Histone Acyl Code in Precision Oncology: Mechanistic Insights from Dietary and Metabolic Factors
Abstract
1. Introduction
2. Histone Acylation
2.1. Mechanisms of Histone Acylation
2.2. Histone Acylation Writers and Erasers
2.3. Histone Acylation Readers
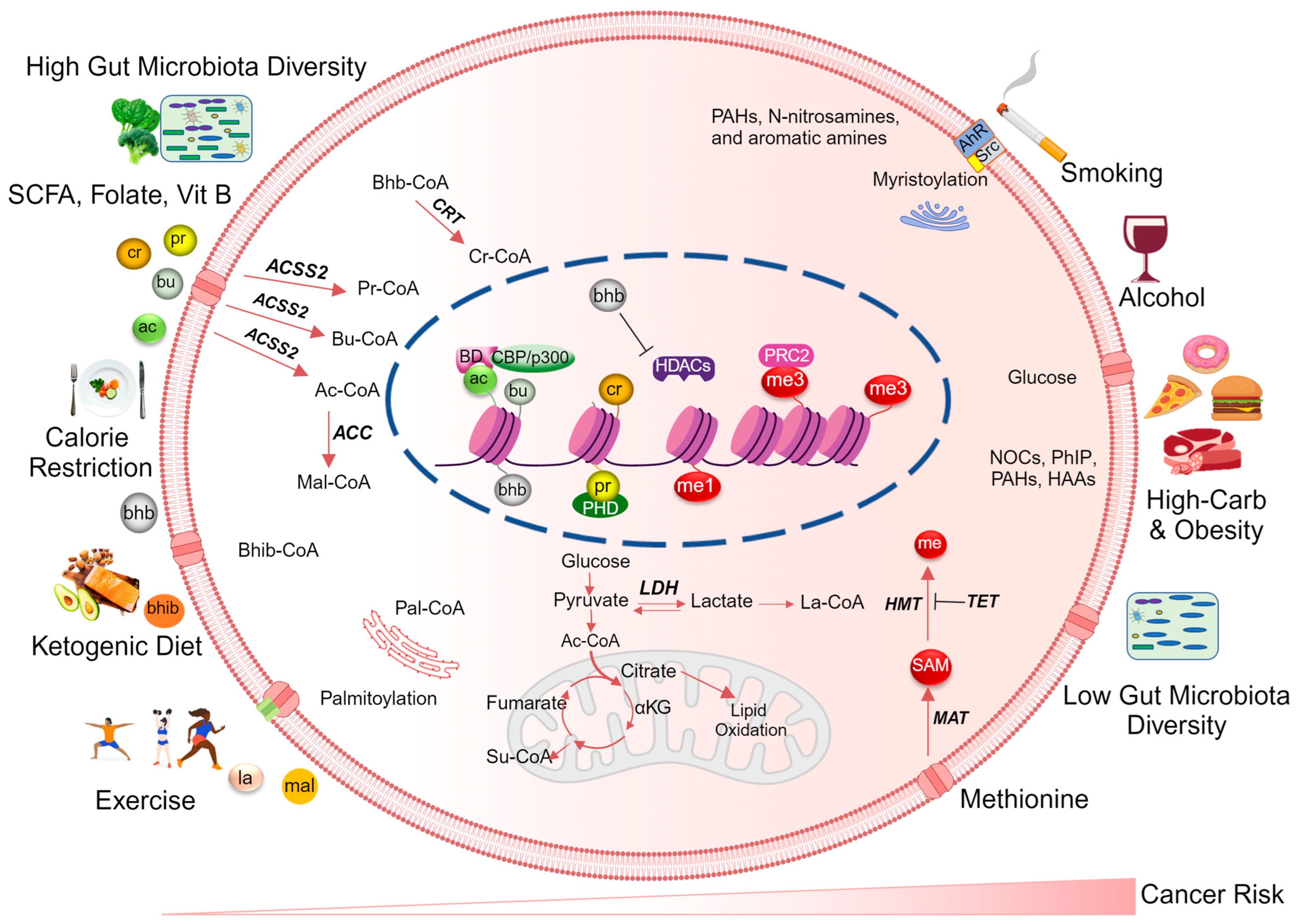
3. Dietary Metabolites Regulating Histone Acylation
- Butyrate, propionate, and acetate are short-chain fatty acids (SCFAs) produced by gut microbiome-mediated fermentation of dietary fiber. These metabolites inhibit HDAC activity and increase histone acetylation status [109];
- Vitamin B3 (niacin) is involved in energy metabolism as a precursor for the coenzyme nicotinamide adenine dinucleotide (NAD+), which is required by SIRTs for deacetylase activity. By affecting NAD+ availability, niacin can indirectly modulate histone acylation [117];
- Glucose utilization, microbiota-derived SCFAs, or dietary fat metabolism can impact acetyl/acyl-CoA ratios, thereby affecting overall histone acetylation patterns [118,119,120,121]. Since most histone acylation competes for the same HATs, the acetyl/acyl-CoA ratios in different cellular pools dictate which acylation pattern occurs on histones [118,121];
- Dietary antioxidants such as vitamins C and E, and certain polyphenols, modulate cellular redox status and signaling pathways involved in histone acetylation [122]. Additionally, nutrient-sensing pathways, such as the mammalian target of rapamycin (mTOR) pathway, can integrate dietary and metabolic signals to influence histone acylation [123]. Among the nutrient-sensing signaling pathways that govern histone PTMs, the sucrose non-fermenting/AMP-activated protein kinase (AMPK/Snf1) and carbohydrate response element binding protein (ChREBP) pathways play pivotal roles. For instance, AMPK/Snf1 acts as a histone kinase [124], not only phosphorylating but also regulating the activity of several HATs and HDACs through enzyme phosphorylation [125]. Moreover, this pathway influences histone acetylation and deacetylation by controlling levels of acetyl CoA and NAD+ levels [125].
