TransHSI: A Hybrid CNN-Transformer Method for Disjoint Sample-Based Hyperspectral Image Classification
Abstract
:1. Introduction
1.1. Literature Review
1.2. Contribution
- (1)
- The TransHSI proposes a new spectral–spatial feature extraction module, in which the spectral feature extraction module combines 3D CNNs with different convolution kernel sizes and Transformer to extract the global and local spectral features of HSIs. In addition, the spatial feature extraction module combines 2D CNNs and Transformer to extract the global and local spatial features of HSIs. The module mentioned above thoroughly considers the disparities between spectral and spatial characteristics in HSIs, facilitating the comprehensive extraction of both the global and local spectral–spatial features in HSIs.
- (2)
- A fusion module is proposed, which first cascades the extracted spectral–spatial features and the original HSIs after dimensionality reduction and captures relevant features from different stages of the network. Secondly, a semantic tokenizer is used to transform the features to enhance the discriminant ability of features. Finally, the features are represented and learned in the Transformer Encode module to fully utilize the image’s shallow and deep features to achieve an efficient fusion classification of spectral–spatial features.
- (3)
- In this paper, the effectiveness of TransHSI is verified using three publicly available datasets, and competitive results are obtained. Crop classification is assessed using the Indian Pines dataset, and urban land cover classification is assessed using the Pavia University dataset and the Data Fusion Contest 2018. These results provide a reference for future research focused on HSI classification.
2. Materials and Methods
2.1. CNNs
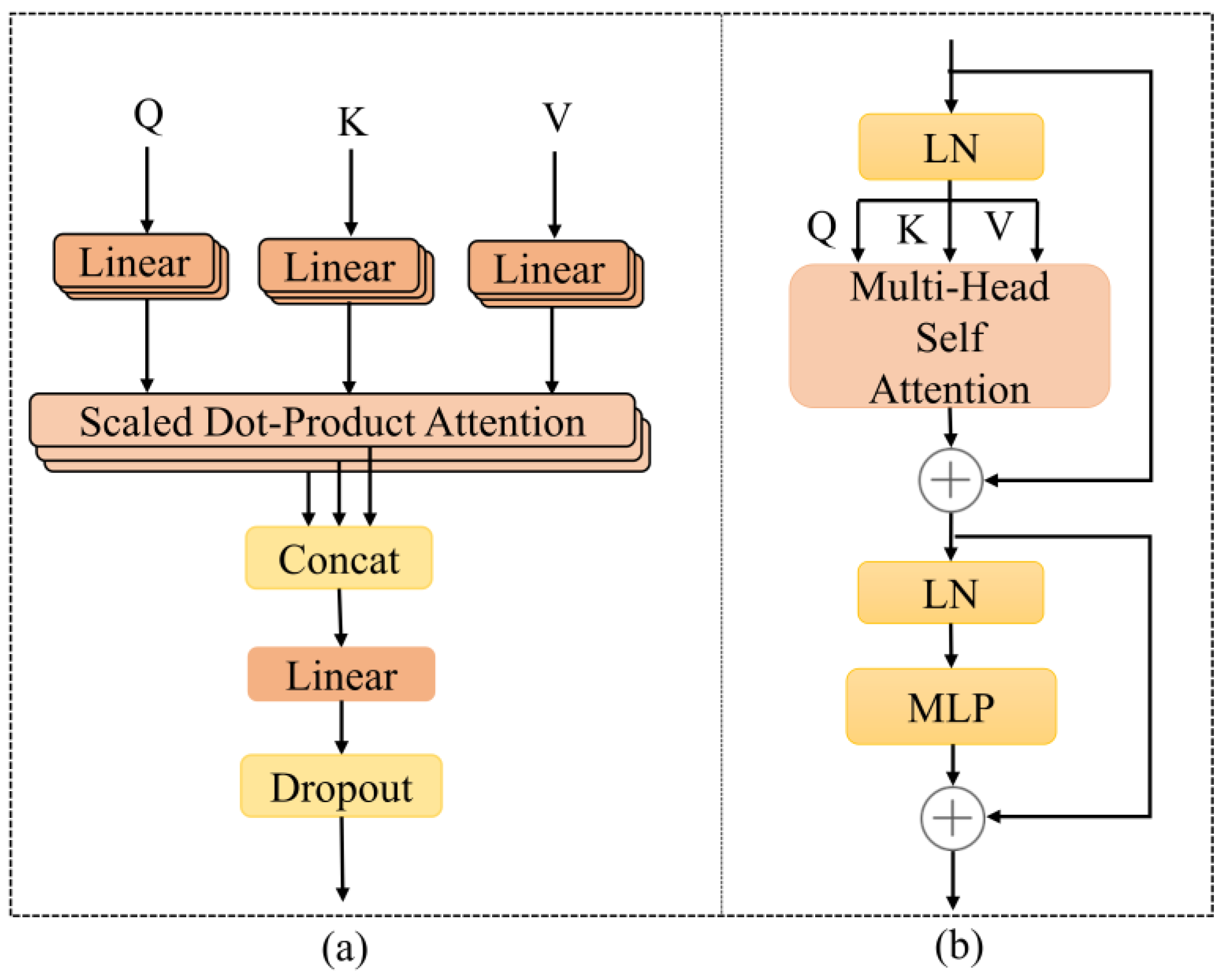
2.2. Transformer Encode
2.3. Proposed Methodology
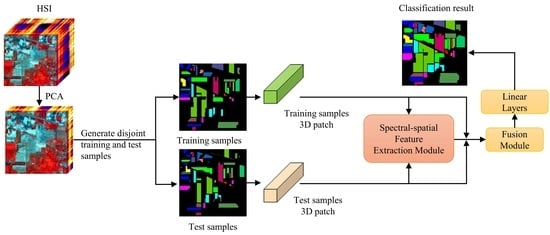
2.3.1. HSI Pretreatment Module
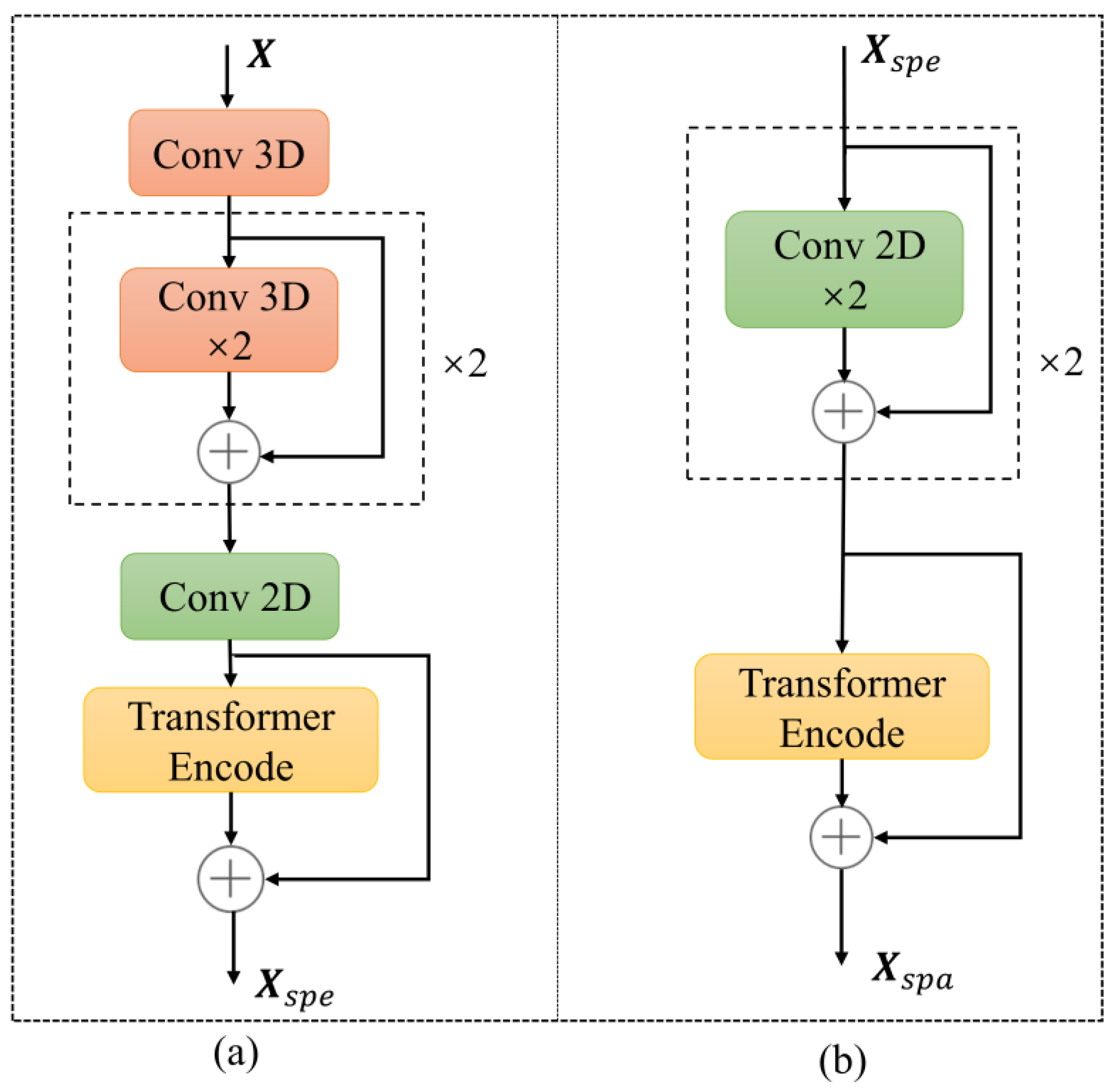
2.3.2. Spectral Feature Extraction Module
2.3.3. Spatial Feature Extraction Module
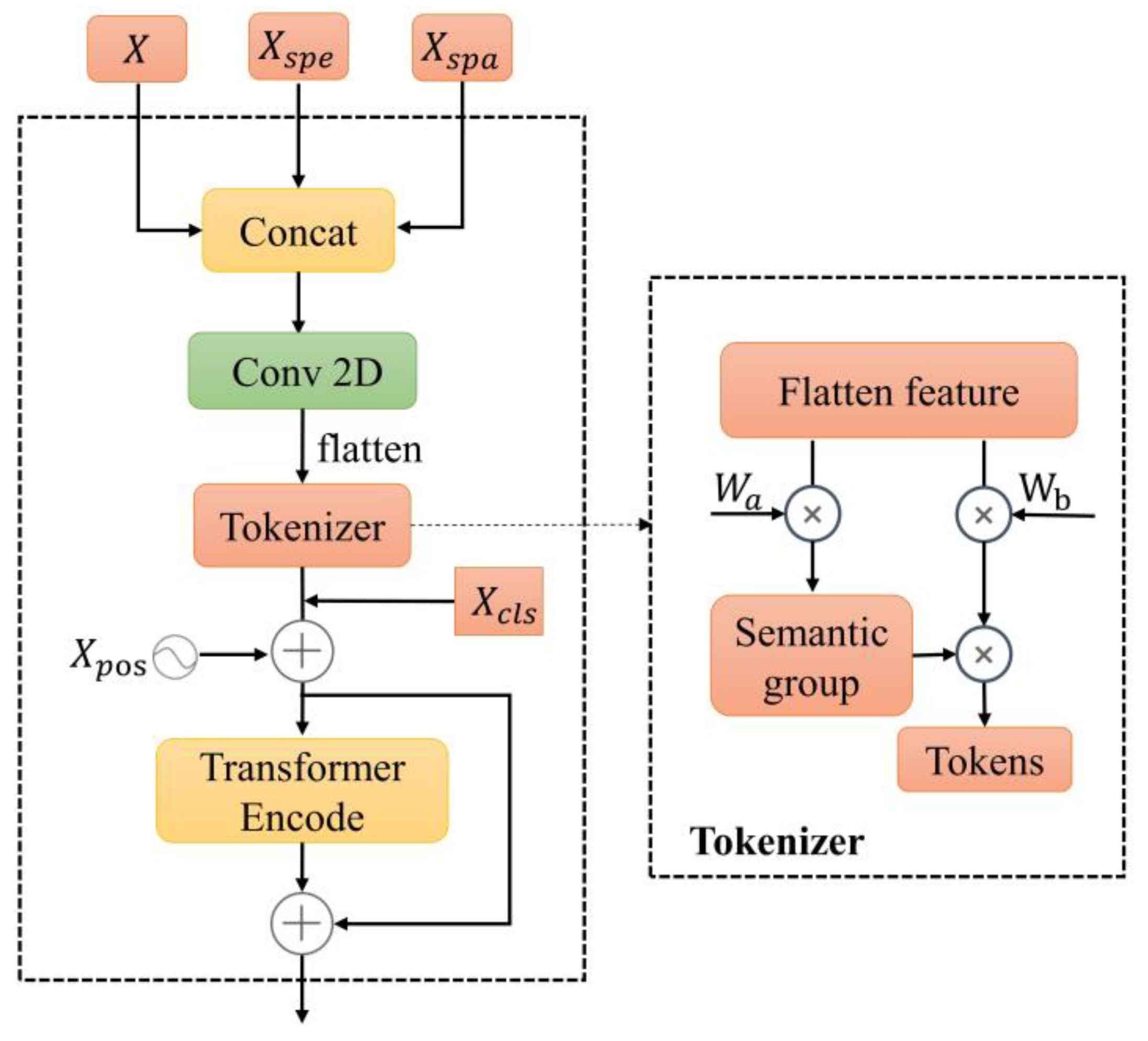
2.3.4. Fusion Module
2.4. Implementation of TransHSI
3. Datasets and Experimental Setup
3.1. Experimental Datasets
3.1.1. Indian Pines Dataset
3.1.2. Pavia University Dataset
3.1.3. Data Fusion Contest 2018 (DFC 2018)
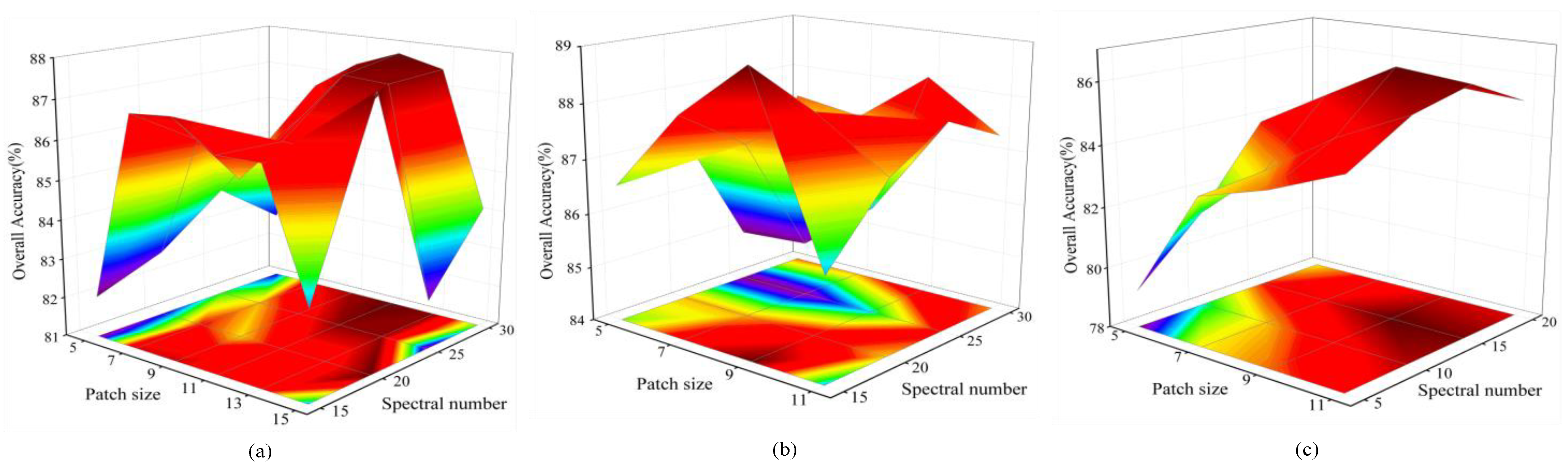
3.2. Experimental Setup
