; Rahim, S.; Holidin; Setiyowati, H.; Nindita, A.; Ritonga, A.W.; Marwiyah, S.; Suwarno, W.B. Stability analysis, agronomic performance, and grain quality of elite new plant type rice lines (Oryza sativa L.) developed for tropical lowland ecosystem. Front. Sustain. Food Syst. 2023, 7, 1147611. [Google Scholar] [CrossRef]
- Awad-Allah, M.M.A. Heterosis and Combining ability Estimates using Line x Tester Analysis to Develop Wide Compatibility and Restorer Lines in Rice. J. Agric. Chem. Biotechnol. 2020, 11, 383–393. [Google Scholar] [CrossRef]
- SC, M.K.; Singh, S.K.; Khaire, A.; Korada, M.; Majhi, P.K.; Singh, D.K. An analytical approach integrating GGE-Biplot and AMMI techniques for assessing genotype-environment interactions and yield stability in rice (Oryza sativa L.) genotypes. Emergent Life Sci. Res. 2023, 9, 168–176. [Google Scholar] [CrossRef]
- El-Malky, M.; Al-Daej, M. Studies of Genetic Parameters and Cluster analysis of some Quantitative Characters through Diallel analysis of rice (Oryza Sativa L.). Vegetos 2018, 2018, 1–10. [Google Scholar]
- El-Mowafi, H.; Reda, M.; Abdallah, R. Combining Ability Analysis for Agronomic and Yield Attributing Traits in Hybrid Rice. Egypt. J. Plant Breed. 2015, 19, 2195–2219. [Google Scholar] [CrossRef]
- El-Mowafi, H.F.; AlKahtani, M.D.; Abdallah, R.M.; Reda, A.M.; Attia, K.A.; El-Hity, M.A.; El-Dabaawy, H.E.; Husnain, L.A.; Al-Ateeq, T.K.; EL-Esawi, M.A. Combining Ability and Gene Action for Yield Characteristics in Novel Aromatic Cytoplasmic Male Sterile Hybrid Rice under Water-Stress Conditions. Agriculture 2021, 11, 226. [Google Scholar] [CrossRef]
- Gaballah, M.M.; Attia, K.A.; Ghoneim, A.M.; Khan, N.; El-Ezz, A.F.; Yang, B.; Xiao, L.; Ibrahim, E.I.; Al-Doss, A.A. Assessment of Genetic Parameters and Gene Action Associated with Heterosis for Enhancing Yield Characters in Novel Hybrid Rice Parental Lines. Plants 2022, 11, 266. [Google Scholar] [CrossRef]
- Ganapati, R.K.; Rasul, G.; Sarker, U.; Singha, A.; Faruquee, M. Gene action of yield and yield contributing traits of submergence tolerant rice (Oryza sativa L.) in Bangladesh. Bull. Natl. Res. Cent. 2020, 44, 8. [Google Scholar] [CrossRef]
- Wricke, G. Uber eine methode zur erfassung der okologischen streubreite in feldversuchen. Z. Pflanzenzucht. 1962, 47, 92–96. [Google Scholar]
- Shukla, G.K. Some statistical aspects of partitioning genotype-environmental components of variability. Heredity 1972, 29, 237–245. [Google Scholar] [CrossRef]
- Francis, T.R.; Kannenberg, L.W. Yield Stability Studies in Short-Season Maize. I. A Descriptive Method for Grouping Genotypes. Can. J. Plant Sci. 1978, 58, 1029–1034. [Google Scholar] [CrossRef]
- Roemer, J. Sinde die ertagdreichen sorten ertagissicherer? Mitt DLG 1917, 32, 87–89. [Google Scholar]
- Plaisted, R.L.; Peterson, L.C. A technique for evaluating the ability of selections to yield consistently in different locations or seasons. Am. J. Potato Res. 1959, 36, 381–385. [Google Scholar] [CrossRef]