3.1. The Role of Dietary and Metabolism-Derived Histone Acylation in Cancer Development and Progression
3.2. Metabolism-Derived Histone Acyl Codes as Cancer Biomarkers
4. Targeting Histone Acylation for Cancer Prevention and Therapy
4.1. Targeting Histone Acylation for Cancer Prevention
4.2. Targeting Histone Acylation for Cancer Therapy
4.2.1. Targeting Acylation Writer and Eraser
4.2.2. Targeting Acylation Readers
4.3. Current Approaches and Future Directions in Targeting Histone Acylation for Cancer Interception
4.4. Challenges in Targeting Histone Acylation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020.
- Ilango, S.; Paital, B.; Jayachandran, P.; Padma, P.R.; Nirmaladevi, R. Epigenetic alterations in cancer. Front. Biosci.-Landmark 2020, 25, 1058–1109. [Google Scholar]
- Baylin, S.B.; Jones, P.A. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.N.; Tan, J.; Chen, Y.X.; Fang, J.Y. Genetic variants in the histone methylation and acetylation pathway and their risks in eight types of cancers. J. Dig. Dis. 2018, 19, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zhou, Y.; Xue, Z.; Hao, N.; Li, Y.; Guo, X.; Wang, D.; Shi, X.; Li, H. Histone benzoylation serves as an epigenetic mark for DPF and YEATS family proteins. Nucleic Acids Res. 2021, 49, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Etchegaray, J.P.; Mostoslavsky, R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol. Cell. 2016, 62, 695–711. [Google Scholar] [CrossRef] [PubMed]
- Simithy, J.; Sidoli, S.; Yuan, Z.-F.; Coradin, M.; Bhanu, N.V.; Marchione, D.M.; Klein, B.J.; Bazilevsky, G.A.; McCullough, C.E.; Magin, R.S. Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun. 2017, 8, 1141. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Z.; Bao, L. An expanding repertoire of protein acylations. Mol. Cell. Proteom. 2022, 21, 100193. [Google Scholar] [CrossRef]
- Liu, X.; Cooper, D.E.; Cluntun, A.A.; Warmoes, M.O.; Zhao, S.; Reid, M.A.; Liu, J.; Lund, P.J.; Lopes, M.; Garcia, B.A.; et al. Acetate Production from Glucose and Coupling to Mitochondrial Metabolism in Mammals. Cell 2018, 175, 502–513.e513. [Google Scholar] [CrossRef]
- Kebede, A.F.; Nieborak, A.; Shahidian, L.Z.; Le Gras, S.; Richter, F.; Gomez, D.A.; Baltissen, M.P.; Meszaros, G.; Magliarelli, H.d.F.; Taudt, A. Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol. 2017, 24, 1048–1056. [Google Scholar] [CrossRef]
- Han, Z.; Wu, H.; Kim, S.; Yang, X.; Li, Q.; Huang, H.; Cai, H.; Bartlett, M.G.; Dong, A.; Zeng, H. Revealing the protein propionylation activity of the histone acetyltransferase MOF (males absent on the first). J. Biol. Chem. 2018, 293, 3410–3420. [Google Scholar] [CrossRef]
- Vollmuth, F.; Geyer, M. Interaction of propionylated and butyrylated histone H3 lysine marks with Brd4 bromodomains. Angew. Chem. 2010, 122, 6920–6924. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, X.; Li, H. Beyond histone acetylation—Writing and erasing histone acylations. Curr. Opin. Struct. Biol. 2018, 53, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Son, M.Y.; Cho, H.S. Anticancer Effects of Gut Microbiota-Derived Short-Chain Fatty Acids in Cancers. J. Microbiol. Biotechnol. 2023, 33, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yu, J.; Li, F.; Ge, S. Oncometabolites drive tumorigenesis by enhancing protein acylation: From chromosomal remodelling to nonhistone modification. J. Exp. Clin. Cancer Res. 2022, 41, 144. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, J.; Chung, M.W.H.; Feng, L.; Sun, H.; Hao, Q. Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation. Proc. Natl. Acad. Sci. USA 2018, 115, 2365–2370. [Google Scholar] [CrossRef] [PubMed]
- Andrews, F.H.; Shinsky, S.A.; Shanle, E.K.; Bridgers, J.B.; Gest, A.; Tsun, I.K.; Krajewski, K.; Shi, X.; Strahl, B.D.; Kutateladze, T.G. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 2016, 12, 396–398. [Google Scholar] [CrossRef]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef]
- Zhao, D.; Guan, H.; Zhao, S.; Mi, W.; Wen, H.; Li, Y.; Zhao, Y.; Allis, C.D.; Shi, X.; Li, H. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016, 26, 629–632. [Google Scholar] [CrossRef]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balázsi, S.; Hajnády, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef]
- Tan, D.; Wei, W.; Han, Z.; Ren, X.; Yan, C.; Qi, S.; Song, X.; Zheng, Y.G.; Wong, J.; Huang, H. HBO1 catalyzes lysine benzoylation in mammalian cells. iScience 2022, 25, 105443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cao, R.; Niu, J.; Yang, S.; Ma, H.; Zhao, S.; Li, H. Molecular basis for hierarchical histone de-β-hydroxybutyrylation by SIRT3. Cell Discov. 2019, 5, 35. [Google Scholar] [CrossRef]
- Gaffney, D.O.; Jennings, E.Q.; Anderson, C.C.; Marentette, J.O.; Shi, T.; Oxvig, A.-M.S.; Streeter, M.D.; Johannsen, M.; Spiegel, D.A.; Chapman, E. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem. Biol. 2020, 27, 206–213.e206. [Google Scholar] [CrossRef]
- Zhou, T.; Cheng, X.; He, Y.; Xie, Y.; Xu, F.; Xu, Y.; Huang, W. Function and mechanism of histone β-hydroxybutyrylation in health and disease. Front. Immunol. 2022, 13, 981285. [Google Scholar] [CrossRef]
- Huang, H.; Luo, Z.; Qi, S.; Huang, J.; Xu, P.; Wang, X.; Gao, L.; Li, F.; Wang, J.; Zhao, W. Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res. 2018, 28, 111–125. [Google Scholar] [CrossRef]
- Zhang, R.; Bons, J.; Scheidemantle, G.; Liu, X.; Bielska, O.; Carrico, C.; Rose, J.; Heckenbach, I.; Scheibye-Knudsen, M.; Schilling, B.; et al. Histone malonylation is regulated by SIRT5 and KAT2A. iScience 2023, 26, 106193. [Google Scholar] [CrossRef]
- Xie, Z.; Dai, J.; Dai, L.; Tan, M.; Cheng, Z.; Wu, Y.; Boeke, J.D.; Zhao, Y. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteom. 2012, 11, 100–107. [Google Scholar] [CrossRef]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.; Skinner, M.E. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef]
- Li, Y.; Sabari, B.R.; Panchenko, T.; Wen, H.; Zhao, D.; Guan, H.; Wan, L.; Huang, H.; Tang, Z.; Zhao, Y. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 2016, 62, 181–193. [Google Scholar] [CrossRef]
- Sakabe, K.; Wang, Z.; Hart, G.W. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. USA 2010, 107, 19915–19920. [Google Scholar] [CrossRef]
- Zou, C.; Ellis, B.M.; Smith, R.M.; Chen, B.B.; Zhao, Y.; Mallampalli, R.K. Acyl-CoA: Lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J. Biol. Chem. 2011, 286, 28019–28025. [Google Scholar] [CrossRef]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, T.; Li, X.; Peng, T.; Hang, H.C.; Li, X.D. Integrative chemical biology approaches to examine ‘erasers’ for protein lysine fatty-acylation. Angew. Chem. (Int. Ed. Engl.) 2015, 54, 1149. [Google Scholar] [CrossRef]
- Kim, E.; H Bisson, W.; V Lohr, C.; E Williams, D.; Ho, E.; H Dashwood, R.; Rajendran, P. Histone and non-histone targets of dietary deacetylase inhibitors. Curr. Top. Med. Chem. 2016, 16, 714–731. [Google Scholar] [CrossRef]
- Kaur, J.; Daoud, A.; Eblen, S.T. Targeting chromatin remodeling for cancer therapy. Curr. Mol. Pharmacol. 2019, 12, 215. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Fayyaz, S.; Poltronieri, P.; Calin, G.; Mallardo, M. Epigenetic deregulation in cancer: Enzyme players and non-coding RNAs. Semin. Cancer Biol. 2022, 83, 197–207. [Google Scholar] [CrossRef]
- Blot, W.J.; Tarone, R.E. Doll and Peto’s quantitative estimates of cancer risks: Holding generally true for 35 years. JNCI J. Natl. Cancer Inst. 2015, 107, djv044. [Google Scholar] [CrossRef]
- Doll, R.; Peto, R. The causes of cancer: Quantitative estimates of avoidable risks of cancer in the United States today. JNCI J. Natl. Cancer Inst. 1981, 66, 1192–1308. [Google Scholar] [CrossRef]
- Guo, D.; Horio, D.T.; Grove, J.S.; Dashwood, R.H. Inhibition by chlorophyllin of 2-amino-3-methylimidazo-[4,5-f] quinoline-induced tumorigenesis in the male F344 rat. Cancer Lett. 1995, 95, 161–165. [Google Scholar] [CrossRef]