4. Experimental Result
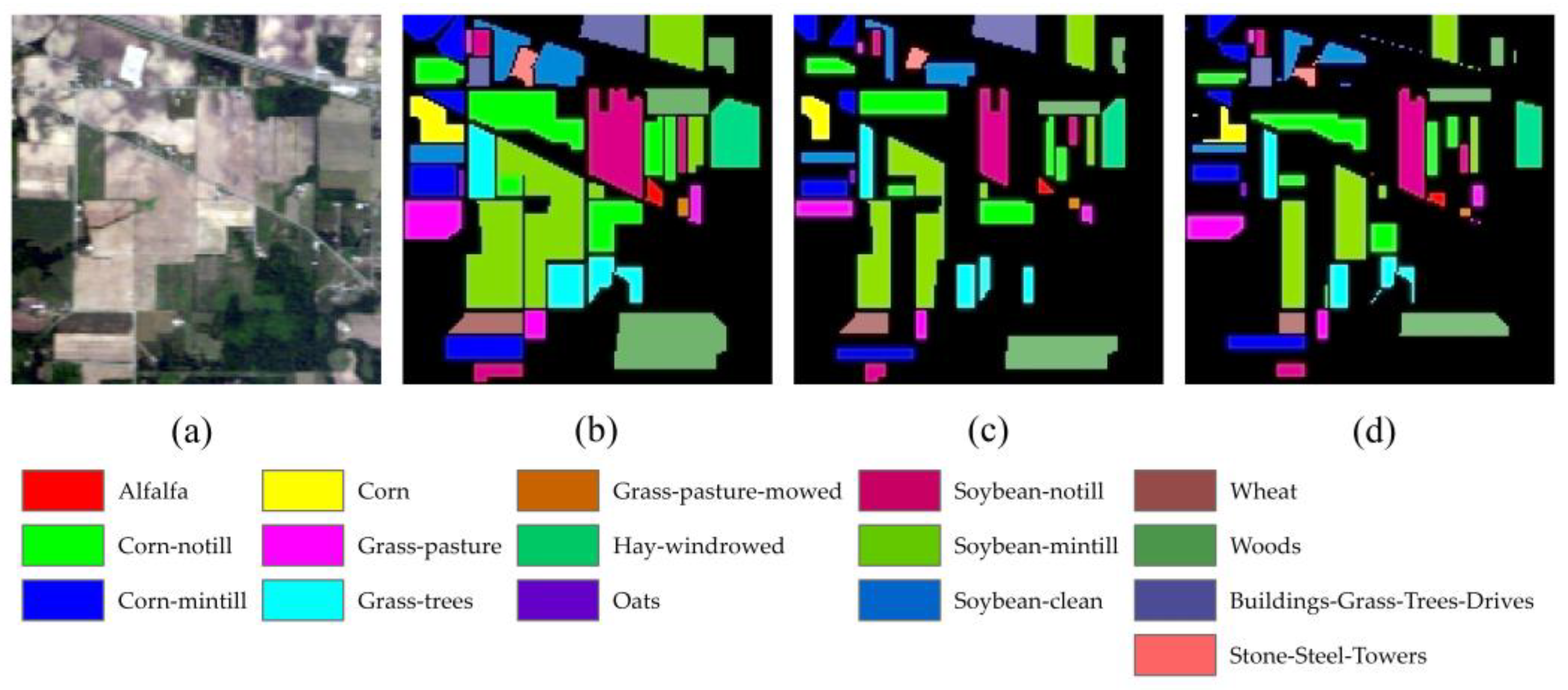
4.1. Classification Results for the Indian Pines Dataset
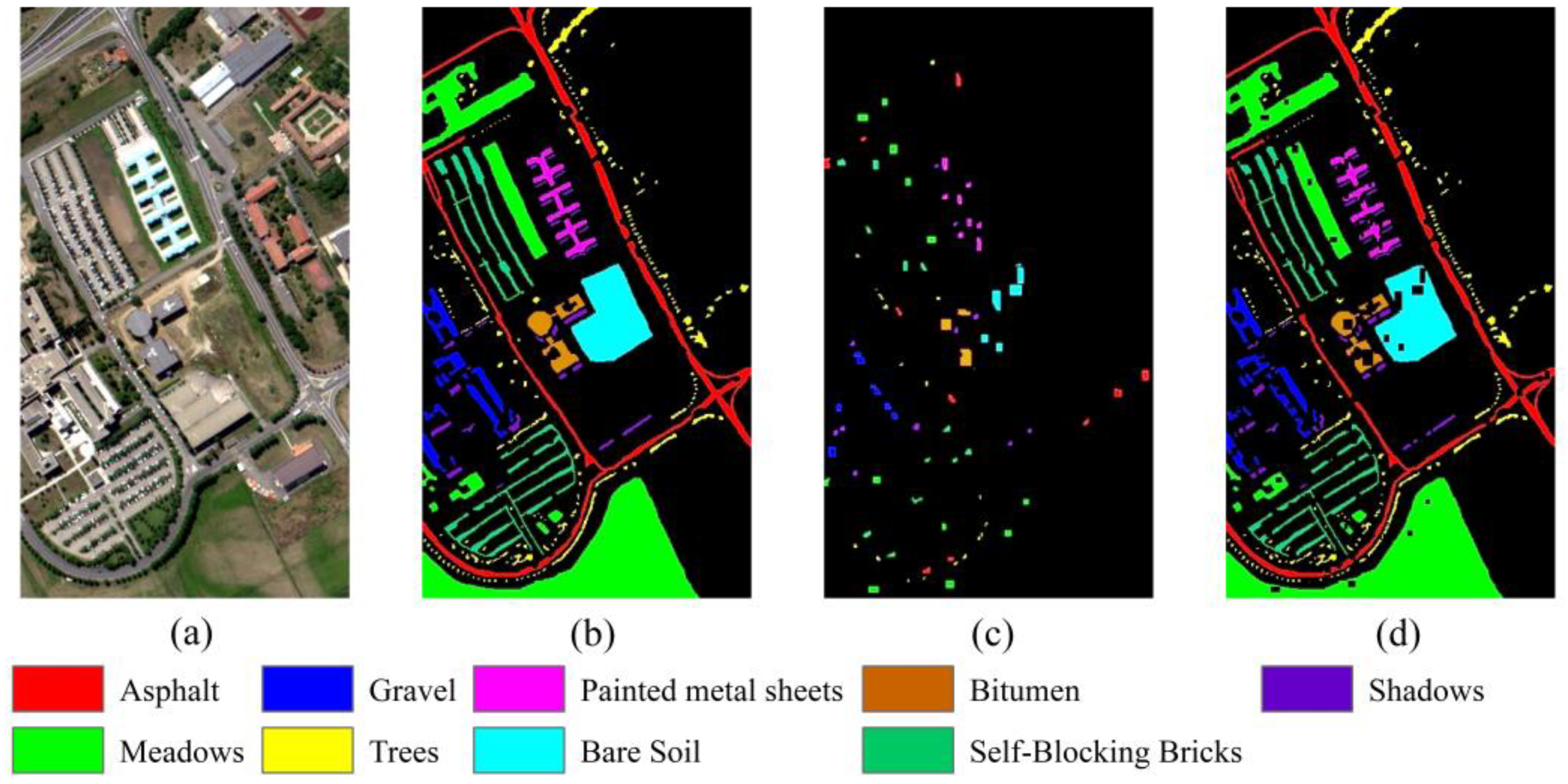
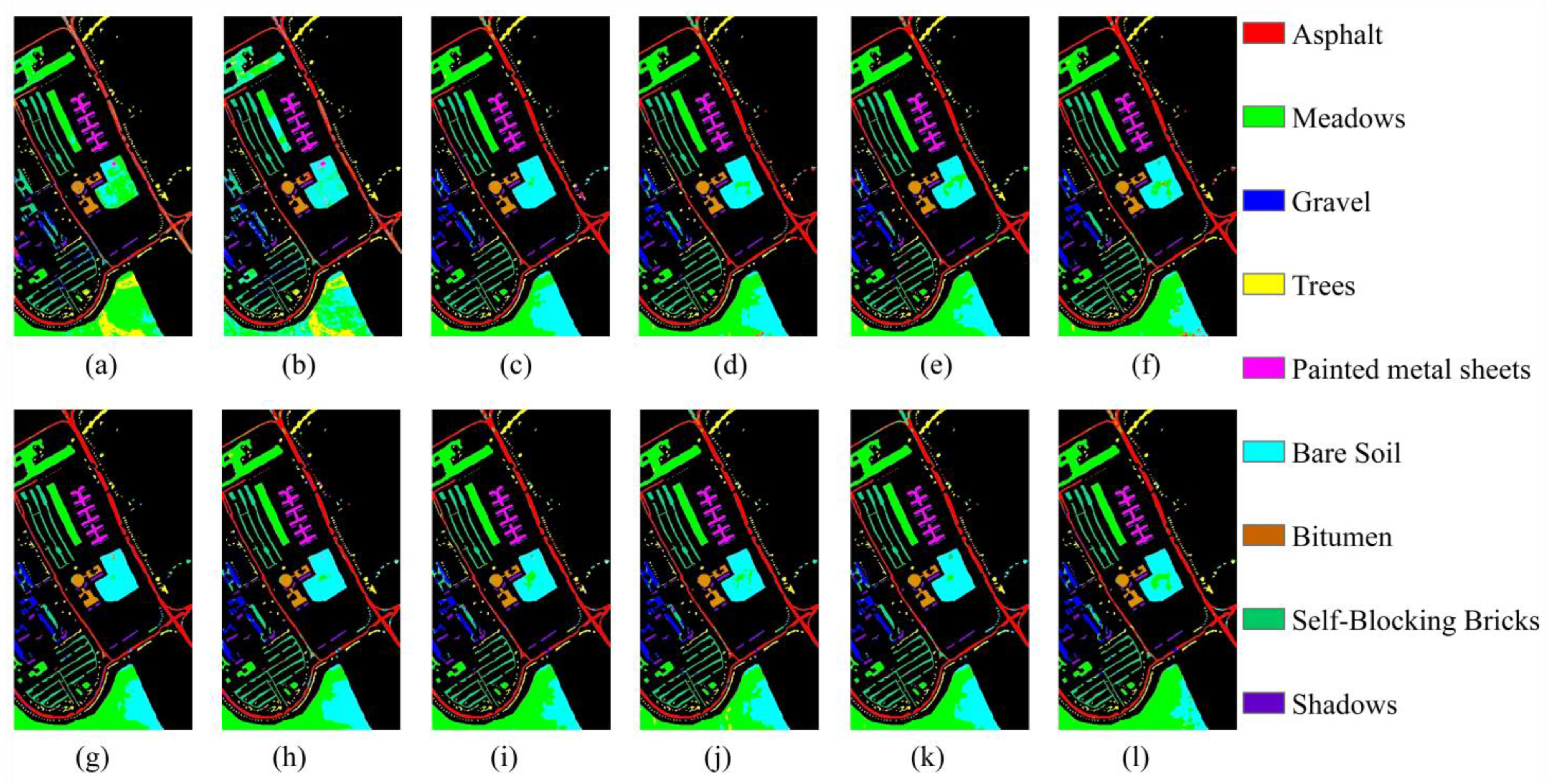
4.2. Classification Results for the Pavia University Dataset
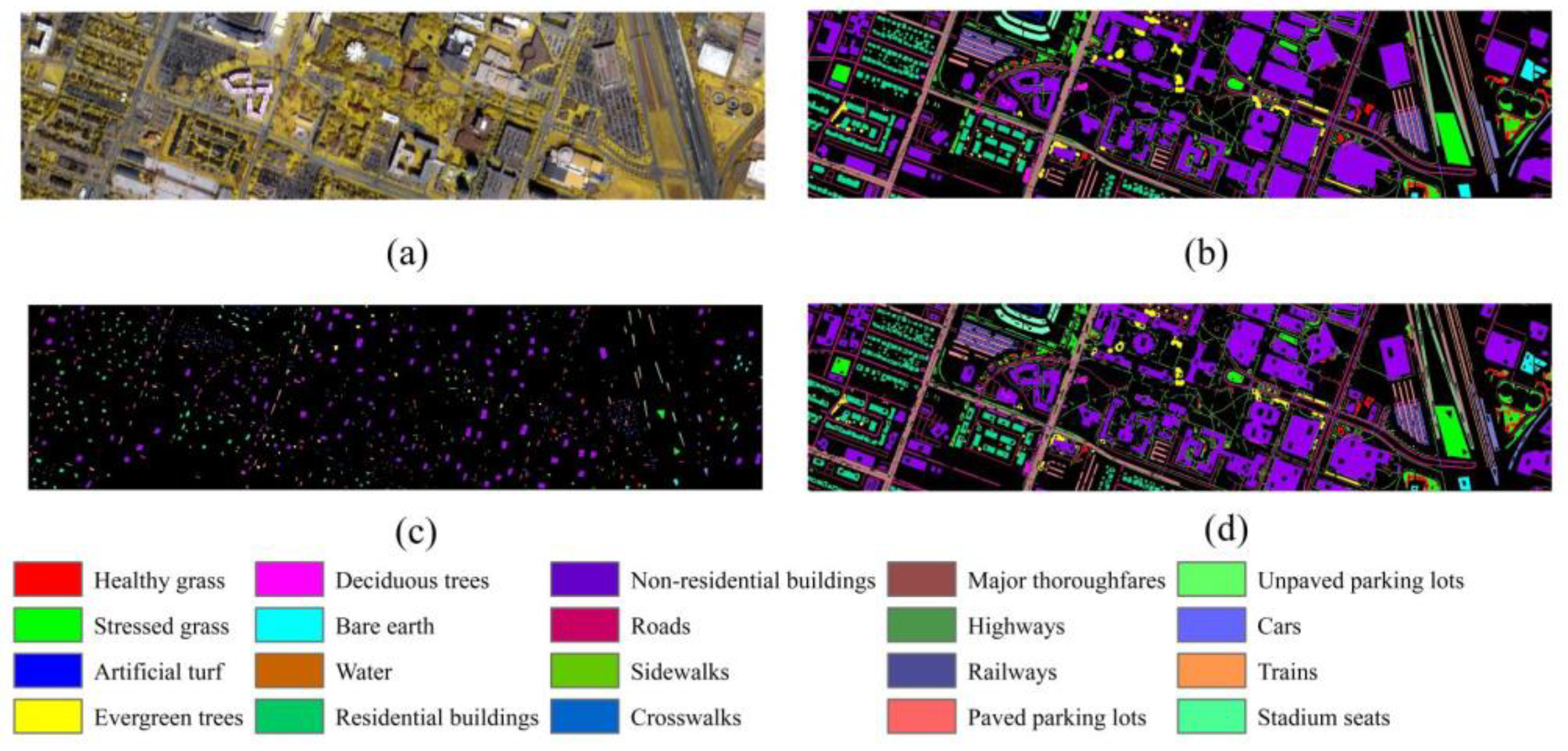
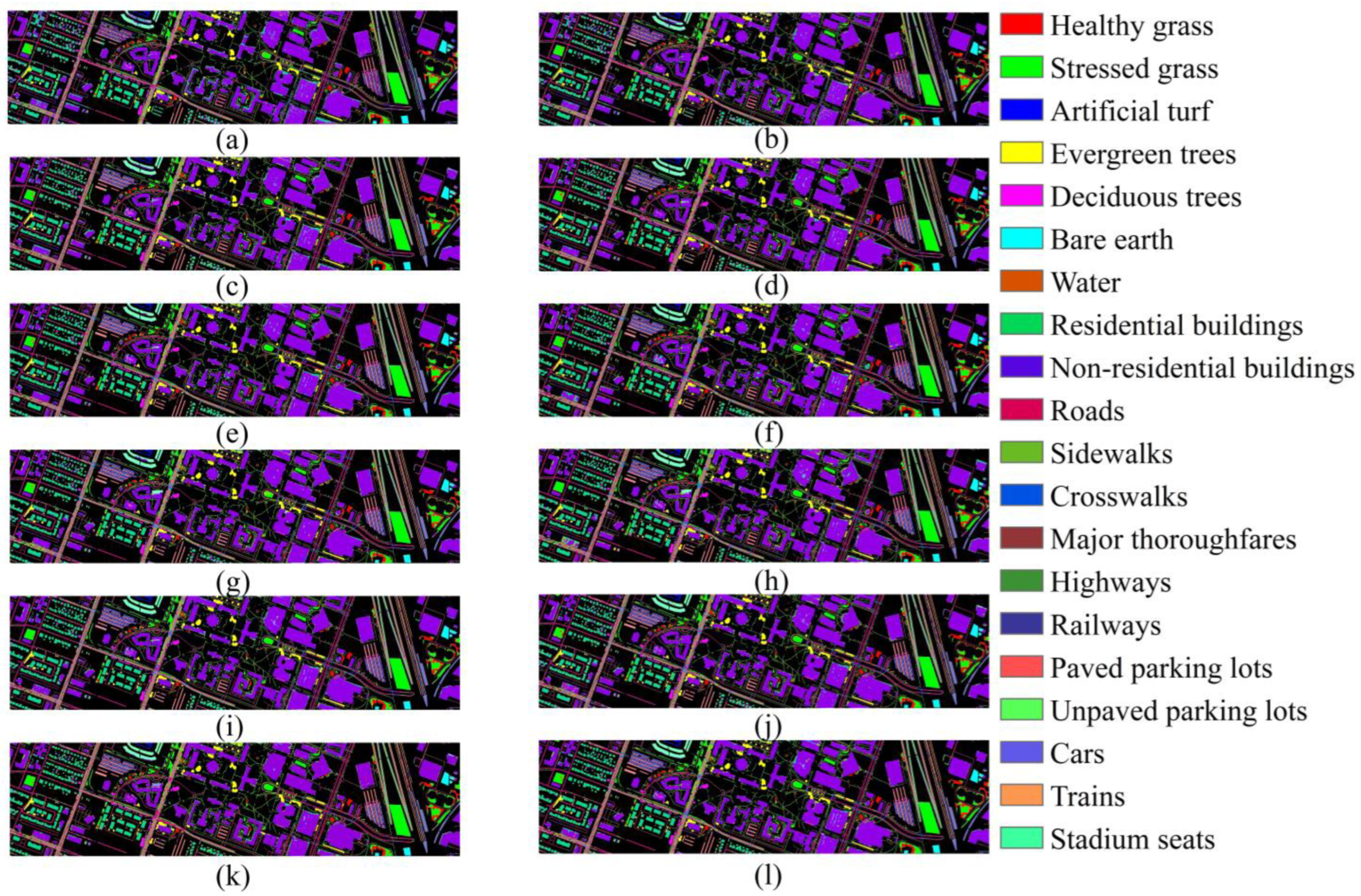
4.3. Classification Results for the DFC 2018
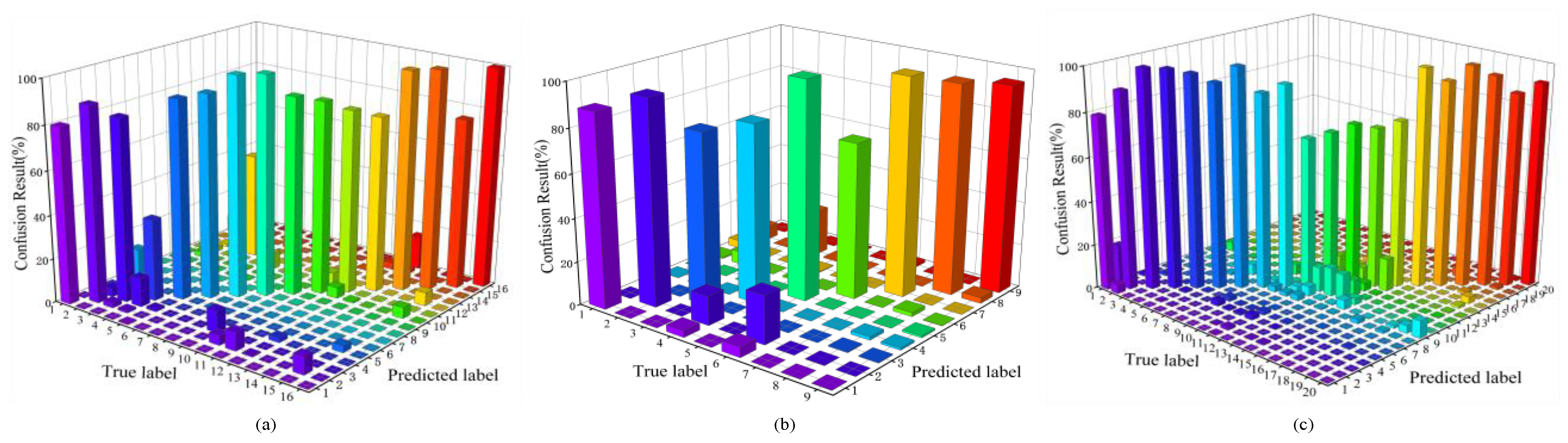
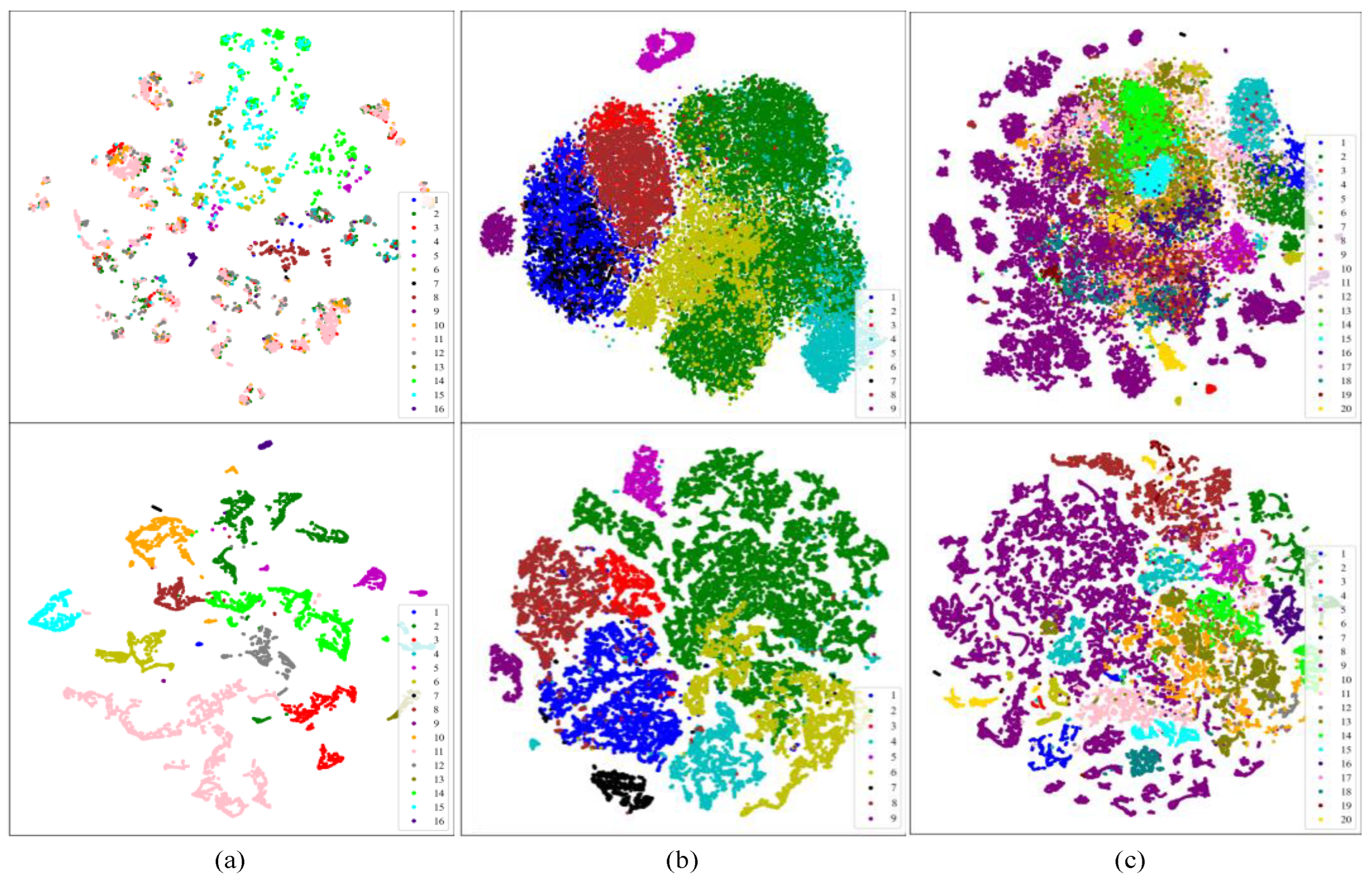
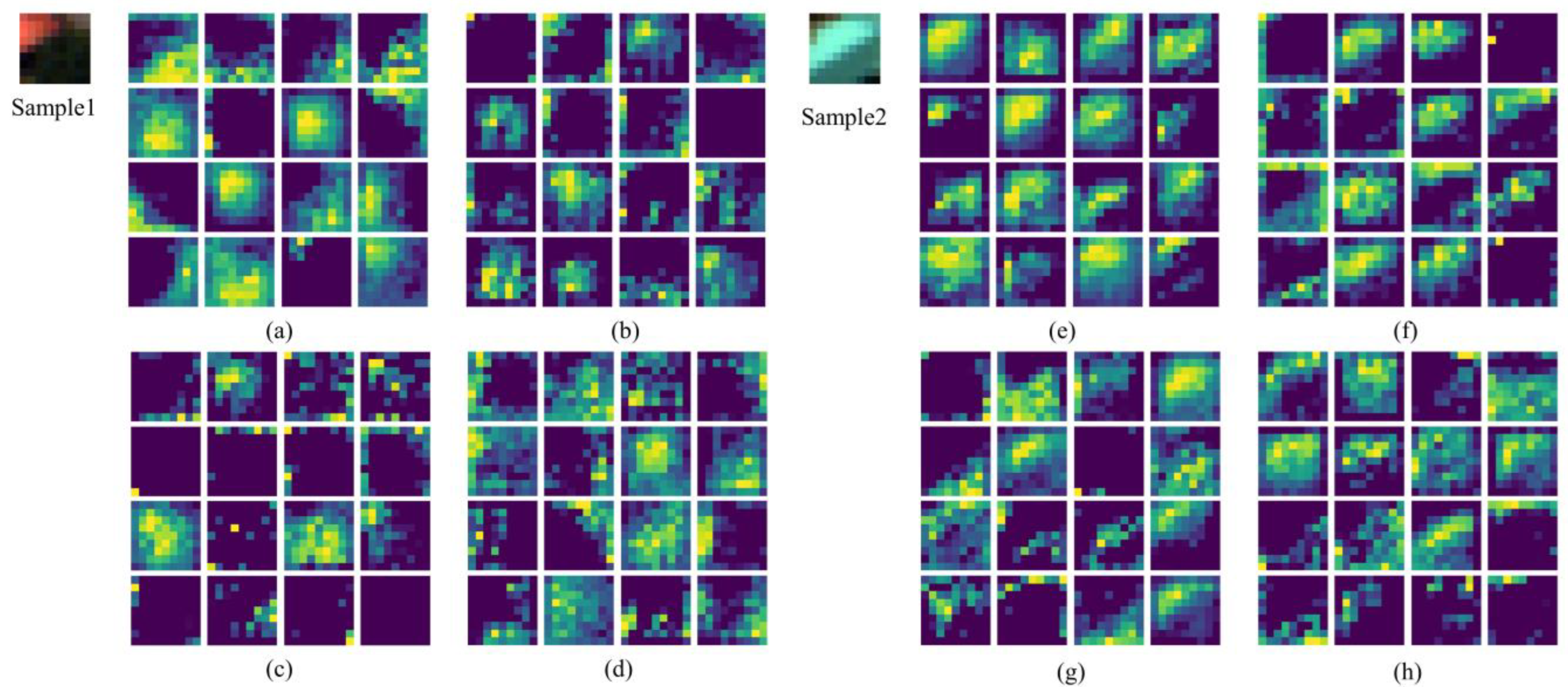
4.4. Visualization Analysis of TransHSI
5. Discussion
5.1. Ablation Experiments
5.1.1. Quantitative Comparison of Classification Results
5.1.2. Activation Maps Visualization
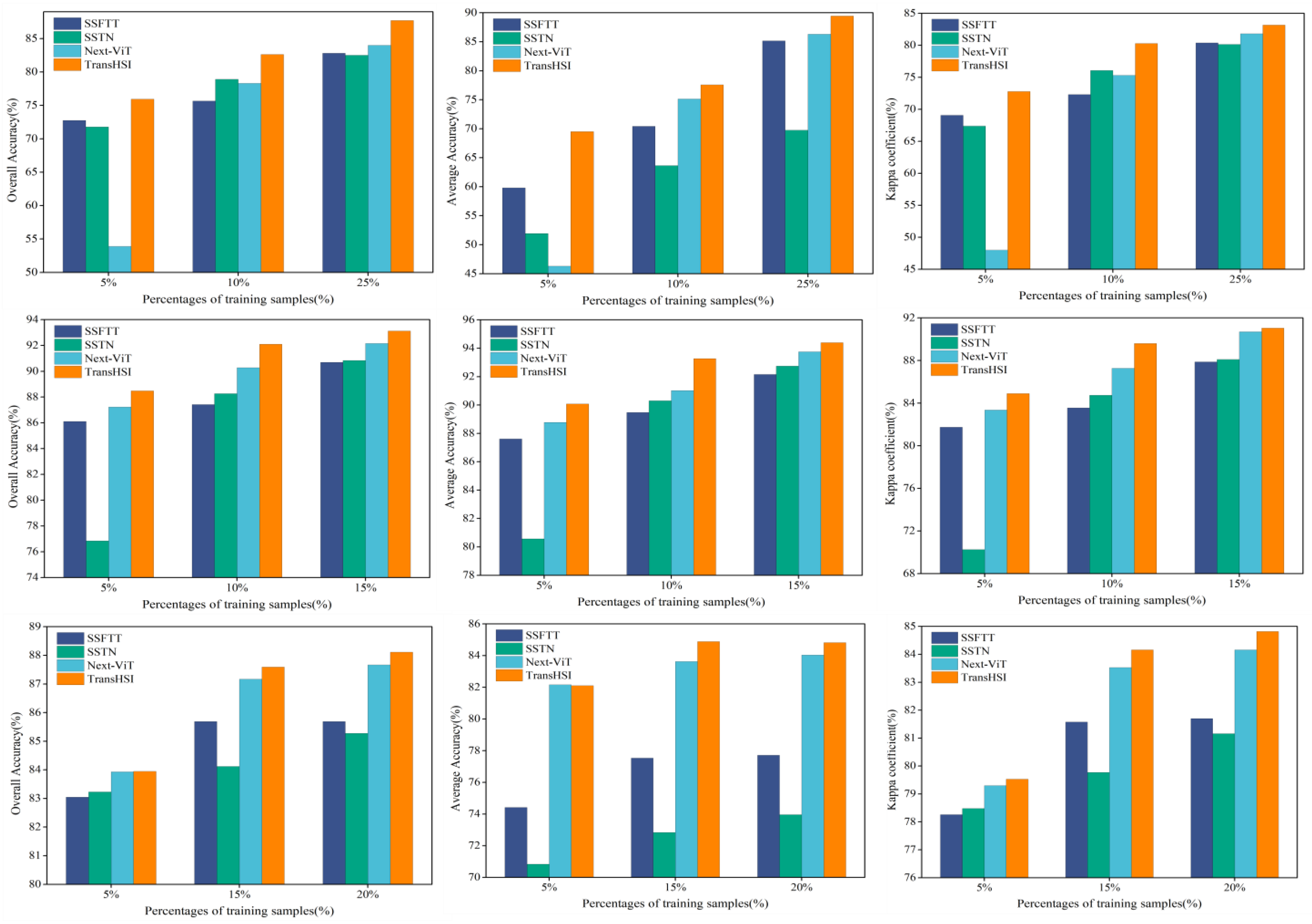
5.2. Effect of Training Sample Percentages on Classification Results
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Audebert, N.; Le Saux, B.; Lefevre, S. Deep Learning for Classification of Hyperspectral Data: A Comparative Review. IEEE Geosci. Remote Sens. Mag. 2019, 7, 159–173. [Google Scholar] [CrossRef]
- Bhosle, K.; Musande, V. Evaluation of Deep Learning CNN Model for Land Use Land Cover Classification and Crop Identification Using Hyperspectral Remote Sensing Images. J. Indian Soc. Remote Sens. 2019, 47, 1949–1958. [Google Scholar] [CrossRef]
- Fong, A.; Shu, G.; McDonogh, B. Farm to Table: Applications for New Hyperspectral Imaging Technologies in Precision Agriculture, Food Quality and Safety. In Proceedings of the Conference on Lasers and Electro-Optics, Washington, DC, USA, 10–15 May 2020; p. AW3K.2. [Google Scholar]
- Lu, B.; Dao, P.D.; Liu, J.G.; He, Y.H.; Shang, J.L. Recent Advances of Hyperspectral Imaging Technology and Applications in Agriculture. Remote Sens. 2020, 12, 2659. [Google Scholar] [CrossRef]
- Wang, Q.; Yuan, Z.; Du, Q.; Li, X. GETNET: A General End-to-End 2-D CNN Framework for Hyperspectral Image Change Detection. IEEE Trans. Geosci. Remote Sens. 2019, 57, 3–13. [Google Scholar] [CrossRef]
- Zhan, T.; Song, B.; Sun, L.; Jia, X.; Wan, M.; Yang, G.; Wu, Z. TDSSC: A Three-Directions Spectral–Spatial Convolution Neural Network for Hyperspectral Image Change Detection. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 377–388. [Google Scholar] [CrossRef]
- Zeng, D.; Zhang, S.; Chen, F.S.; Wang, Y.M. Multi-Scale CNN Based Garbage Detection of Airborne Hyperspectral Data. IEEE Access 2019, 7, 104514–104527. [Google Scholar] [CrossRef]