- Plaisted, R.I. A shorter methods for evaluating the ability of selections to yield consistently over locations. Am. J. Potato Res. 1960, 37, 166172. [Google Scholar] [CrossRef]
- Finlay, K.W.; Wilkinson, G.N. The analysis of adaptation in a plant-breeding programme. Aust. J. Agric. Res. 1963, 14, 742–754. [Google Scholar] [CrossRef]
- Gauch, H.G.; Zobel, R.W. Predictive and postdictive success of statistical analyses of yield trials. Theor. Appl. Genet. 1988, 76, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yan, W. GGEbiplot—A Windows Application for Graphical Analysis of Multienvironment Trial Data and Other Types of Two-Way Data. Agron. J. 2001, 93, 1111–1118. [Google Scholar] [CrossRef]
- Gauch, H.G.; Piepho, H.; Annicchiarico, P. Statistical Analysis of Yield Trials by AMMI and GGE: Further Considerations. Crop. Sci. 2008, 48, 866–889. [Google Scholar] [CrossRef]
- Page, A.L.; Miller, R.H.; Keeney, D.R. Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties; American Society of Agronomy: Madison, WI, USA, 1982; Volume 1159. [Google Scholar]
- Freeman, G.H.; Gomez, K.A.; Gomez, A.A. Statistical Procedures for Agricultural Research with Emphasis on Rice. Biometrics 1978, 34, 721. [Google Scholar] [CrossRef]
- Herwibawa, B.; Sakhidin; Haryanto, T.A.D. Agronomic performances of aromatic and non-aromatic M1 rice under drought stress. Open Agric. 2019, 4, 575–584. [Google Scholar] [CrossRef]
- Parimala, K.; Raju, C.S.; Kumar, S.S.; Reddy, S.N. Stability analysis over different environments for grain yield and its components in hybrid rice (Oryza sativa L.). Electron. J. Plant Breed. 2019, 10, 389–399. [Google Scholar] [CrossRef]
- Kumar, C.P.S.; Sathiyabama, R.; Suji, D.B.; Muraleedharan, A. Estimation of heterosis for earliness and certain growth characters in rice (Oryza sativa L.). Plant Arch. 2020, 20, 1429–1432. [Google Scholar]
- Kumari, J.; Mahatman, K.K.; Sharma, S.; Singh, A.K.; Adhikari, S.; Bansal, R.; Kaur, V.; Kumar, S.; Yadav, M.C. Recent Advances in Different Omics Mechanism for Drought Stress Tolerance in Rice. Russ. J. Plant Physiol. 2022, 69, 18. [Google Scholar] [CrossRef]
- Kunnam, J.; Pinta, W.; Ruttanaprasert, R.; Bunphan, D.; Thabthimtho, T.; Aninbon, C. Stability of Phenols, Antioxidant Capacity and Grain Yield of Six Rice Genotypes. Plants 2023, 12, 2787. [Google Scholar] [CrossRef]
- El-Nashart, A.B.; El-Nwehy, S.S.; Rezk, A.E.-H.I.; Ibrahim, O.M. Improving seed and oil yield of sunflower grown in calcareous soil under saline stress conditions. Asian J. Crop. Sci. 2017, 9, 35–39. [Google Scholar] [CrossRef]
- Manjunath, K.; Chandramohan, Y.; Shankar, V.; Balram, M.; Lavuri, K. Genetic and molecular studies on fertility restoration in rice (Oryza sativa L.). Genotypes 2020, 10, 944–950. [Google Scholar]
- Ranjith, S.A.; Ram, R.S.; Saravanan, K.R.; Karthikeyan, P.G.; Anbananthan, V.; Sathiyanarayanan, G. Studies on heterosis breeding for qualitative and quantitative traits in rice (Oryza Sativa L.). Plant Arch. 2020, 20, 1349–1353. [Google Scholar]