- Wang, R.; Dashwood, W.M.; Nian, H.; Löhr, C.V.; Fischer, K.A.; Tsuchiya, N.; Nakagama, H.; Ashktorab, H.; Dashwood, R.H. NADPH oxidase overexpression in human colon cancers and rat colon tumors induced by 2-amino-1-methyl-6-phenylimidazo [4, 5-b] pyridine (PhIP). Int. J. Cancer 2011, 128, 2581–2590. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Nian, H.; Rajendran, P.; Kim, E.; Dashwood, W.; Pinto, J.; Boardman, L.; Thibodeau, S.; Limburg, P.; Löhr, C. HDAC8 and STAT3 repress BMF gene activity in colon cancer cells. Cell Death Dis. 2014, 5, e1476. [Google Scholar] [CrossRef]
- Blum, C.A.; Xu, M.; Orner, G.A.; Fong, A.T.; Bailey, G.S.; Stoner, G.D.; Horio, D.T.; Dashwood, R.H. β-Catenin mutation in rat colon tumors initiated by 1, 2-dimethylhydrazine and 2-amino-3-methylimidazo [4, 5-f] quinoline, and the effect of post-initiation treatment with chlorophyllin and indole-3-carbinol. Carcinogenesis 2001, 22, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, C.M.; Lynch, T.P.; Sodi, V.L.; Falcone, J.N.; Schwab, L.P.; Peacock, D.L.; Vocadlo, D.J.; Seagroves, T.N.; Reginato, M.J. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell 2014, 54, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-A.; Yeom, Y.I. Metabolic signaling to epigenetic alterations in cancer. Biomol. Ther. 2018, 26, 69. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G.; McKnight, S.L. Influence of metabolism on epigenetics and disease. Cell 2013, 153, 56–69. [Google Scholar] [CrossRef]
- Trefely, S.; Lovell, C.D.; Snyder, N.W.; Wellen, K.E. Compartmentalised acyl-CoA metabolism and roles in chromatin regulation. Mol. Metab. 2020, 38, 100941. [Google Scholar] [CrossRef]
- Lund, P.J.; Gates, L.A.; Leboeuf, M.; Smith, S.A.; Chau, L.; Lopes, M.; Friedman, E.S.; Saiman, Y.; Kim, M.S.; Shoffler, C.A.; et al. Stable isotope tracing in vivo reveals a metabolic bridge linking the microbiota to host histone acetylation. Cell Rep. 2022, 41, 111809. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Y.; Chen, X.-F.; Tang, X. Acylations in cardiovascular biology and diseases, what’s beyond acetylation. Ebiomedicine 2023, 87, 104418. [Google Scholar] [CrossRef]
- Trub, A.G.; Hirschey, M.D. Reactive acyl-CoA species modify proteins and induce carbon stress. Trends Biochem. Sci. 2018, 43, 369–379. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Singh, B.N.; Zhang, G.; Hwa, Y.L.; Li, J.; Dowdy, S.C.; Jiang, S.-W. Nonhistone protein acetylation as cancer therapy targets. Expert Rev. Anticancer. Ther. 2010, 10, 935–954. [Google Scholar] [CrossRef]
- Li, T.; Zhang, C.; Hassan, S.; Liu, X.; Song, F.; Chen, K.; Zhang, W.; Yang, J. Histone deacetylase 6 in cancer. J. Hematol. Oncol. 2018, 11, 111. [Google Scholar] [CrossRef]
- Deakin, N.O.; Turner, C.E. Paxillin inhibits HDAC6 to regulate microtubule acetylation, Golgi structure, and polarized migration. J. Cell Biol. 2014, 206, 395–413. [Google Scholar] [CrossRef]
- Klein, B.J.; Jang, S.M.; Lachance, C.; Mi, W.; Lyu, J.; Sakuraba, S.; Krajewski, K.; Wang, W.W.; Sidoli, S.; Liu, J. Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nat. Commun. 2019, 10, 4724. [Google Scholar] [CrossRef]
- Dai, L.; Peng, C.; Montellier, E.; Lu, Z.; Chen, Y.; Ishii, H.; Debernardi, A.; Buchou, T.; Rousseaux, S.; Jin, F.; et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014, 10, 365–370. [Google Scholar] [CrossRef]
- Goudarzi, A.; Zhang, D.; Huang, H.; Barral, S.; Kwon, O.K.; Qi, S.; Tang, Z.; Buchou, T.; Vitte, A.-L.; He, T.; et al. Dynamic Competing Histone H4 K5K8 Acetylation and Butyrylation Are Hallmarks of Highly Active Gene Promoters. Mol. Cell 2016, 62, 169–180. [Google Scholar] [CrossRef]
- Sabari, B.R.; Tang, Z.; Huang, H.; Yong-Gonzalez, V.; Molina, H.; Kong, H.E.; Dai, L.; Shimada, M.; Cross, J.R.; Zhao, Y.; et al. Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Mol. Cell 2015, 58, 203–215. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, D.; Chung, D.; Tang, Z.; Huang, H.; Dai, L.; Qi, S.; Li, J.; Colak, G.; Chen, Y.; et al. Metabolic Regulation of Gene Expression by Histone Lysine β-Hydroxybutyrylation. Mol. Cell 2016, 62, 194–206. [Google Scholar] [CrossRef]
- Smestad, J.; Erber, L.; Chen, Y.; Maher, L.J. Chromatin succinylation correlates with active gene expression and is perturbed by defective TCA cycle metabolism. Iscience 2018, 2, 63–75. [Google Scholar] [CrossRef]
- Bao, X.; Liu, Z.; Zhang, W.; Gladysz, K.; Fung, Y.M.E.; Tian, G.; Xiong, Y.; Wong, J.W.H.; Yuen, K.W.Y.; Li, X.D. Glutarylation of Histone H4 Lysine 91 Regulates Chromatin Dynamics. Mol. Cell 2019, 76, 660–675.e669. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, D.; Wang, Y.; Perez-Neut, M.; Han, Z.; Zheng, Y.G.; Hao, Q.; Zhao, Y. Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun. 2018, 9, 3374. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Jo, C.; Park, S.; Oh, S.; Choi, J.; Kim, E.-K.; Youn, H.-D.; Cho, E.-J. Histone acylation marks respond to metabolic perturbations and enable cellular adaptation. Exp. Mol. Med. 2020, 52, 2005–2019. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, J.; Jiang, S. Role of histone acetyltransferases and histone deacetylases in adipocyte differentiation and adipogenesis. Eur. J. Cell Biol. 2014, 93, 170–177. [Google Scholar] [CrossRef]
- Peserico, A.; Simone, C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011, 2011, 371832. [Google Scholar] [CrossRef]
- Wapenaar, H.; Dekker, F.J. Histone acetyltransferases: Challenges in targeting bi-substrate enzymes. Clin. Epigenetics 2016, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Rodger, E.J.; Eccles, M.R. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin. Cancer Biol. 2018, 51, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Abmayr, S.M.; Workman, J.L. Diverse Activities of Histone Acylations Connect Metabolism to Chromatin Function. Mol. Cell 2016, 63, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, V. The expanding constellation of histone post-translational modifications in the epigenetic landscape. Genes 2021, 12, 1596. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Cat, A.; Zheng, Y.G. New Histone Lysine Acylation Biomarkers and Their Roles in Epigenetic Regulation. Curr. Protoc. 2023, 3, e746. [Google Scholar] [CrossRef]
- Grevengoed, T.J.; Klett, E.L.; Coleman, R.A. Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 2014, 34, 1–30. [Google Scholar] [CrossRef]
- Morrison, A.J. Cancer cell metabolism connects epigenetic modifications to transcriptional regulation. FEBS J. 2022, 289, 1302–1314. [Google Scholar] [CrossRef]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef]
- Dai, Z.; Ramesh, V.; Locasale, J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020, 21, 737–753. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Y.R.; Liu, K.; Yin, Z.; Liu, R.; Xia, Y.; Tan, L.; Yang, P.; Lee, J.-H.; Li, X.-j. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 2017, 552, 273–277. [Google Scholar] [CrossRef]
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Ntorla, A.; Burgoyne, J.R. The regulation and function of histone crotonylation. Front. Cell Dev. Biol. 2021, 9, 624914. [Google Scholar] [CrossRef] [PubMed]