- Lowe, A.; Harrison, N.; French, A.P. Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 2017, 13, 80. [Google Scholar] [CrossRef]
- Peyghambari, S.; Zhang, Y. Hyperspectral remote sensing in lithological mapping, mineral exploration, and environmental geology: An updated review. J. Appl. Remote Sens. 2021, 15, 031501. [Google Scholar] [CrossRef]
- Ma, L.; Crawford, M.M.; Tian, J. Local Manifold Learning-Based k-Nearest-Neighbor for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4099–4109. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Kang, X.; Li, S.; Fang, L.; Li, M.; Benediktsson, J.A. Extended random walker-based classification of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2014, 53, 144–153. [Google Scholar] [CrossRef]
- Farrell, M.D.; Mersereau, R.M. On the impact of PCA dimension reduction for hyperspectral detection of difficult targets. IEEE Geosci. Remote Sens. Lett. 2005, 2, 192–195. [Google Scholar] [CrossRef]
- Menon, V.; Du, Q.; Fowler, J.E. Fast SVD With Random Hadamard Projection for Hyperspectral Dimensionality Reduction. IEEE Geosci. Remote Sens. Lett. 2016, 13, 1275–1279. [Google Scholar] [CrossRef]
- Jia, S.; Zhao, Q.; Zhuang, J.; Tang, D.; Long, Y.; Xu, M.; Zhou, J.; Li, Q. Flexible Gabor-Based Superpixel-Level Unsupervised LDA for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 10394–10409. [Google Scholar] [CrossRef]
- Falco, N.; Benediktsson, J.A.; Bruzzone, L. A Study on the Effectiveness of Different Independent Component Analysis Algorithms for Hyperspectral Image Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2183–2199. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, W.; Fang, Z. Multiple Kernel-Based SVM Classification of Hyperspectral Images by Combining Spectral, Spatial, and Semantic Information. Remote Sens. 2020, 12, 120. [Google Scholar] [CrossRef]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep Convolutional Neural Networks for Hyperspectral Image Classification. J. Sens. 2015, 2015, 258619. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, Y.-Q.; Chan, J.C.-W. Learning and Transferring Deep Joint Spectral–Spatial Features for Hyperspectral Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4729–4742. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep Feature Extraction and Classification of Hyperspectral Images Based on Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef]
