Development and Application of Image-Based High-Throughput Phenotyping Methodology for Salt Tolerance in Lentils
Abstract
:1. Introduction
2. Materials and Methods
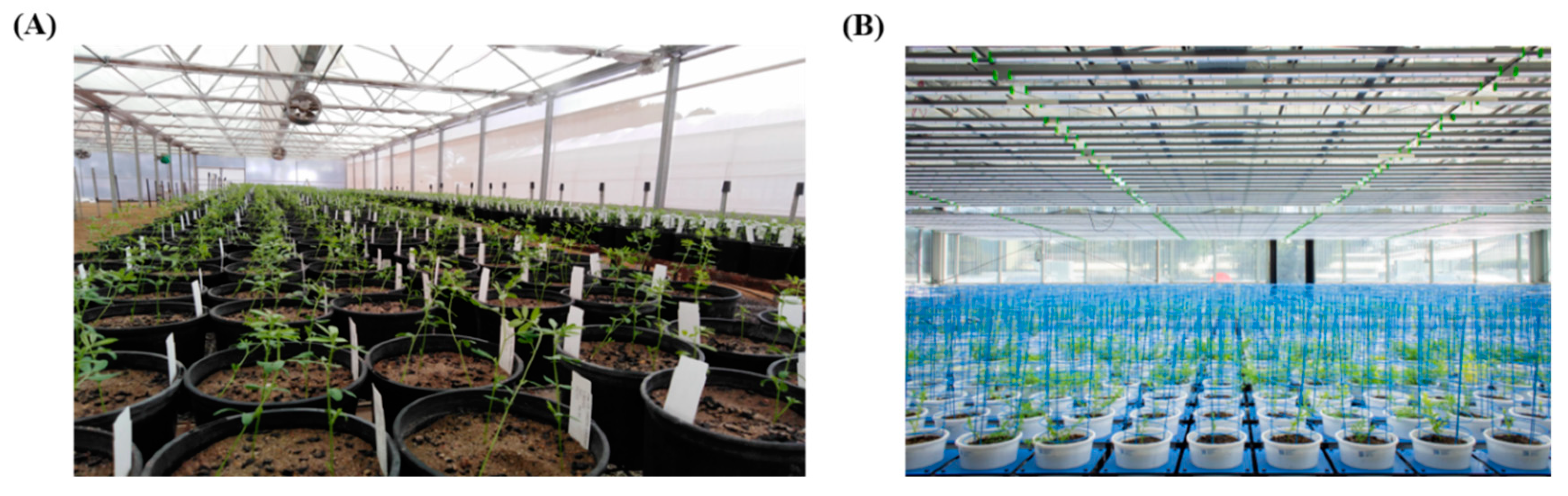
2.1. Experiment 1: Salt Tolerance Screening of Lentil Accessions Using Conventional Phenotyping
2.2. Experiment 2: Optimization of Salt Toxicity Screening Using Image-Based Phenotyping
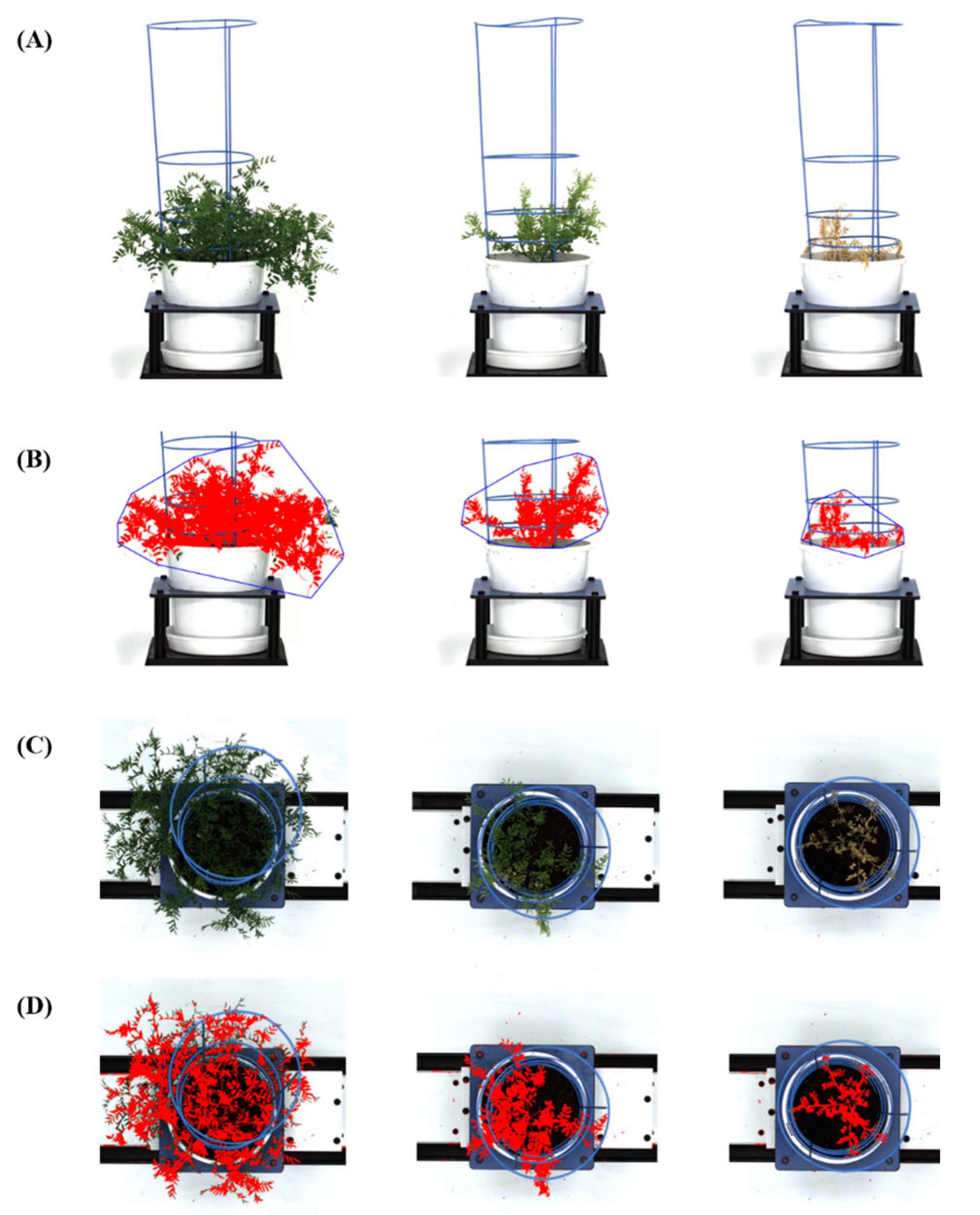
2.3. Experiment 3: Evaluation of Lentil Accessions for Salt Tolerance Using Image-Based Phenotyping
2.4. Data Analysis
2.4.1. Conventional Phenotyping
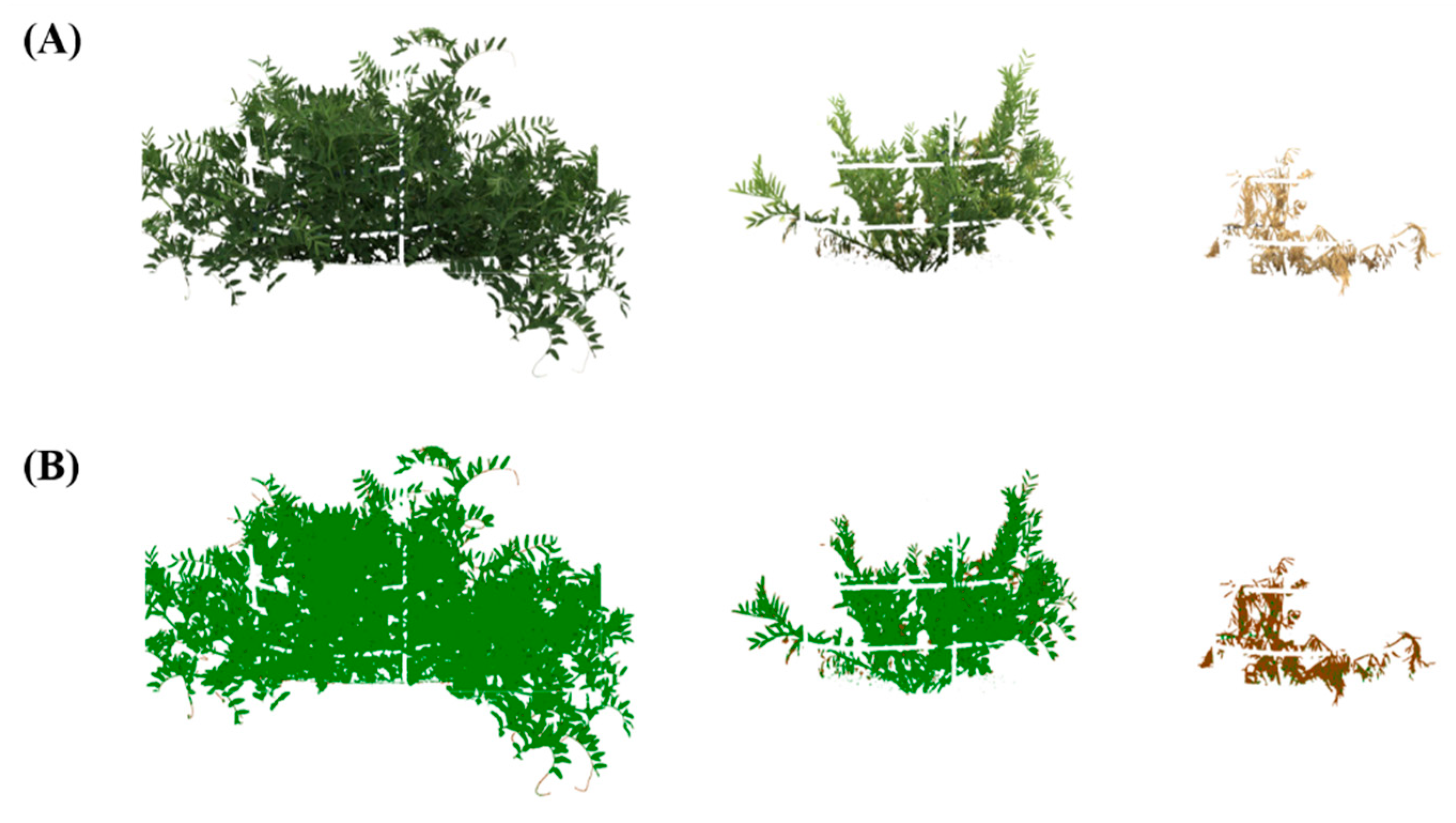