- Zorro Shahidian, L.; Haas, M.; Le Gras, S.; Nitsch, S.; Mourão, A.; Geerlof, A.; Margueron, R.; Michaelis, J.; Daujat, S.; Schneider, R. Succinylation of H3K122 destabilizes nucleosomes and enhances transcription. EMBO Rep. 2021, 22, e51009. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sprung, R.; Tang, Y.; Ball, H.; Sangras, B.; Kim, S.C.; Falck, J.R.; Peng, J.; Gu, W.; Zhao, Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteom. 2007, 6, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhang, D.; Weng, Y.; Delaney, K.; Tang, Z.; Yan, C.; Qi, S.; Peng, C.; Cole, P.A.; Roeder, R.G. The regulatory enzymes and protein substrates for the lysine β-hydroxybutyrylation pathway. Sci. Adv. 2021, 7, eabe2771. [Google Scholar] [CrossRef] [PubMed]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; McCaffery, J.M.; Irizarry, R.A.; Boeke, J.D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 2006, 23, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.V.; Carrer, A.; Shah, S.; Snyder, N.W.; Wei, S.; Venneti, S.; Worth, A.J.; Yuan, Z.-F.; Lim, H.-W.; Liu, S. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014, 20, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Sutter, B.M.; Li, B.; Tu, B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 2011, 42, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Moffett, J.R.; Puthillathu, N.; Vengilote, R.; Jaworski, D.M.; Namboodiri, A.M. Acetate revisited: A key biomolecule at the nexus of metabolism, epigenetics and oncogenesis—Part 1: Acetyl-CoA, acetogenesis and acyl-CoA short-chain synthetases. Front. Physiol. 2020, 11, 580167. [Google Scholar] [CrossRef]
- Legube, G.; Trouche, D. Regulating histone acetyltransferases and deacetylases. EMBO Rep 2003, 4, 944–947. [Google Scholar] [CrossRef]
- Roessler, C.; Tüting, C.; Meleshin, M.; Steegborn, C.; Schutkowski, M. A novel continuous assay for the deacylase sirtuin 5 and other deacetylases. J. Med. Chem. 2015, 58, 7217–7223. [Google Scholar] [CrossRef]
- Yang, L.; Ma, X.; He, Y.; Yuan, C.; Chen, Q.; Li, G.; Chen, X. Sirtuin 5: A review of structure, known inhibitors and clues for developing new inhibitors. Sci. China Life Sci. 2017, 60, 249–256. [Google Scholar] [CrossRef]
- Andrews, A.J.; Chen, X.; Zevin, A.; Stargell, L.A.; Luger, K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 2010, 37, 834–842. [Google Scholar] [CrossRef]
- Manohar, M.; Mooney, A.M.; North, J.A.; Nakkula, R.J.; Picking, J.W.; Edon, A.; Fishel, R.; Poirier, M.G.; Ottesen, J.J. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J. Biol. Chem. 2009, 284, 23312–23321. [Google Scholar] [CrossRef] [PubMed]
- Khochbin, S.; Verdel, A.; Lemercier, C.; Seigneurin-Berny, D. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 2001, 11, 162–166. [Google Scholar] [CrossRef]
- Baldensperger, T.; Glomb, M.A. Pathways of non-enzymatic lysine acylation. Front. Cell Dev. Biol. 2021, 9, 664553. [Google Scholar] [CrossRef]
- Baldensperger, T.; Eggen, M.; Kappen, J.; Winterhalter, P.R.; Pfirrmann, T.; Glomb, M.A. Comprehensive analysis of posttranslational protein modifications in aging of subcellular compartments. Sci. Rep. 2020, 10, 7596. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Sano, N.; Muto, S.; Horikoshi, M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief. Funct. Genom. 2006, 5, 190–208. [Google Scholar] [CrossRef]
- Kaczmarska, Z.; Ortega, E.; Goudarzi, A.; Huang, H.; Kim, S.; Márquez, J.A.; Zhao, Y.; Khochbin, S.; Panne, D. Structure of p300 in complex with acyl-CoA variants. Nat. Chem. Biol. 2017, 13, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Madiraju, S.R.M.; Aumais, A.; Joly, É.; Prentki, M. A Role for ATP-Citrate Lyase, Malic Enzyme, and Pyruvate/Citrate Cycling in Glucose-induced Insulin Secretion*. J. Biol. Chem. 2007, 282, 35657–35665. [Google Scholar] [CrossRef]
- Koren, D.; Palladino, A. Chapter 3—Hypoglycemia. In Genetic Diagnosis of Endocrine Disorders, 2nd ed.; Weiss, R.E., Refetoff, S., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 31–75. [Google Scholar] [CrossRef]
- Blasl, A.-T.; Schulze, S.; Qin, C.; Graf, L.G.; Vogt, R.; Lammers, M. Post-translational lysine ac (et) ylation in health, ageing and disease. Biol. Chem. 2022, 403, 151–194. [Google Scholar] [CrossRef]
- Miller, T.L.; Jenesel, S.E. Enzymology of butyrate formation by Butyrivibrio fibrisolvens. J. Bacteriol. 1979, 138, 99–104. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Khan, A.; Bridgers, J.B.; Strahl, B.D. Expanding the Reader Landscape of Histone Acylation. Structure 2017, 25, 571–573. [Google Scholar] [CrossRef]
- Kim, J.; Daniel, J.; Espejo, A.; Lake, A.; Krishna, M.; Xia, L.; Zhang, Y.; Bedford, M.T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006, 7, 397–403. [Google Scholar] [CrossRef]
- Scheid, R.; Chen, J.; Zhong, X. Biological role and mechanism of chromatin readers in plants. Curr. Opin. Plant Biol. 2021, 61, 102008. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.L.; Licht, J.D. Targeting epigenetics in cancer. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 187–207. [Google Scholar] [CrossRef]
- Josling, G.A.; Selvarajah, S.A.; Petter, M.; Duffy, M.F. The role of bromodomain proteins in regulating gene expression. Genes 2012, 3, 320–343. [Google Scholar] [CrossRef] [PubMed]
- Huber, F.M.; Greenblatt, S.M.; Davenport, A.M.; Martinez, C.; Xu, Y.; Vu, L.P.; Nimer, S.D.; Hoelz, A. Histone-binding of DPF2 mediates its repressive role in myeloid differentiation. Proc. Natl. Acad. Sci. USA 2017, 114, 6016–6021. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Abdelsalam, S.A.; Renu, K.; Veeraraghavan, V.; Ben Ammar, R.; Ahmed, E.A. Polyphenols as potent epigenetics agents for cancer. Int. J. Mol. Sci. 2022, 23, 11712. [Google Scholar] [CrossRef] [PubMed]
- Bonkowski, M.S.; Sinclair, D.A. Slowing ageing by design: The rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 2016, 17, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Milne, J.C.; Denu, J.M. The Sirtuin family: Therapeutic targets to treat diseases of aging. Curr. Opin. Chem. Biol. 2008, 12, 11–17. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Song, N.-Y.; Suh, J.; Kim, D.-H.; Kim, W.; Ann, J.; Lee, J.; Baek, J.-H.; Na, H.-K.; Surh, Y.-J. Curcumin suppresses oncogenicity of human colon cancer cells by covalently modifying the cysteine 67 residue of SIRT1. Cancer Lett. 2018, 431, 219–229. [Google Scholar] [CrossRef]
- Isaac, A.R.; da Silva, E.A.N.; de Matos, R.J.B.; Augusto, R.L.; Moreno, G.M.M.; Mendonça, I.P.; de Souza, R.F.; Cabral-Filho, P.E.; Rodrigues, C.G.; Gonçalves-Pimentel, C.; et al. Low omega-6/omega-3 ratio in a maternal protein-deficient diet promotes histone-3 changes in progeny neural cells and favors leukemia inhibitory factor gene transcription. J. Nutr. Biochem. 2018, 55, 229–242. [Google Scholar] [CrossRef]
- Sadli, N.; Ackland, M.L.; De Mel, D.; Sinclair, A.J.; Suphioglu, C. Effects of zinc and DHA on the epigenetic regulation of human neuronal cells. Cell. Physiol. Biochem. 2012, 29, 87–98. [Google Scholar] [CrossRef]
- Bakhshi, T.J.; Way, T.; Muncy, B.; Georgel, P.T. Effects of the omega-3 fatty acid DHA on histone and p53 acetylation in diffuse large B cell lymphoma. Biochem. Cell Biol. 2023, 101, 172–191. [Google Scholar] [CrossRef]
- Merzvinskyte, R.; Treigyte, G.; Savickiene, J.; Magnusson, K.E.; Navakauskiene, R. Effects of histone deacetylase inhibitors, sodium phenyl butyrate and vitamin B3, in combination with retinoic acid on granulocytic differentiation of human promyelocytic leukemia HL-60 cells. Ann. N. Y. Acad. Sci. 2006, 1091, 356–367. [Google Scholar] [CrossRef]
- Fellows, R.C. Microbiota Regulate Short Chain Fatty Acids and Influence Histone Acylations in Intestinal Epithelial Cells. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2020. [Google Scholar]
- Bradshaw, P.C. Acetyl-CoA Metabolism and Histone Acetylation in the Regulation of Aging and Lifespan. Antioxidants 2021, 10, 572. [Google Scholar] [CrossRef] [PubMed]
- Sidoli, S.; Trefely, S.; Garcia, B.A.; Carrer, A. Integrated Analysis of Acetyl-CoA and Histone Modification via Mass Spectrometry to Investigate Metabolically Driven Acetylation. Methods Mol. Biol. 2019, 1928, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Carrer, A.; Parris, J.L.; Trefely, S.; Henry, R.A.; Montgomery, D.C.; Torres, A.; Viola, J.M.; Kuo, Y.-M.; Blair, I.A.; Meier, J.L. Impact of a high-fat diet on tissue Acyl-CoA and histone acetylation levels. J. Biol. Chem. 2017, 292, 3312–3322. [Google Scholar] [CrossRef] [PubMed]