- Zhong, Z.; Li, J.; Luo, Z.; Chapman, M. Spectral–Spatial Residual Network for Hyperspectral Image Classification: A 3-D Deep Learning Framework. IEEE Trans. Geosci. Remote Sens. 2018, 56, 847–858. [Google Scholar] [CrossRef]
- Roy, S.K.; Krishna, G.; Dubey, S.R.; Chaudhuri, B.B. HybridSN: Exploring 3-D–2-D CNN Feature Hierarchy for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2020, 17, 277–281. [Google Scholar] [CrossRef]
- Hang, R.L.; Liu, Q.S.; Hong, D.F.; Ghamisi, P. Cascaded Recurrent Neural Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 5384–5394. [Google Scholar] [CrossRef]
- Mou, L.C.; Ghamisi, P.; Zhu, X.X. Deep Recurrent Neural Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef]
- Hao, S.Y.; Wang, W.; Salzmann, M. Geometry-Aware Deep Recurrent Neural Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 2448–2460. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, Z.; Zhao, X.; Hong, D.; Cai, W.; Yu, C.; Yang, N.; Cai, W. Multi-feature fusion: Graph neural network and CNN combining for hyperspectral image classification. Neurocomputing 2022, 501, 246–257. [Google Scholar] [CrossRef]
- Hong, D.; Gao, L.; Yao, J.; Zhang, B.; Plaza, A.; Chanussot, J. Graph Convolutional Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 5966–5978. [Google Scholar] [CrossRef]
- Mou, L.; Lu, X.; Li, X.; Zhu, X.X. Nonlocal Graph Convolutional Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 8246–8257. [Google Scholar] [CrossRef]
- Wan, S.; Gong, C.; Zhong, P.; Du, B.; Zhang, L.; Yang, J. Multiscale Dynamic Graph Convolutional Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3162–3177. [Google Scholar] [CrossRef]
- Wang, J.; Guo, S.; Huang, R.; Li, L.; Zhang, X.; Jiao, L. Dual-Channel Capsule Generation Adversarial Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5501016. [Google Scholar] [CrossRef]
- Zhan, Y.; Hu, D.; Wang, Y.; Yu, X. Semisupervised Hyperspectral Image Classification Based on Generative Adversarial Networks. IEEE Geosci. Remote Sens. Lett. 2018, 15, 212–216. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Generative Adversarial Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5046–5063. [Google Scholar] [CrossRef]
- He, J.; Zhao, L.; Yang, H.; Zhang, M.; Li, W. HSI-BERT: Hyperspectral Image Classification Using the Bidirectional Encoder Representation from Transformers. IEEE Trans. Geosci. Remote Sens. 2020, 58, 165–178. [Google Scholar] [CrossRef]