2.4.2. Image-Based Phenotyping
3. Results
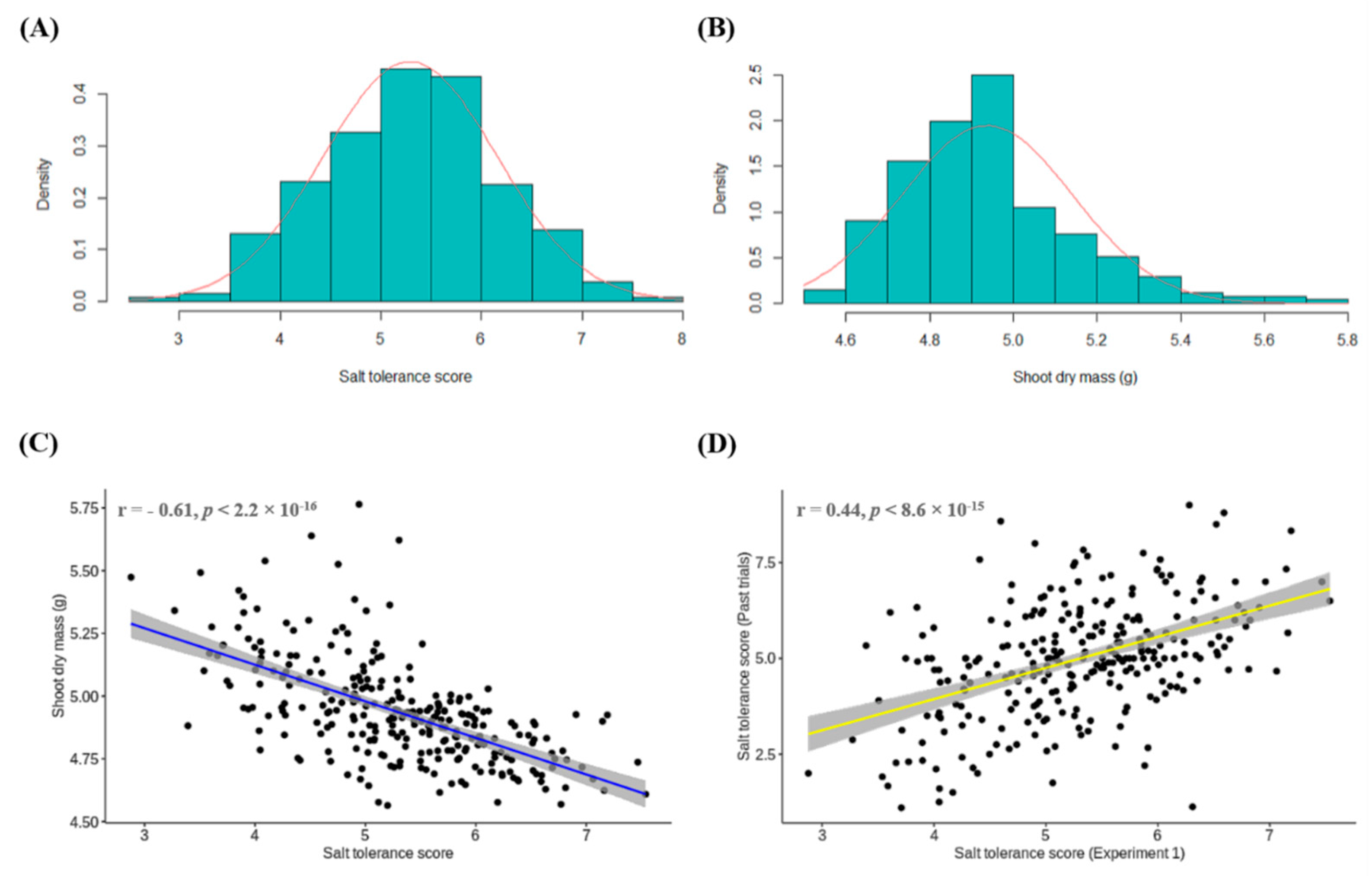
3.1. Experiment 1: Conventional Phenotyping of Lentil Accessions for Salt Toxicity Screening
3.2. Experiment 2: Establishment of High-Throughput Phenotyping Protocol for Salt Tolerance
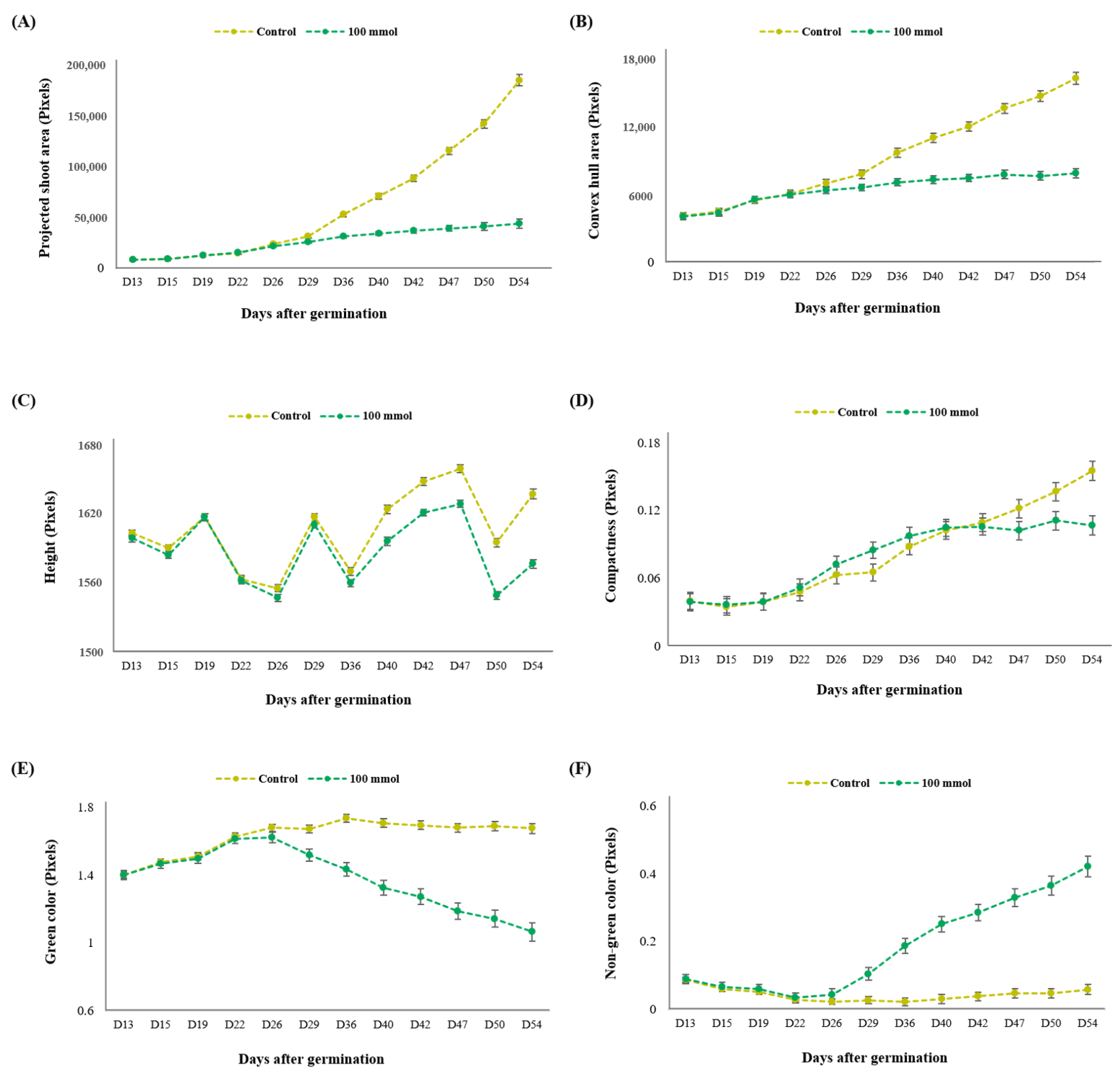
3.2.1. Identification of the Optimal Salt Concentration for the Image-Based Assay
3.2.2. Determination of Growth Stage for Distinguishing Salt Tolerance in Lentil
3.2.3. Relationship between Destructive and Non-Destructive Measurement
3.3. Experiment 3: Application of High-Throughput Phenotyping Protocol for Salt Tolerance