- Khajebishak, Y.; Alivand, M.; Faghfouri, A.H.; Moludi, J.; Payahoo, L. The effects of vitamins and dietary pattern on epigenetic modification of non-communicable diseases. Int. J. Vitam. Nutr. Res. 2021, 93, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Rohde, J.R.; Cardenas, M.E. The Tor Pathway Regulates Gene Expression by Linking Nutrient Sensing to Histone Acetylation. Mol. Cell. Biol. 2003, 23, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Bungard, D.; Fuerth, B.J.; Zeng, P.Y.; Faubert, B.; Maas, N.L.; Viollet, B.; Carling, D.; Thompson, C.B.; Jones, R.G.; Berger, S.L. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 2010, 329, 1201–1205. [Google Scholar] [CrossRef]
- Salminen, A.; Kauppinen, A.; Kaarniranta, K. AMPK/Snf1 signaling regulates histone acetylation: Impact on gene expression and epigenetic functions. Cell. Signal. 2016, 28, 887–895. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Sundaram, C.; Harikumar, K.B.; Tharakan, S.T.; Lai, O.S.; Sung, B.; Aggarwal, B.B. Cancer is a preventable disease that requires major lifestyle changes. Pharm. Res. 2008, 25, 2097–2116. [Google Scholar] [CrossRef] [PubMed]
- Irigaray, P.; Newby, J.A.; Clapp, R.; Hardell, L.; Howard, V.; Montagnier, L.; Epstein, S.; Belpomme, D. Lifestyle-related factors and environmental agents causing cancer: An overview. Biomed. Pharmacother. 2007, 61, 640–658. [Google Scholar] [CrossRef] [PubMed]
- Dey, P.; Kimmelman, A.C.; DePinho, R.A. Metabolic Codependencies in the Tumor Microenvironment. Cancer Discov. 2021, 11, 1067–1081. [Google Scholar] [CrossRef]
- Campbell, S.L.; Wellen, K.E. Metabolic signaling to the nucleus in cancer. Mol. Cell 2018, 71, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Chen, Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 2015, 368, 7–13. [Google Scholar] [CrossRef]
- Thakur, C.; Chen, F. Connections between metabolism and epigenetics in cancers. Semin. Cancer Biol. 2019, 57, 52–58. [Google Scholar] [CrossRef]
- Min, H.Y.; Lee, H.Y. Oncogene-Driven Metabolic Alterations in Cancer. Biomol. Ther. 2018, 26, 45–56. [Google Scholar] [CrossRef]
- Ge, T.; Gu, X.; Jia, R.; Ge, S.; Chai, P.; Zhuang, A.; Fan, X. Crosstalk between metabolic reprogramming and epigenetics in cancer: Updates on mechanisms and therapeutic opportunities. Cancer Commun. 2022, 42, 1049–1082. [Google Scholar] [CrossRef]
- Kim, Y.I. Folate and colorectal cancer: An evidence-based critical review. Mol. Nutr. Food Res. 2007, 51, 267–292. [Google Scholar]
- Ulrich, C.M.; Potter, J.D. Folate and cancer—Timing is everything. JAMA 2007, 297, 2408–2409. [Google Scholar] [CrossRef] [PubMed]
- Loedin, A.K.; Speijer, D. Is There a Carcinogenic Risk Attached to Vitamin B(12) Deficient Diets and What Should We Do About It? Reviewing the Facts. Mol. Nutr. Food Res. 2021, 65, e2000945. [Google Scholar] [CrossRef] [PubMed]
- Aksan, A.; Farrag, K.; Aksan, S.; Schroeder, O.; Stein, J. Flipside of the coin: Iron deficiency and colorectal cancer. Front. Immunol. 2021, 12, 635899. [Google Scholar] [CrossRef] [PubMed]
- Wanders, D.; Hobson, K.; Ji, X. Methionine Restriction and Cancer Biology. Nutrients 2020, 12, 684. [Google Scholar] [CrossRef] [PubMed]
- Bulanda, S.; Janoszka, B. Consumption of Thermally Processed Meat Containing Carcinogenic Compounds (Polycyclic Aromatic Hydrocarbons and Heterocyclic Aromatic Amines) versus a Risk of Some Cancers in Humans and the Possibility of Reducing Their Formation by Natural Food Additives-A Literature Review. Int. J. Environ. Res. Public Health 2022, 19, 4781. [Google Scholar] [CrossRef]
- Li, Y.; Hecht, S.S. Carcinogenic components of tobacco and tobacco smoke: A 2022 update. Food Chem. Toxicol. 2022, 165, 113179. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free. Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- García-Giménez, J.-L.; Garcés, C.; Romá-Mateo, C.; Pallardó, F.V. Oxidative stress-mediated alterations in histone post-translational modifications. Free. Radic. Biol. Med. 2021, 170, 6–18. [Google Scholar] [CrossRef]
- McDonald, O.G.; Li, X.; Saunders, T.; Tryggvadottir, R.; Mentch, S.J.; Warmoes, M.O.; Word, A.E.; Carrer, A.; Salz, T.H.; Natsume, S. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 2017, 49, 367–376. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the intersections between metabolism and cancer biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Hitosugi, T.; Chung, T.-W.; Xie, J.; Ge, Q.; Gu, T.-L.; Polakiewicz, R.D.; Chen, G.Z.; Boggon, T.J.; Lonial, S. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD+ redox homeostasis in cancer cells. Mol. Cell. Biol. 2011, 31, 4938–4950. [Google Scholar] [CrossRef]
- Chen, A.-N.; Luo, Y.; Yang, Y.-H.; Fu, J.-T.; Geng, X.-M.; Shi, J.-P.; Yang, J. Lactylation, a novel metabolic reprogramming code: Current status and prospects. Front. Immunol. 2021, 12, 688910. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Osthus, R.C.; Shim, H.; Kim, S.; Li, Q.; Reddy, R.; Mukherjee, M.; Xu, Y.; Wonsey, D.; Lee, L.A.; Dang, C.V. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000, 275, 21797–21800. [Google Scholar] [CrossRef] [PubMed]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.-Y.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef] [PubMed]
- Hitosugi, T.; Kang, S.; Vander Heiden, M.G.; Chung, T.-W.; Elf, S.; Lythgoe, K.; Dong, S.; Lonial, S.; Wang, X.; Chen, G.Z. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009, 2, ra73. [Google Scholar] [CrossRef]
- Miranda-Gonçalves, V.; Lameirinhas, A.; Henrique, R.; Jerónimo, C. Metabolism and Epigenetic Interplay in Cancer: Regulation and Putative Therapeutic Targets. Front. Genet. 2018, 9, 427. [Google Scholar] [CrossRef]
- Ling, R.; Chen, G.; Tang, X.; Liu, N.; Zhou, Y.; Chen, D. Acetyl-CoA synthetase 2(ACSS2): A review with a focus on metabolism and tumor development. Discov. Oncol. 2022, 13, 58. [Google Scholar] [CrossRef]
- Schug, Z.T.; Peck, B.; Jones, D.T.; Zhang, Q.; Grosskurth, S.; Alam, I.S.; Goodwin, L.M.; Smethurst, E.; Mason, S.; Blyth, K. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 2015, 27, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Lin, S.H.; Ren, F.; Li, J.T.; Chen, J.J.; Yao, C.B.; Yang, H.B.; Jiang, S.X.; Yan, G.Q.; Wang, D.; et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun. 2016, 7, 11960. [Google Scholar] [CrossRef]
- McDonnell, E.; Crown, S.B.; Fox, D.B.; Kitir, B.; Ilkayeva, O.R.; Olsen, C.A.; Grimsrud, P.A.; Hirschey, M.D. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016, 17, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Rabhi, N.; Hannou, S.A.; Froguel, P.; Annicotte, J.-S. Cofactors as metabolic sensors driving cell adaptation in physiology and disease. Front. Endocrinol. 2017, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012, 15, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Thaiss, C.A.; Elinav, E. Metabolites: Messengers between the microbiota and the immune system. Genes Dev. 2016, 30, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Rossi, T.; Vergara, D.; Fanini, F.; Maffia, M.; Bravaccini, S.; Pirini, F. Microbiota-derived metabolites in tumor progression and metastasis. Int. J. Mol. Sci. 2020, 21, 5786. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Li, J.; Menon, R.; Jayaraman, A.; Lee, K.; Huang, Y.; Dashwood, W.M.; Zhang, K.; Sun, D.; Dashwood, R.H. Dietary spinach reshapes the gut microbiome in an Apc-mutant genetic background: Mechanistic insights from integrated multi-omics. Gut Microbes 2021, 13, 1972756. [Google Scholar] [CrossRef]
- Hanus, M.; Parada-Venegas, D.; Landskron, G.; Wielandt, A.M.; Hurtado, C.; Alvarez, K.; Hermoso, M.A.; López-Köstner, F.; De la Fuente, M. Immune system, microbiota, and microbial metabolites: The unresolved triad in colorectal cancer microenvironment. Front. Immunol. 2021, 12, 612826. [Google Scholar] [CrossRef]