- Rasheed, A.; Hassan, M.; Aamer, M.; Bian, J.; Xu, Z.; He, X.; Yan, G.; Wu, Z. Iron toxicity, tolerance and quantitative trait loci mapping in rice—A review. Appl. Ecol. Environ. Res. 2020, 18, 7483–7498. [Google Scholar] [CrossRef]
- Shrestha, J.; Kushwaha, U.K.S.; Maharjan, B.; Kandel, M.; Gurung, S.B.; Poudel, A.P.; Karna, M.K.L.; Acharya, R. Grain Yield Stability of Rice Genotypes. Indones. J. Agric. Res. 2020, 3, 116–126. [Google Scholar] [CrossRef]
- Suvi, W.T.; Shimelis, H.; Laing, M.; Mathew, I.; Shayanowako, A.I.T. Determining the Combining Ability and Gene Action for Rice Yellow Mottle Virus Disease Resistance and Agronomic Traits in Rice (Oryza sativa L.). Agronomy 2020, 11, 12. [Google Scholar] [CrossRef]
- Chandramohan, Y.; Krishna, L.; Srinivas, B.; Rukmini, K.; Sreedhar, S.; Prasad, K.S.; Kishore, N.S.; Rani, C.V.D.; Singh, T.V.J.; Jagadeeshwar, R. Stability analysis of short duration rice genotypes in Telangana using AMMI and GGE Bi-plot models. Environ. Conserv. J. 2023, 24, 243–252. [Google Scholar] [CrossRef]
- Thirumalai, R.; Anbananthan, V.; Azmath, S. Genetic Effects and Combining Ability for Yield and its Component Traits in Rice (Oryza sativa L.) Using Line X Tester. Int. J. Sci. Res. 2018, 7, 1478–1483. [Google Scholar]
- Zewdu, Z.; Abebe, T.; Mitiku, T.; Worede, F.; Dessie, A.; Berie, A.; Atnaf, M. Performance evaluation and yield stability of upland rice (Oryza sativa L.) varieties in Ethiopia. Cogent Food Agric. 2020, 6, 1842679. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, S. Heterosis and combining ability analysis for yield and its components in rice (Oryza sativa L.). J. Pharmacogn. Phytochem. 2019, 8, 3172–3181. [Google Scholar] [CrossRef]
- El-Hadi, A.H.A.; Kash, K.S.; El-Mowafi, H.F.; Anis, G.B. The utilization of cytoplasmic male sterile (cms) and restorer lines in the developing of hybrid rice. J. Agric. Chem. Biotechnol. 2013, 4, 263–274. [Google Scholar] [CrossRef]
- Hasan, M.K.; Kulsum, M.U.; Hossain, E.; Hossain, M.M.; Rahman, M.M.; Rahmat, N.M.F. Combining ability analysis for iden-tifying elite parents for heterotic rice hybrids. Acad. J. Agric. Res. 2015, 3, 70–75. [Google Scholar]
- Ali, M.A.; Ghazy, A.I.; Alotaibi, K.D.; Ibrahim, O.M.; Al-Doss, A.A. Nitrogen efficiency indexes association with nitrogen recovery, utilization, and use efficiency in spring barley at various nitrogen application rates. Agron. J. 2022, 114, 2290–2309. [Google Scholar] [CrossRef]
- Wu, C.; Cui, K.; Li, Q.; Li, L.; Wang, W.; Hu, Q.; Ding, Y.; Li, G.; Fahad, S.; Huang, J.; et al. Estimating the yield stability of heat-tolerant rice genotypes under various heat conditions across reproductive stages: A 5-year case study. Sci. Rep. 2021, 11, 13604. [Google Scholar] [CrossRef]
- Maulana, H.; Solihin, E.; Trimo, L.; Hidayat, S.; Wijaya, A.A.; Hariadi, H.; Amien, S.; Ruswandi, D.; Karuniawan, A. Genotype-by-environment interactions (GEIs) and evaluate superior sweet potato (Ipomoea batatas [L.] Lam) using combined analysis and GGE biplot. Heliyon 2023, 9, e20203. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).