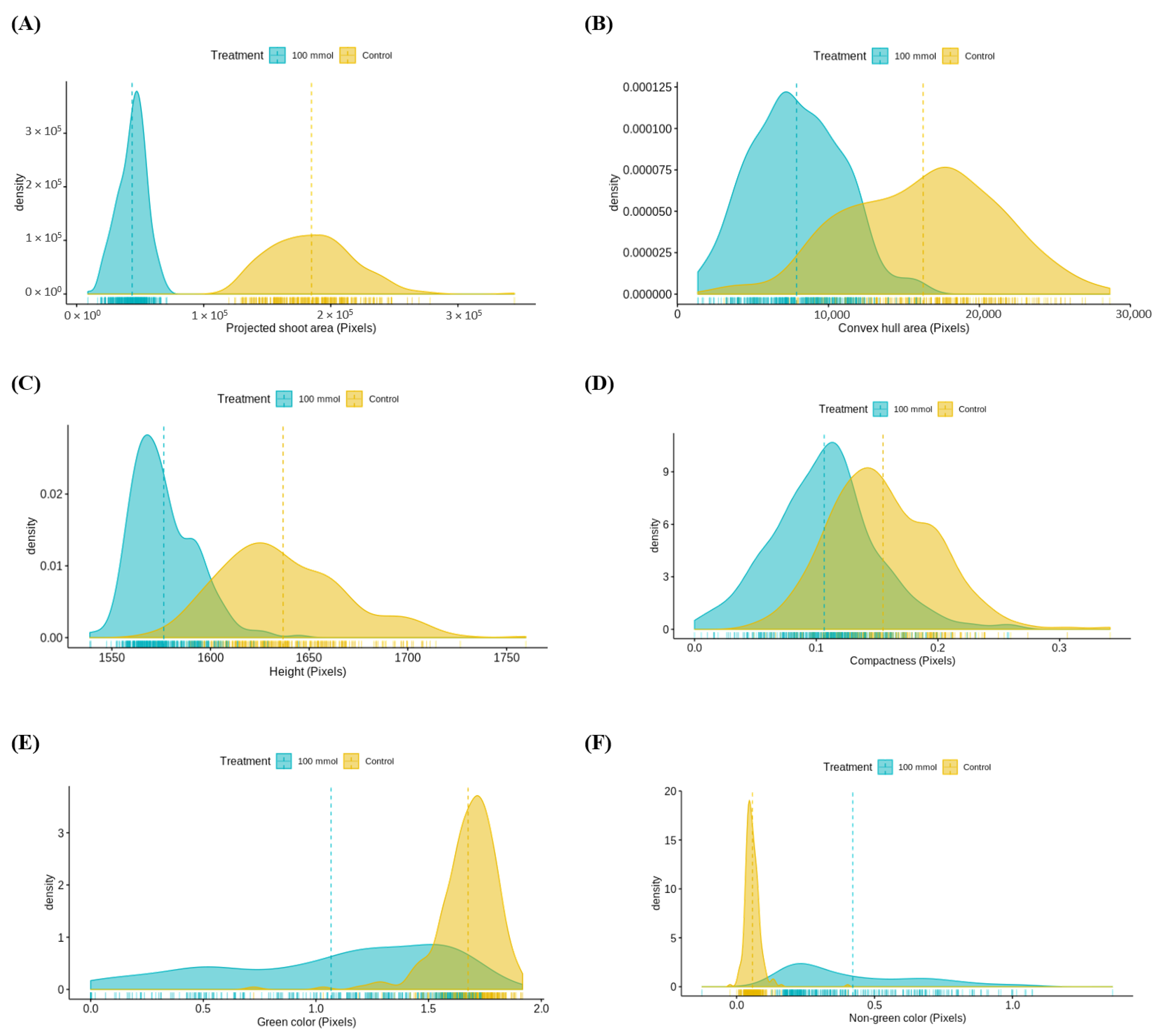
3.3.1. Assessment of Multiple Non-Destructive Measurements for Salt Tolerance in Lentils
3.3.2. Relationship between Destructive and Non-Destructive Measurements
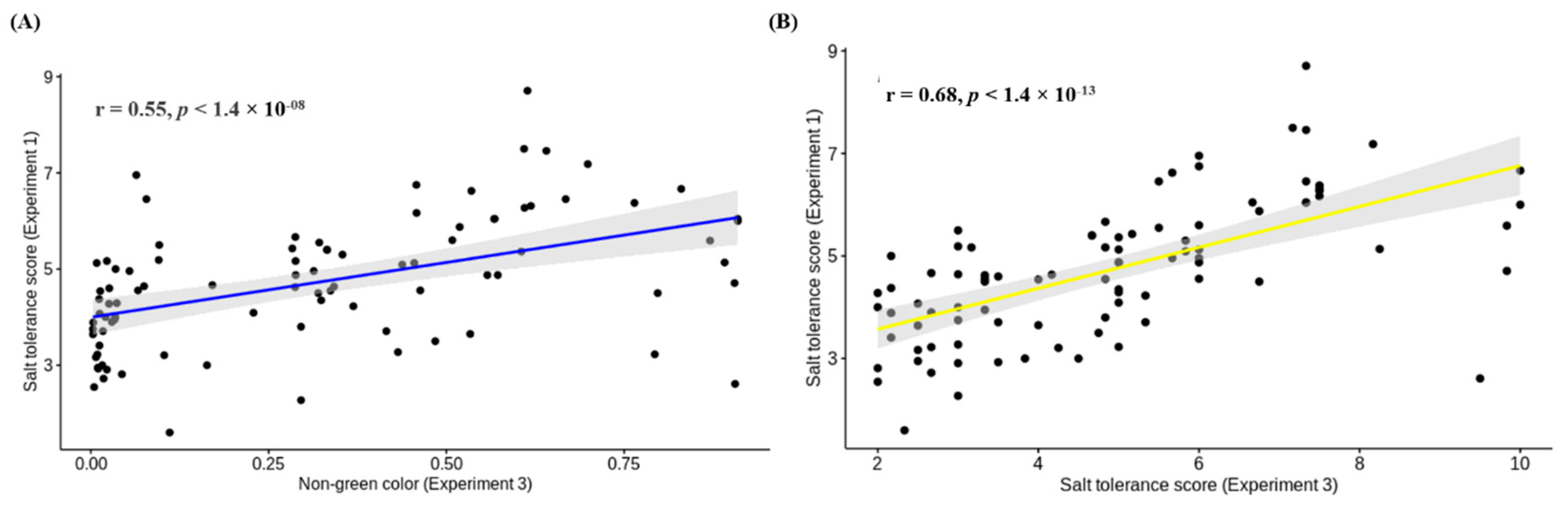
3.4. Comparison of Salt Tolerance Levels between Conventional and HTP Methods for Validation
4. Discussion
4.1. Comparison of a High-Throughput Phenotyping Protocol to the Conventional Method for Salt Screening in Lentils
4.2. Development of a High-Throughput Image-Based Method for Salt Toxicity Screening in Lentils
4.3. Application of Optimized High-Throughput Phenotyping Protocol
4.4. Validation of High-Throughput Phenotyping Protocol for Salt Tolerance in Lentils
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kokten, K.; Karakoy, T.; Bakoglu, A.; Akcura, M. Determination of salinity tolerance of some lentil (Lens culinaris M.) varieties. J. Food Agric. Environ. 2010, 8, 140–143. [Google Scholar]
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrào, S.; Tester, M. Salt stress under the scalpel—Dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef] [Green Version]
- Kumawat, K.R.; Gothwal, D.K.; Singh, D. Salinity tolerance of lentil genotypes based on stress tolerance indices. J. Pharmacogn. Phytochem. 2017, 6, 1368–1372. [Google Scholar]
- Maher, L.; Armstrong, R.; Connor, D. Salt Tolerant Lentils—A Possibility for the Future? In Solutions for a Better Environment, Proceedings of the 11th Australian Agronomy Conference, Geelong, Victoria, Australia, 2–6 February 2003; Unkovich, M., O’Leary, G., Eds.; The Australian Society of Agronomy Inc.: Geelong, Australia, 2003; Available online: http://www.agronomyaustraliaproceedings.org/images/sampledata/2003/c/17/maher.pdf (accessed on 21 October 2019).