- Kespohl, M.; Vachharajani, N.; Luu, M.; Harb, H.; Pautz, S.; Wolff, S.; Sillner, N.; Walker, A.; Schmitt-Kopplin, P.; Boettger, T. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. Front. Immunol. 2017, 8, 1036. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-S.; Li, J.; Neja, S.; Kapoor, S.; Tovar Perez, J.E.; Tripathi, C.; Menon, R.; Jayaraman, A.; Lee, K.; Dashwood, W.M. Metabolomics of Acute vs. Chronic Spinach Intake in an Apc–Mutant Genetic Background: Linoleate and Butanoate Metabolites Targeting HDAC Activity and IFN–γ Signaling. Cells 2022, 11, 573. [Google Scholar] [CrossRef] [PubMed]
- Luu, M.; Weigand, K.; Wedi, F.; Breidenbend, C.; Leister, H.; Pautz, S.; Adhikary, T.; Visekruna, A. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 2018, 8, 14430. [Google Scholar] [CrossRef] [PubMed]
- Kiweler, N.; Wünsch, D.; Wirth, M.; Mahendrarajah, N.; Schneider, G.; Stauber, R.H.; Brenner, W.; Butter, F.; Krämer, O.H. Histone deacetylase inhibitors dysregulate DNA repair proteins and antagonize metastasis-associated processes. J. Cancer Res. Clin. Oncol. 2020, 146, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhou, H.; Wurtele, H.; Davies, B.; Horazdovsky, B.; Verreault, A.; Zhang, Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 2008, 134, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wang, J.; Rousseaux, S.; Tan, M.; Pan, L.; Peng, L.; Wang, S.; Xu, W.; Ren, J.; Liu, Y. Metabolically controlled histone H4K5 acylation/acetylation ratio drives BRD4 genomic distribution. Cell Rep. 2021, 36, 109460. [Google Scholar] [CrossRef] [PubMed]
- Nitsch, S.; Zorro Shahidian, L.; Schneider, R. Histone acylations and chromatin dynamics: Concepts, challenges, and links to metabolism. EMBO Rep. 2021, 22, e52774. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Liu, J.; Hua, F. Protein acylation: Mechanisms, biological functions and therapeutic targets. Signal Transduct. Target. Ther. 2022, 7, 396. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Chen, L.; Huang, L.; Gu, Y.; Cang, W.; Sun, P.; Xiang, Y. Lactate-lactylation hands between metabolic reprogramming and immunosuppression. Int. J. Mol. Sci. 2022, 23, 11943. [Google Scholar] [CrossRef]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef]
- Kamińska, K.; Nalejska, E.; Kubiak, M.; Wojtysiak, J.; Żołna, Ł.; Kowalewski, J.; Lewandowska, M.A. Prognostic and predictive epigenetic biomarkers in oncology. Mol. Diagn. Ther. 2019, 23, 83–95. [Google Scholar] [CrossRef]
- Seligson, D.B.; Horvath, S.; Shi, T.; Yu, H.; Tze, S.; Grunstein, M.; Kurdistani, S.K. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005, 435, 1262–1266. [Google Scholar] [CrossRef]
- Wan, L.; Wen, H.; Li, Y.; Lyu, J.; Xi, Y.; Hoshii, T.; Joseph, J.K.; Wang, X.; Loh, Y.-H.E.; Erb, M.A.; et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017, 543, 265–269. [Google Scholar] [CrossRef]
- Chai, X.; Guo, J.; Dong, R.; Yang, X.; Deng, C.; Wei, C.; Xu, J.; Han, W.; Lu, J.; Gao, C. Quantitative acetylome analysis reveals histone modifications that may predict prognosis in hepatitis B-related hepatocellular carcinoma. Clin. Transl. Med. 2021, 11, e313. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Rousseau, J.; Machol, K.; Cross, L.A.; Agre, K.E.; Gibson, C.F.; Goverde, A.; Engleman, K.L.; Verdin, H.; De Baere, E. Deficient histone H3 propionylation by BRPF1-KAT6 complexes in neurodevelopmental disorders and cancer. Sci. Adv. 2020, 6, eaax0021. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hamor, C.; An, Y.; Zhu, L.; Gong, Y.; Toh, Y.; Guo, Y.R. Biological functions and therapeutic potential of acylation by histone acetyltransferases. Acta Mater. Medica 2023, 2, 228–254. [Google Scholar] [CrossRef]
- Wan, J.; Liu, H.; Ming, L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed. Pharmacother. 2019, 111, 976–982. [Google Scholar] [CrossRef]
- Xu, X.; Zhu, X.; Liu, F.; Lu, W.; Wang, Y.; Yu, J. The effects of histone crotonylation and bromodomain protein 4 on prostate cancer cell lines. Transl. Androl. Urol. 2021, 10, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chang, Z.; Qin, L.-n.; Liang, B.; Han, J.-x.; Qiao, K.-l.; Yang, C.; Liu, Y.-r.; Zhou, H.-g.; Sun, T. MTA2 triggered R-loop trans-regulates BDH1-mediated β-hydroxybutyrylation and potentiates propagation of hepatocellular carcinoma stem cells. Signal Transduct. Target. Ther. 2021, 6, 135. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Zhao, K.; Qiu, P.; Deng, Z.; Yao, W.; Wang, J. Global landscape of 2-hydroxyisobutyrylation in human pancreatic cancer. Front. Oncol. 2022, 12, 1001807. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Li, F.; Sun, Q.; Lin, N.; Han, H.; You, K.; Tian, F.; Mao, Z.; Li, T.; Tong, T. p53 β-hydroxybutyrylation attenuates p53 activity. Cell Death Dis. 2019, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Feng, F.; Wu, J.; Fan, S.; Han, J.; Wang, S.; Yang, L.; Liu, W.; Wang, C.; Xu, K. Demethylzeylasteral targets lactate by inhibiting histone lactylation to suppress the tumorigenicity of liver cancer stem cells. Pharmacol. Res. 2022, 181, 106270. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Lin, Y.; Darwanto, A.; Song, X.; Xu, G.; Zhang, K. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 2009, 284, 32288–32295. [Google Scholar] [CrossRef] [PubMed]
- Ježek, P. 2-Hydroxyglutarate in Cancer Cells. Antioxid Redox Signal 2020, 33, 903–926. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Janke, R.; Iavarone, A.T.; Rine, J. Oncometabolite D-2-Hydroxyglutarate enhances gene silencing through inhibition of specific H3K36 histone demethylases. eLife 2017, 6, e22451. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, E.M.; Koeppe, E.; Everett, J.; Ulintz, P.; Kiel, M.; Osborne, J.; Williams, L.; Hanson, K.; Gruber, S.B.; Rozek, L.S. Germline genetic features of young individuals with colorectal cancer. Gastroenterology 2018, 154, 897–905.e891. [Google Scholar] [CrossRef]
- Stoffel, E.M.; Murphy, C.C. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology 2020, 158, 341–353. [Google Scholar] [CrossRef]
- Vilar, E.; Stoffel, E.M. Universal genetic testing for younger patients with colorectal cancer. JAMA Oncol. 2017, 3, 448–449. [Google Scholar] [CrossRef]
- Stoffel, E.M. Colorectal Cancer in Young Individuals: Opportunities for Prevention. J. Clin. Oncol. 2015, 33, 3525–3527. [Google Scholar] [CrossRef] [PubMed]
- Kanherkar, R.R.; Bhatia-Dey, N.; Csoka, A.B. Epigenetics across the human lifespan. Front. Cell Dev. Biol. 2014, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Busch, C.; Burkard, M.; Leischner, C.; Lauer, U.M.; Frank, J.; Venturelli, S. Epigenetic activities of flavonoids in the prevention and treatment of cancer. Clin. Epigenetics 2015, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Pehlivan, F.E. Diet-Epigenome interactions: Epi-drugs modulating the epigenetic machinery during cancer prevention. In Epigenetics to Optogenetics—A New Paradigm in the Study of Biology; IntechOpen: London, UK, 2021. [Google Scholar]
- Szarc vel Szic, K.; Ndlovu, M.N.; Haegeman, G.; Vanden Berghe, W. Nature or nurture: Let food be your epigenetic medicine in chronic inflammatory disorders. Biochem. Pharmacol. 2010, 80, 1816–1832. [Google Scholar] [CrossRef] [PubMed]
- Vahid, F.; Zand, H.; Nosrat–Mirshekarlou, E.; Najafi, R.; Hekmatdoost, A. The role dietary of bioactive compounds on the regulation of histone acetylases and deacetylases: A review. Gene 2015, 562, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.W.; Ferguson, B.S. Food Bioactive HDAC Inhibitors in the Epigenetic Regulation of Heart Failure. Nutrients 2018, 10, 1120. [Google Scholar] [CrossRef] [PubMed]
- Shankar, E.; Kanwal, R.; Candamo, M.; Gupta, S. Dietary phytochemicals as epigenetic modifiers in cancer: Promise and challenges. Semin. Cancer Biol. 2016, 40–41, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Özyalçin, B.; Sanlier, N. The effect of diet components on cancer with epigenetic mechanisms. Trends Food Sci. Technol. 2020, 102, 138–145. [Google Scholar] [CrossRef]
- Lévesque, S.; Pol, J.G.; Ferrere, G.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Trial watch: Dietary interventions for cancer therapy. Oncoimmunology 2019, 8, 1591878. [Google Scholar] [CrossRef]