- Hong, D.; Han, Z.; Yao, J.; Gao, L.; Zhang, B.; Plaza, A.; Chanussot, J. SpectralFormer: Rethinking Hyperspectral Image Classification with Transformers. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5518615. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Fırat, H.; Asker, M.E.; Hanbay, D. Classification of hyperspectral remote sensing images using different dimension reduction methods with 3D/2D CNN. Remote Sens. Appl. Soc. Environ. 2022, 25, 100694. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, G.; Jia, X.; Wu, L.; Zhang, A.; Ren, J.; Fu, H.; Yao, Y. Spectral-Spatial Self-Attention Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5512115. [Google Scholar] [CrossRef]
- Ge, H.M.; Wang, L.G.; Liu, M.Q.; Zhu, Y.X.; Zhao, X.Y.; Pan, H.Z.; Liu, Y.Z. Two-Branch Convolutional Neural Network with Polarized Full Attention for Hyperspectral Image Classification. Remote Sens. 2023, 15, 848. [Google Scholar] [CrossRef]
- Sun, H.; Zheng, X.T.; Lu, X.Q.; Wu, S.Y. Spectral-Spatial Attention Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3232–3245. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Roy, S.K.; Deria, A.; Hong, D.; Rasti, B.; Plaza, A.; Chanussot, J. Multimodal Fusion Transformer for Remote Sensing Image Classification. arXiv 2022, arXiv:2203.16952. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Y.; Yang, J.; Zhao, N.; Wu, L.; Wang, L.; Wang, T. FusionNet: A Convolution-Transformer Fusion Network for Hyperspectral Image Classification. Remote Sens. 2022, 14, 4066. [Google Scholar] [CrossRef]
- He, X.; Chen, Y.; Lin, Z. Spatial-Spectral Transformer for Hyperspectral Image Classification. Remote Sens. 2021, 13, 498. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, G.; Zheng, Y.; Wu, Z. Spectral–Spatial Feature Tokenization Transformer for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5522214. [Google Scholar] [CrossRef]
- Zhong, Z.; Li, Y.; Ma, L.; Li, J.; Zheng, W.-S. Spectral–Spatial Transformer Network for Hyperspectral Image Classification: A Factorized Architecture Search Framework. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5514715. [Google Scholar] [CrossRef]
- Li, J.; Xia, X.; Li, W.; Li, H.; Wang, X.; Xiao, X.; Wang, R.; Zheng, M.; Pan, X. Next-ViT: Next Generation Vision Transformer for Efficient Deployment in Realistic Industrial Scenarios. arXiv 2022, arXiv:2207.05501. [Google Scholar] [CrossRef]