- Jayasundara, H.P.S.; Thomson, B.D.; Tang, C. Responses of cool season grain legumes to soil abiotic stresses. Adv. Agron. 1998, 63, 77–151. [Google Scholar]
- Nadeem, M.; Li, J.; Yahya, M.; Wang, M.; Ali, A.; Cheng, A.; Wang, X.; Ma, C. Grain legumes and fear of salt stress: Focus on mechanisms and management strategies. IJMS 2019, 20, 799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, J.; Pratap, A.; Kumar, S. (Eds.) Plant phenomics: An overview. In Phenomics in Crop Plants: Trends, Options and Limitations; Springer: London, UK, 2015; pp. 1–10. [Google Scholar]
- Rousseau, C.; Belin, E.; Bove, E.; Rousseau, D.; Fabre, F.; Berruyer, R.; Guillaumès, J.; Manceau, C.; Jacques, M.; Boureau, T. High throughput quantitative phenotyping of plant resistance using chlorophyll fluorescence image analysis. Plant Methods 2013, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Newman, I.; Zhou, M.; Mendham, N.; Zhang, G.; Shabala, S. Screening plants for salt tolerance by measuring K+ flux: A case study for barley. Plant Cell Environ. 2005, 28, 1230–1246. [Google Scholar] [CrossRef]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. A comparison of hydroponic and soil-based screening methods to identify salt tolerance in the field in barley. J. Exp. Bot. 2012, 63, 3853–3868. [Google Scholar]
- Singh, D.; Singh, C.K.; Kumari, S.; Tomar, R.S.S.; Karwa, S.; Singh, R.; Singh, R.B.; Sarkar, S.K.; Pal, M. Discerning morpho-anatomical, physiological and molecular multiformity in cultivated and wild genotypes of lentil with reconciliation to salinity stress. PLoS ONE 2017, 12, e0177465. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics-technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Snider, J.L.; Chee, P.W. In-field high throughput phenotyping and cotton plant growth analysis using LiDAR. Front. Plant Sci. 2018, 9, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Badenhorst, P.; Phelan, A.; Pembleton, L.; Shi, F.; Cogan, N.; Spangenberg, G.; Smith, K. Using sensors and unmanned aircraft systems for high-throughput phenotyping of biomass in perennial ryegrass breeding trials. Front. Plant Sci. 2019, 10, 1381. [Google Scholar] [CrossRef] [PubMed]
- Gebremedhin, A.; Badenhorst, P.; Wang, J.; Giri, K.; Spangenberg, G.; Smith, K. Development and validation of a model to combine NDVI and plant height for high-throughput phenotyping of herbage yield in a perennial ryegrass breeding program. Remote Sens. 2019, 11, 2494. [Google Scholar] [CrossRef] [Green Version]
- Ghamkhar, K.; Irie, K.; Hagedorn, M.; Hsiao, J.; Fourie, J.; Gebbie, S.; Hoyos-Villegas, V.; George, R.; Stewart, A.; Inch, C.; et al. Real-time, non-destructive and in-field foliage yield and growth rate measurement in perennial ryegrass (Lolium perenne L.). Plant Methods 2019, 15, 72. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dimech, A.M.; Spangenberg, G.; Smith, K.; Badenhorst, P. Rapid screening of nitrogen use efficiency in perennial ryegrass (Lolium perenne L.) using automated image-based phenotyping. Front. Plant Sci. 2020, 11, 565361. [Google Scholar] [CrossRef]
- Qiu, Q.; Sun, N.; Bai, H.; Wang, N.; Fan, Z.; Wang, Y.; Meng, Z.; Li, B.; Cong, Y. Field-based high-throughput phenotyping for maize plant using 3D LiDAR point cloud generated with a “Phenomobile”. Front. Plant Sci. 2019, 10, 554. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Huang, C.; Wu, D.; Qiao, F.; Li, W.; Duan, L.; Wang, K.; Xiao, Y.; Chen, G.; Liu, Q.; et al. High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiol. 2017, 173, 1554–1564. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Knapp, S.; Schmidhalter, U. Advancing high-throughput phenotyping of wheat in early selection cycles. Remote Sens. 2020, 12, 574. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Wang, X.; Kumar, U.; Gao, L.; Noor, M.; Imtiaz, M.; Singh, R.P.; Poland, J. High-throughput phenotyping enabled genetic dissection of crop lodging in wheat. Front. Plant Sci. 2019, 10, 394. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, G.N.; Maharjan, P.; Maphosa, L.; Vakani, J.; Thoday-Kennedy, E.; Kant, S. A robust automated image-based phenotyping method for rapid vegetative screening of wheat germplasm for nitrogen use efficiency. Front Plant Sci. 2019, 10, 1372. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Shabala, L.; Zhou, M.; Shabala, S. Developing a high-throughput phenotyping method for oxidative stress tolerance in barley roots. Plant Methods 2019, 15, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hairmansis, A.; Berger, B.; Tester, M.; Roy, S.J. Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice 2014, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atieno, J.; Li, Y.; Langridge, P.; Dowling, K.; Brien, C.; Berger, B. Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci. Rep. 2017, 7, 1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhanga, C.; Chenb, W.; Sankarana, S. High-throughput field phenotyping of Ascochyta blight disease severity in chickpea. Crop Prot. 2019, 125, 104885. [Google Scholar] [CrossRef]
- Humplík, J.F.; Lazár, D.; Fürst, T.; Husičková, A.; Hýbl, M.; Spíchal, L. Automated integrative high-throughput phenotyping of plant shoots: A case study of the cold-tolerance of pea (Pisum sativum L.). Plant Methods 2015, 11, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzougui, A.; Ma, Y.; Zhang, C.; McGee, R.J.; Coyne, C.J.; Main, D.; Sankaran, S. Advanced imaging for quantitative evaluation of aphanomyces root rot resistance in lentil. Front. Plant Sci. 2019, 10, 383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, I.; Eramian, M.; Ovsyannikov, I.; van der Kamp, W.; Nielsen, K.; Duddu, H.S.; Rumali, A.; Shirtliffe, S.; Bett, K. Automatic Detection and Segmentation of Lentil Crop Breeding Plots from Multi-spectral Images Captured by UAV-mounted Camera. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa Village, HI, USA, 7–11 January 2019; pp. 1673–1681. [Google Scholar] [CrossRef]
- Marko, D.; Briglia, N.; Summerer, S.; Petrozza, A.; Cellini, F.; Iannacone, R. High-throughput phenotyping in plant stress response: Methods and potential applications to polyamine field. In Polyamines: Methods and Protocols, Methods in Molecular Biology; Alcàzar, R., Tiburcio, A.F., Eds.; Humana Press: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, N. Imaging technologies for plant high-throughput phenotyping: A review. Front. Agric. Sci. Eng. 2018, 5, 406–419. [Google Scholar] [CrossRef] [Green Version]
- Pratap, A.; Gupta, S.; Nair, R.M.; Gupta, S.K.; Schafleitner, R.; Basu, P.S.; Singh, C.M.; Prajapati, U.; Gupta, A.K.; Nayyar, H.; et al. Using plant phenomics to exploit the gains of genomics. Agronomy 2019, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Yol, E.; Toker, C.; Uzun, B. Traits for phenotyping. In Phenomics in Crop Plants: Trends, Options and Limitations; Kumar, J., Pratap, A., Kumar, S., Eds.; Springer: London, UK, 2015; pp. 11–26. [Google Scholar]
- Berger, B.; de Regt, B.; Tester, M. High-throughput phenotyping of plant shoots. In High-Throughput Phenotyping in Plants. Methods in Molecular Biology; Normanly, J., Ed.; Humana Press: New York, NY, USA, 2012; pp. 9–20. [Google Scholar]
- Awlia, M.; Nigro, A.; Fajkus, J.; Schmoeckel, S.M.; Negrão, S.; Santelia, D.; Trtílek, M.; Tester, M.; Julkowska, M.M.; Panzarová, K. High-throughput non-destructive phenotyping of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1414. [Google Scholar] [CrossRef] [Green Version]
- Al-Tamimi, N.; Brien, C.; Oakey, H.; Berger, B.; Saade, S.; Ho, Y.S.; Schmockel, S.M.; Tester, M.; Negrao, S. Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat. Commun. 2016, 7, 13342. [Google Scholar] [CrossRef] [Green Version]
- GRDC. Available online: https://grdc.com.au/research/projects/project?id=4297 (accessed on 29 October 2020).
- Gilmour, A.R.; Gogel, B.J.; Cullis, B.R.; Welham, S.J.; Thompson, R. ASReml User Guide Release 4.1 Functional Specification; VSN International Ltd.: Hemel Hempstead, UK, 2015; pp. 1–329. Available online: www.vsni.co.uk (accessed on 9 August 2018).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 13 July 2019).
- Kassambara, A. ggpubr. 2020. Available online: https://rdrr.io/cran/ggpubr/ (accessed on 10 September 2020).
- Klukas, C.; Chen, D.; Pape, J.M. Integrated Analysis Platform: An open-source information system for high-throughput plant phenotyping. Plant Physiol. 2014, 165, 506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulse Australia. Available online: http://www.pulseaus.com.au/growing-pulses/bmp/lentil (accessed on 11 November 2018).