- Mercier, B.D.; Tizpa, E.; Philip, E.J.; Feng, Q.; Huang, Z.; Thomas, R.M.; Pal, S.K.; Dorff, T.B.; Li, Y.R. Dietary Interventions in Cancer Treatment and Response: A Comprehensive Review. Cancers 2022, 14, 5149. [Google Scholar] [CrossRef]
- Mottamal, M.; Zheng, S.; Huang, T.L.; Wang, G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules 2015, 20, 3898–3941. [Google Scholar] [CrossRef]
- Pal, D.; Raj, K.; Nandi, S.S.; Sinha, S.; Mishra, A.; Mondal, A.; Lagoa, R.; Burcher, J.T.; Bishayee, A. Potential of Synthetic and Natural Compounds as Novel Histone Deacetylase Inhibitors for the Treatment of Hematological Malignancies. Cancers 2023, 15, 2808. [Google Scholar] [CrossRef]
- Sauter, E.R.; Mohammed, A. Natural Products for Cancer Prevention and Interception: Preclinical and Clinical Studies and Funding Opportunities. Pharmaceuticals 2024, 17, 136. [Google Scholar] [CrossRef]
- Taylor, E.M.; Jones, A.D.; Henagan, T.M. A review of mitochondrial-derived fatty acids in epigenetic regulation of obesity and type 2 diabetes. J. Nutr. Health Food Sci. 2014, 2, 1. [Google Scholar]
- McGee, S.L.; Fairlie, E.; Garnham, A.P.; Hargreaves, M. Exercise-induced histone modifications in human skeletal muscle. J. Physiol. 2009, 587, 5951–5958. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Bullo, M.M.H.; Ahmed, Z.; Imtiaz, A.; Yaqoob, E.; Jadoon, M.; Ahmed, H.; Afreen, A.; Yaqoob, S. Nutrigenomics: Epigenetics and cancer prevention: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.M.; Tollefsbol, T.O. Epigenetic diet: Impact on the epigenome and cancer. Epigenomics 2011, 3, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Ho, E.; Beaver, L.M.; Williams, D.E.; Dashwood, R.H. Dietary factors and epigenetic regulation for prostate cancer prevention. Adv. Nutr. 2011, 2, 497–510. [Google Scholar] [CrossRef]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef] [PubMed]
- Nieborak, A.; Schneider, R. Metabolic intermediates–cellular messengers talking to chromatin modifiers. Mol. Metab. 2018, 14, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Pérez, M.V.; Sainero-Alcolado, L.; Oliynyk, G.; Matuschek, I.; Balboni, N.; Ubhayasekera, S.; Snaebjornsson, M.T.; Makowski, K.; Aaltonen, K.; Bexell, D.; et al. Inhibition of fatty acid synthesis induces differentiation and reduces tumor burden in childhood neuroblastoma. iScience 2021, 24, 102128. [Google Scholar] [CrossRef]
- Sena, L.A.; Denmeade, S.R. Fatty Acid Synthesis in Prostate Cancer: Vulnerability or Epiphenomenon? Cancer Res. 2021, 81, 4385–4393. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bollu, L.R.; Tozzi, F.; Ye, X.; Bhattacharya, R.; Gao, G.; Dupre, E.; Xia, L.; Lu, J.; Fan, F. ATP citrate lyase mediates resistance of colorectal cancer cells to SN38. Mol. Cancer Ther. 2013, 12, 2782–2791. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.L.; Zhu, S.N.; Lu, S.L.; Dai, Z.S.; Jin, Y.L. Inhibitor of fatty acid synthase induced apoptosis in human colonic cancer cells. World J. Gastroenterol. 2000, 6, 295. [Google Scholar] [PubMed]
- Shiragami, R.; Murata, S.; Kosugi, C.; Tezuka, T.; Yamazaki, M.; Hirano, A.; Yoshimura, Y.; Suzuki, M.; Shuto, K.; Koda, K. Enhanced antitumor activity of cerulenin combined with oxaliplatin in human colon cancer cells. Int. J. Oncol. 2013, 43, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. HDAC inhibitors as epigenetic regulators of the immune system: Impacts on cancer therapy and inflammatory diseases. BioMed Res. Int. 2016, 2016, 8797206. [Google Scholar] [CrossRef]
- Su, M.; Gong, X.; Liu, F. An update on the emerging approaches for histone deacetylase (HDAC) inhibitor drug discovery and future perspectives. Expert Opin. Drug Discov. 2021, 16, 745–761. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Tian, Y.; Zhu, W.-G. The roles of histone deacetylases and their inhibitors in cancer therapy. Front. Cell Dev. Biol. 2020, 8, 576946. [Google Scholar] [CrossRef]
- Bose, P.; Dai, Y.; Grant, S. Histone deacetylase inhibitor (HDACI) mechanisms of action: Emerging insights. Pharmacol. Ther. 2014, 143, 323–336. [Google Scholar] [CrossRef]
- Bassett, S.A.; Barnett, M.P. The role of dietary histone deacetylases (HDACs) inhibitors in health and disease. Nutrients 2014, 6, 4273–4301. [Google Scholar] [CrossRef]
- Dashwood, R.H.; Ho, E. Dietary histone deacetylase inhibitors: From cells to mice to man. Semin. Cancer Biol. 2007, 17, 363–369. [Google Scholar] [CrossRef]
- Rajendran, P.; Ho, E.; Williams, D.E.; Dashwood, R.H. Dietary phytochemicals, HDAC inhibition, and DNA damage/repair defects in cancer cells. Clin. Epigenetics 2011, 3, 4. [Google Scholar] [CrossRef]
- Fiorentino, F.; Castiello, C.; Mai, A.; Rotili, D. Therapeutic Potential and Activity Modulation of the Protein Lysine Deacylase Sirtuin 5. J. Med. Chem. 2022, 65, 9580–9606. [Google Scholar] [CrossRef] [PubMed]
- Xiangyun, Y.; Xiaomin, N.; Yunhua, X.; Ziming, L.; Yongfeng, Y.; Zhiwei, C.; Shun, L. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget 2017, 8, 6984. [Google Scholar] [CrossRef] [PubMed]
- Madsen, A.S.; Olsen, C.A. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angew. Chem. (Int. Ed. Engl.) 2012, 51, 9083–9087. [Google Scholar] [CrossRef]
- Du, W.; Hua, F.; Li, X.; Zhang, J.; Li, S.; Wang, W.; Zhou, J.; Wang, W.; Liao, P.; Yan, Y. Loss of optineurin drives cancer immune evasion via palmitoylation-dependent IFNGR1 lysosomal sorting and degradation. Cancer Discov. 2021, 11, 1826–1843. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Nihira, N.T.; Bu, X.; Chu, C.; Zhang, J.; Kolodziejczyk, A.; Fan, Y.; Chan, N.T.; Ma, L.; Liu, J. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat. Cell Biol. 2020, 22, 1064–1075. [Google Scholar] [CrossRef]
- Ma, L.; Liu, J.; Liu, L.; Duan, G.; Wang, Q.; Xu, Y.; Xia, F.; Shan, J.; Shen, J.; Yang, Z. Overexpression of the Transcription Factor MEF2D in Hepatocellular Carcinoma Sustains Malignant Character by Suppressing G2–M Transition GenesPromotion of Cancer Cell Growth by MEF2D in HCC. Cancer Res. 2014, 74, 1452–1462. [Google Scholar] [CrossRef]
- Yao, H.; Lan, J.; Li, C.; Shi, H.; Brosseau, J.-P.; Wang, H.; Lu, H.; Fang, C.; Zhang, Y.; Liang, L. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 2019, 3, 306–317. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, J.-M.; Sun, L.; Chan, L.-C.; Li, C.-W.; Hsu, J.L.; Wei, Y.; Xia, W.; Hou, J.; Qiu, Y. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019, 29, 83–86. [Google Scholar] [CrossRef]
- Fang, H.; Huang, Y.; Luo, Y.; Tang, J.; Yu, M.; Zhang, Y.; Zhong, M. SIRT1 induces the accumulation of TAMs at colorectal cancer tumor sites via the CXCR4/CXCL12 axis. Cell. Immunol. 2022, 371, 104458. [Google Scholar] [CrossRef]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550, 128–132. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, B.; Liu, X.; Zhang, X.D.; Zhang, L.; Liu, T. Histone acetyltransferases CBP/p300 in tumorigenesis and CBP/p300 inhibitors as promising novel anticancer agents. Theranostics 2022, 12, 4935. [Google Scholar] [CrossRef] [PubMed]
- Waddell, A.R.; Huang, H.; Liao, D. CBP/p300: Critical Co-Activators for Nuclear Steroid Hormone Receptors and Emerging Therapeutic Targets in Prostate and Breast Cancers. Cancers 2021, 13, 2872. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.H.; Smolik, S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000, 14, 1553–1577. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, S.; Gustafson, T.; Zhang, M.; Chen, Y.-S.; Li, J.; Nguyen, N.; Perez, J.E.T.; Dashwood, W.M.; Rajendran, P.; Dashwood, R.H. Deacetylase plus bromodomain inhibition downregulates ERCC2 and suppresses the growth of metastatic colon cancer cells. Cancers 2021, 13, 1438. [Google Scholar] [CrossRef] [PubMed]
- Mazur, P.K.; Herner, A.; Mello, S.S.; Wirth, M.; Hausmann, S.; Sánchez-Rivera, F.J.; Lofgren, S.M.; Kuschma, T.; Hahn, S.A.; Vangala, D. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 2015, 21, 1163–1171. [Google Scholar] [CrossRef]