- Firat, H.; Asker, M.E.; Bayindir, M.İ.; Hanbay, D. 3D residual spatial–spectral convolution network for hyperspectral remote sensing image classification. Neural Comput. Appl. 2022, 35, 4479–4497. [Google Scholar] [CrossRef]
- Ahmad, M.; Ghous, U.; Hong, D.; Khan, A.M.; Yao, J.; Wang, S.; Chanussot, J. A Disjoint Samples-Based 3D-CNN With Active Transfer Learning for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5539616. [Google Scholar] [CrossRef]
- Zhang, F.; Yan, M.; Hu, C.; Ni, J.; Zhou, Y. Integrating Coordinate Features in CNN-Based Remote Sensing Imagery Classification. IEEE Geosci. Remote Sens. Lett. 2022, 19, 5502505. [Google Scholar] [CrossRef]
- Cao, X.; Liu, Z.; Li, X.; Xiao, Q.; Feng, J.; Jiao, L. Nonoverlapped Sampling for Hyperspectral Imagery: Performance Evaluation and a Cotraining-Based Classification Strategy. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5506314. [Google Scholar] [CrossRef]
- Geib, C.; Aravena Pelizari, P.; Schrade, H.; Brenning, A.; Taubenbock, H. On the Effect of Spatially Non-Disjoint Training and Test Samples on Estimated Model Generalization Capabilities in Supervised Classification with Spatial Features. IEEE Geosci. Remote Sens. Lett. 2017, 14, 2008–2012. [Google Scholar] [CrossRef]
- Liang, J.; Zhou, J.; Qian, Y.; Wen, L.; Bai, X.; Gao, Y. On the Sampling Strategy for Evaluation of Spectral-Spatial Methods in Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 862–880. [Google Scholar] [CrossRef]
- Wang, W.; Dai, J.; Chen, Z.; Huang, Z.; Li, Z.; Zhu, X.; Hu, X.-h.; Lu, T.; Lu, L.; Li, H.; et al. InternImage: Exploring Large-Scale Vision Foundation Models with Deformable Convolutions. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 19–24 June 2022; pp. 14408–14419. [Google Scholar]
- Ahmad, M.; Shabbir, S.; Roy, S.K.; Hong, D.; Wu, X.; Yao, J.; Khan, A.M.; Mazzara, M.; Distefano, S.; Chanussot, J. Hyperspectral Image Classification-Traditional to Deep Models: A Survey for Future Prospects. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 968–999. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Liu, M.; Pan, H.; Ge, H.; Wang, L. MS3Net: Multiscale stratified-split symmetric network with quadra-view attention for hyperspectral image classification. Signal Process. 2023, 212, 109153. [Google Scholar] [CrossRef]