- Spies, B.; Woodgate, P. Salinity and Hydrogeology. In Salinity Mapping Methods in the Australian Context; Department of the Environment and Heritage and Agriculture, Fisheries and Forestry: Canberra, Australia, 2005; p. 17. [Google Scholar]
- Mutka, A.M.; Bart, R.S. Image-based phenotyping of plant disease symptoms. Front. Plant Sci. 2014, 5, 734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Shi, R.; Pape, J.M.; Neumann, K.; Arend, D.; Graner, A.; Chen, M.; Klukas, C. Predicting plant biomass accumulation from image-derived parameters. GigaScience 2018, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Salt Treatment (mmol) | Df | Sum of Squares | Mean Squares | F-Value | p-Value |
|---|---|---|---|---|---|
| 0.0 (Control) | 5 | 3.88 × 1022 | 7.76 × 1021 | 1.852 | 0.10 |
| 42.5 | 5 | 9.66 × 1021 | 1.93 × 1021 | 0.811 | 0.54 |
| 85.0 | 5 | 1.25 × 1022 | 2.51 × 1021 | 2.974 | 0.05 * |
| 127.5 | 5 | 4.96 × 1021 | 9.91 × 1020 | 5.913 | 0.001 *** |
| Projected Shoot Area Variation | Df | Sum of Squares | Mean Squares | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| T2 vs. T1 | Genotype | 5 | 5.21 × 1022 | 1.04 × 1022 | 2.314 | 0.06 |
| Treatment | 1 | 3.44 × 1022 | 3.44 × 1022 | 7.639 | 0.01 ** | |
| Genotype:Treatment | 5 | 3.31 × 1022 | 6.61 × 1021 | 1.468 | 0.22 | |
| T3 vs. T1 | Genotype | 5 | 8.10 × 1022 | 1.62 × 1022 | 4.689 | 0.01 ** |
| Treatment | 1 | 2.02 × 1023 | 2.02 × 1023 | 58.61 | 0.001 *** | |
| Genotype:Treatment | 5 | 1.82 × 1022 | 3.64 × 1021 | 1.054 | 0.40 | |
| T4 vs. T1 | Genotype | 5 | 2.87 × 1022 | 5.73 × 1021 | 2.352 | 0.06 |
| Treatment | 1 | 4.09 × 1023 | 4.09 × 1023 | 167.732 | 0.001 *** | |
| Genotype:Treatment | 5 | 5.42 × 1022 | 1.08 × 1022 | 4.446 | 0.01 ** |
| Dry Mass | Salt Tolerance Scores | |||||||
|---|---|---|---|---|---|---|---|---|
| VSummary | VIAP | VLemnaTec | VKeygene | VSummary | VIAP | VLemnaTec | VKeygene | |
| Projected shoot area | 0.92 | 0.95 | 0.95 | 0.96 | −0.24 | −0.32 | −0.32 | −0.43 |
| Convex hull area | 0.86 | 0.91 | 0.91 | 0.90 | −0.20 | −0.44 | −0.44 | −0.45 |
| Compactness | 0.63 | 0.90 | 0.90 | 0.82 | −0.04 | −0.32 | −0.33 | −0.35 |
| Height | 0.74 | 0.71 | 0.72 | 0.70 | −0.39 | −0.54 | −0.58 | −0.54 |
| Green color | 0.48 | 0.45 | 0.45 | 0.44 | −0.90 | −0.91 | −0.91 | −0.92 |
| Non-green color | −0.40 | −0.41 | −0.41 | −0.54 | 0.92 | 0.92 | 0.92 | 0.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dissanayake, R.; Kahrood, H.V.; Dimech, A.M.; Noy, D.M.; Rosewarne, G.M.; Smith, K.F.; Cogan, N.O.I.; Kaur, S. Development and Application of Image-Based High-Throughput Phenotyping Methodology for Salt Tolerance in Lentils. Agronomy 2020, 10, 1992. https://doi.org/10.3390/agronomy10121992
Dissanayake R, Kahrood HV, Dimech AM, Noy DM, Rosewarne GM, Smith KF, Cogan NOI, Kaur S. Development and Application of Image-Based High-Throughput Phenotyping Methodology for Salt Tolerance in Lentils. Agronomy. 2020; 10(12):1992. https://doi.org/10.3390/agronomy10121992
Chicago/Turabian StyleDissanayake, Ruwani, Hossein V. Kahrood, Adam M. Dimech, Dianne M. Noy, Garry M. Rosewarne, Kevin F. Smith, Noel O. I. Cogan, and Sukhjiwan Kaur. 2020. "Development and Application of Image-Based High-Throughput Phenotyping Methodology for Salt Tolerance in Lentils" Agronomy 10, no. 12: 1992. https://doi.org/10.3390/agronomy10121992
APA StyleDissanayake, R., Kahrood, H. V., Dimech, A. M., Noy, D. M., Rosewarne, G. M., Smith, K. F., Cogan, N. O. I., & Kaur, S. (2020). Development and Application of Image-Based High-Throughput Phenotyping Methodology for Salt Tolerance in Lentils. Agronomy, 10(12), 1992. https://doi.org/10.3390/agronomy10121992