- Rajendran, P.; Johnson, G.; Li, L.; Chen, Y.-S.; Dashwood, M.; Nguyen, N.; Ulusan, A.; Ertem, F.; Zhang, M.; Li, J. Acetylation of CCAR2 establishes a BET/BRD9 acetyl switch in response to combined deacetylase and bromodomain inhibition. Cancer Res. 2019, 79, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wei, T.; Cai, Y.; Jin, J. Small molecules targeting the specific domains of histone-mark readers in cancer therapy. Molecules 2020, 25, 578. [Google Scholar] [CrossRef] [PubMed]
- Shorstova, T.; Foulkes, W.D.; Witcher, M. Achieving clinical success with BET inhibitors as anti-cancer agents. Br. J. Cancer 2021, 124, 1478–1490. [Google Scholar] [CrossRef]
- Kapoor, S.; Damiani, E.; Wang, S.; Dharmanand, R.; Tripathi, C.; Tovar Perez, J.E.; Dashwood, W.M.; Rajendran, P.; Dashwood, R.H. BRD9 Inhibition by Natural Polyphenols Targets DNA Damage/Repair and Apoptosis in Human Colon Cancer Cells. Nutrients 2022, 14, 4317. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Panchenko, T.; Yang, S.; Zhao, S.; Yan, P.; Zhang, W.; Xie, W.; Li, Y.; Zhao, Y.; Allis, C.D. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 2016, 12, 1111–1118. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Neja, S.A. Site-Specific DNA Demethylation as a Potential Target for Cancer Epigenetic Therapy. Epigenetics Insights 2020, 13, 2516865720964808. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Zhao, Y.T.; Lamonica, J.M.; Zhou, Z. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat. Commun. 2017, 8, 15315. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Eom, G.H. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam. Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef]
- Stimson, L.; Rowlands, M.G.; Newbatt, Y.M.; Smith, N.F.; Raynaud, F.I.; Rogers, P.; Bavetsias, V.; Gorsuch, S.; Jarman, M.; Bannister, A.; et al. Isothiazolones as inhibitors of PCAF and p300 histone acetyltransferase activity. Mol. Cancer Ther. 2005, 4, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, J.; Zhai, L.; Zhang, T.; Yin, H.; Gao, H.; Zhao, F.; Wang, Z.; Yang, X.; Jin, M.; et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell 2024, 187, 294–311.e221. [Google Scholar] [CrossRef]
- Ma, W.; Sun, Y.; Yan, R.; Zhang, P.; Shen, S.; Lu, H.; Zhou, Z.; Jiang, Z.; Ye, L.; Mao, Q.; et al. OXCT1 functions as a succinyltransferase, contributing to hepatocellular carcinoma via succinylating LACTB. Mol. Cell, 2024; in press. [Google Scholar] [CrossRef]
- Ozsvari, B.; Sotgia, F.; Simmons, K.; Trowbridge, R.; Foster, R.; Lisanti, M.P. Mitoketoscins: Novel mitochondrial inhibitors for targeting ketone metabolism in cancer stem cells (CSCs). Oncotarget 2017, 8, 78340–78350. [Google Scholar] [CrossRef]
- Tu, B.; Zhang, M.; Liu, T.; Huang, Y. Nanotechnology-based histone deacetylase inhibitors for cancer therapy. Front. Cell Dev. Biol. 2020, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ren, Y.; Weng, S.; Xu, H.; Li, L.; Han, X. A New Trend in Cancer Treatment: The Combination of Epigenetics and Immunotherapy. Front. Immunol. 2022, 13, 809761. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.; Tabernacki, T.; Kobyra, J.; Roberts, P.; Chiappinelli, K.B. Combining epigenetic and immune therapy to overcome cancer resistance. Semin. Cancer Biol. 2020, 65, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, L.; Álvarez-Errico, D.; Esteller, M. The contribution of epigenetics to cancer immunotherapy. Trends Immunol. 2020, 41, 676–691. [Google Scholar] [CrossRef]
- Huang, H.; Lin, S.; Garcia, B.A.; Zhao, Y. Quantitative proteomic analysis of histone modifications. Chem. Rev. 2015, 115, 2376–2418. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef]
- Dragic, H.; Chaveroux, C.; Cosset, E.; Manie, S.N. Modelling cancer metabolism in vitro: Current improvements and future challenges. FEBS J. 2022. early view. [Google Scholar] [CrossRef]
| Type of Acylation | Chemical Nature | Dietary/Metabolic Source | Writers | Readers | Erasers | References |
|---|---|---|---|---|---|---|
| Acetylation (Ac) | Hydrophobic | CHO & SCFA from gut microbes, glycolysis, TCA | p300/CBP, HAT, GNATs | BRD3, BRD4, PBRM1 | All HDAC family | [8,9,10] |
| Propionylation (Pr) | Hydrophobic | SCFA from dietary fiber & gut microbes, TCA | p300/CBP, GNATs, MYSTs | YEATS, DPF | SIRT1,2,3 | [11,12,13,14,15] |
| Butyrylation (Bu) | Hydrophobic | SCFA from dietary fiber & gut microbes, TCA | p300/CBP, GNATs, HBO1 | YEATS, DPF | SIRT1,2,3 | [9,15,16,17] |
| Crotonylation (Cr) | Hydrophobic | SCFA from dietary fiber & gut microbes, TCA | p300/CBP | YEATS, DPF | SIRT1,2,3, HDAC3 | [9,18,19,20,21] |
| Benzoylation (Bz) | Hydrophobic | N/A | HBO1 | YEATS, DPF | [5,9,22] | |
| β-Hydroxybutyrylation (Bhb) | Polar | Ketogenic diet, starvation, | p300/CBP | YEATS, DPF | SIRT3, HDAC1,2,3 | [23,24,25] |
| 2-Hydroxyisobutyrylation (Bhib) | Polar | SCFA, Amino acid metabolism | P300, MYSTs | YEATS, DPF | N/A | [16,26] |
| Lactylation (La) | Acidic | Glycolysis, lactate from exercise, LGSH | p300 | N/A | HDAC1,3 | [16] |
| Malonylation (Mal) | Acidic | Citrate metabolism, FAO | N/A | N/A | SIRT2,5 | [27,28] |
| Succinylation (Succ) | Acidic | TCA | p300/CBP, GNATs, CPT1A, GCN5 | YEATS | SIRT5, 7 | [17,29,30] |
| Glutarylation (Glu) | Acidic | TCA, amino acid metabolism | p300, GCN5 | N/A | N/A | [9,26] |
| O-GlcNacylation (GlcNac) | Polar | Pentose–phosphate pathway | N/A | N/A | N/A | [31] |
| Palmitoylation (Pal) | Hydrophilic | Edible oils, HFD | LPCAT1 | N/A | APT, PPT SIRT6 | [32,33] |
| Myristoylation (Myr) | Hydrophilic | Edible oils, HFD | N/A | N/A | SIRT2, 6 | [33,34] |
| Histone Acylation Type | Cancer Type | Association with Cancer | References |
|---|---|---|---|
| Global H3K18ac, H3K9ac, H3K12ac | Prostate | Elevated levels correlate with prostate cancer risk | [175] |
| Global losses of H3K16ac | Leukemia, lymphoma, breast, colorectal, lung, prostate, cervical | A hallmark of human tumor cells | [173] |
| H3K23pr | Medulloblastoma, leukemia, glioma, colorectal | Low H3K23pr contributes to cancer development | [178] |
| Global histone Kcr | Esophageal, colon, pancreatic, lung | Low Kcr is associated with cancer | [179] |
| HCC | Kcr levels correlate with HCC progression | [180] | |
| Prostate | Kcr levels correlate with prostate cancer malignancy | [181] | |
| H3K9bhb | HCC | High H3K9bhb correlates with HCC progression | [182] |
| Global Khib | Pancreatic | Khib is a tumor promoter in pancreatic cancer | [183] |
| H3K18la | Melanoma | High H3K18la enhances melanoma | [184] |
| H3K9la and H3K56la | HCC | High H3K9la and H3K56la increase the proliferation and migration of liver cancer stem cells | [185] |
| H3K79succ, H3K122succ | Glioblastoma | High H3K79succ promotes the proliferation and development of glioma cells | [77] |
| Global histone Kbz | HCC | Kbz is involved in HCC progression | [22] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neja, S.; Dashwood, W.M.; Dashwood, R.H.; Rajendran, P. Histone Acyl Code in Precision Oncology: Mechanistic Insights from Dietary and Metabolic Factors. Nutrients 2024, 16, 396. https://doi.org/10.3390/nu16030396
Neja S, Dashwood WM, Dashwood RH, Rajendran P. Histone Acyl Code in Precision Oncology: Mechanistic Insights from Dietary and Metabolic Factors. Nutrients. 2024; 16(3):396. https://doi.org/10.3390/nu16030396
Chicago/Turabian StyleNeja, Sultan, Wan Mohaiza Dashwood, Roderick H. Dashwood, and Praveen Rajendran. 2024. "Histone Acyl Code in Precision Oncology: Mechanistic Insights from Dietary and Metabolic Factors" Nutrients 16, no. 3: 396. https://doi.org/10.3390/nu16030396
APA StyleNeja, S., Dashwood, W. M., Dashwood, R. H., & Rajendran, P. (2024). Histone Acyl Code in Precision Oncology: Mechanistic Insights from Dietary and Metabolic Factors. Nutrients, 16(3), 396. https://doi.org/10.3390/nu16030396