- Mei, X.; Pan, E.; Ma, Y.; Dai, X.; Huang, J.; Fan, F.; Du, Q.; Zheng, H.; Ma, J. Spectral-Spatial Attention Networks for Hyperspectral Image Classification. Remote Sens. 2019, 11, 963. [Google Scholar] [CrossRef]


| Reference | Deep Learning Techniques | Strengths | Limitations |
|---|---|---|---|
| [18,19,20,21,22] | CNN |
|
|
| [23,24,25] | RNN |
|
|
| [26,27,28,29] | GCN |
|
|
| [30,31,32] | GAN |
|
|
| [33,34,35] | Transformer |
|
|
| Layer (Type) | Output Shape | Parameter |
|---|---|---|
| Input_1 (InputLayer) | (1, 15, 9, 9) | 0 |
| 3D CNN_1 (3D CNN) | (32, 15, 9, 9) | 960 |
| 3D CNN_2 (3D CNN) | (64, 15, 9, 9) | 92,352 |
| 3D CNN_3 (3D CNN) | (32, 15, 9, 9) | 129,120 |
| 3D CNN_4 (3D CNN) | (64, 15, 9, 9) | 92,352 |
| 3D CNN_5 (3D CNN) | (32, 15, 9, 9) | 129,120 |
| 2D CNN_1 (2D CNN) | (64, 9, 9) | 30,912 |
| Flat_1 (Flatten) | (81, 64) | 128 |
| Trans_1 (Transformer Encoder) | (81, 64) | 17,864 |
| 2D CNN_2 (2D CNN) | (128, 9, 9) | 74,112 |
| 2D CNN_3 (2D CNN) | (64, 9, 9) | 73,920 |
| 2D CNN_4 (2D CNN) | (128, 9, 9) | 74,112 |
| 2D CNN_5 (2D CNN) | (64, 9, 9) | 73,920 |
| Flat_2 (Flatten) | (81, 64) | 128 |
| Trans_2 (Transformer Encoder) | (81, 64) | 17,864 |
| Cascade layer (2D CNN) | (128, 9, 9) | 165,120 |
| Tokenizer layer (Flatten) | (5, 128) | 256 |
| Trans_3 (Transformer Encoder) | (5, 128) | 68,488 |
| Cls_token (Identity) | (128) | 0 |
| Linear_1 (Linear layer) | (64) | 8256 |
| Linear_2 (Linear layer) | (9) | 585 |
| Total Trainable parameters: 1,049,569 | ||
| No. | Indian Pines Dataset | Pavia University Dataset | Data Fusion Contest 2018 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Class | Training | Test | Class | Training | Test | Class | Training | Test | |
| 1 | Alfalfa | 21 | 25 | Asphalt | 327 | 6304 | Healthy grass | 858 | 8941 |
| 2 | Corn-notill | 753 | 675 | Meadows | 503 | 18,146 | Stressed grass | 1954 | 30,548 |
| 3 | Corn-mintill | 426 | 404 | Gravel | 284 | 1815 | Artificial turf | 126 | 558 |
| 4 | Corn | 138 | 99 | Trees | 152 | 2912 | Evergreen trees | 1810 | 11,785 |
| 5 | Grass-pasture | 209 | 274 | Painted metal sheets | 232 | 1113 | Deciduous trees | 1073 | 3948 |
| 6 | Grass-trees | 376 | 354 | Bare Soil | 457 | 4572 | Bare earth | 600 | 3916 |
| 7 | Grass-pasture-mowed | 16 | 12 | Bitumen | 349 | 981 | Water | 74 | 192 |
| 8 | Hay-windrowed | 228 | 250 | Self-Blocking Bricks | 318 | 3364 | Residential buildings | 3633 | 36,139 |
| 9 | Oats | 10 | 10 | Shadows | 152 | 795 | Non-residential buildings | 18,571 | 205,181 |
| 10 | Soybean-notill | 469 | 503 | Roads | 2899 | 42,967 | |||
| 11 | Soybean-mintill | 1390 | 1065 | Sidewalks | 1962 | 32,067 | |||
| 12 | Soybean-clean | 311 | 282 | Crosswalks | 417 | 1101 | |||
| 13 | Wheat | 125 | 80 | Major thoroughfares | 3189 | 43,159 | |||
| 14 | Woods | 720 | 545 | Highways | 1518 | 8347 | |||
| 15 | Buildings-Grass-Trees-Drives | 287 | 99 | Railways | 708 | 6229 | |||
| 16 | Stone-Steel-Towers | 49 | 44 | Paved parking lots | 799 | 10,701 | |||
| 17 | Unpaved parking lots | 51 | 95 | ||||||
| 18 | Cars | 575 | 5972 | ||||||
| 19 | Trains | 155 | 5214 | ||||||
| 20 | Stadium seats | 556 | 6268 | ||||||
| Total | 5528 | 4721 | 2774 | 40002 | 41,528 | 463,328 | |||
| Class | SVM | RF | 2D CNN | 3D CNN | HybirdSN | SSRN | InternImage | ViT | Next -ViT | SSFTT | SSTN | TransHSI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.00 | 18.67 | 4.00 | 66.67 | 81.33 | 89.33 | 75.27 | 80.00 | 80.00 | 76.00 | 13.33 | 88.00 |
| 2 | 61.78 | 64.55 | 76.54 | 76.45 | 79.16 | 74.27 | 88.45 | 68.59 | 77.58 | 79.55 | 70.86 | 90.82 |
| 3 | 48.43 | 43.48 | 70.54 | 85.81 | 79.79 | 76.57 | 78.71 | 82.92 | 79.46 | 76.98 | 81.68 | 79.87 |
| 4 | 22.22 | 23.57 | 90.24 | 34.68 | 51.52 | 60.61 | 71.52 | 65.66 | 71.38 | 37.71 | 60.60 | 59.59 |
| 5 | 78.71 | 71.53 | 82.36 | 88.93 | 88.56 | 88.93 | 88.03 | 83.94 | 90.88 | 89.17 | 87.10 | 91.61 |
| 6 | 93.50 | 96.80 | 96.33 | 99.06 | 98.12 | 89.27 | 90.96 | 97.18 | 86.53 | 97.55 | 92.37 | 92.37 |
| 7 | 38.89 | 58.33 | 11.11 | 86.11 | 80.56 | 33.33 | 59.44 | 33.33 | 75.00 | 86.11 | 0.00 | 97.22 |
| 8 | 100.00 | 100.00 | 99.87 | 98.67 | 99.60 | 100.00 | 99.85 | 100.00 | 99.87 | 98.67 | 100.00 | 96.93 |
| 9 | 0.00 | 26.67 | 6.67 | 50.00 | 50.00 | 0.00 | 65.92 | 0.00 | 96.67 | 83.33 | 0.00 | 96.67 |
| 10 | 37.84 | 27.70 | 62.29 | 67.40 | 78.99 | 90.26 | 71.28 | 85.29 | 74.55 | 77.47 | 78.80 | 85.15 |
| 11 | 84.63 | 83.38 | 80.66 | 76.90 | 89.36 | 82.82 | 81.08 | 76.43 | 80.00 | 82.47 | 87.80 | 83.44 |
| 12 | 55.67 | 52.60 | 58.75 | 68.32 | 81.20 | 83.09 | 68.15 | 79.79 | 72.93 | 81.44 | 95.74 | 83.33 |
| 13 | 92.92 | 93.75 | 95.83 | 97.50 | 89.58 | 90.00 | 82.59 | 83.75 | 88.33 | 92.92 | 90.42 | 99.17 |
| 14 | 94.56 | 92.54 | 95.05 | 96.39 | 99.27 | 100.00 | 99.30 | 99.27 | 98.53 | 99.45 | 99.88 | 98.84 |
| 15 | 60.95 | 66.33 | 59.94 | 43.09 | 37.37 | 10.10 | 47.87 | 27.27 | 35.35 | 47.14 | 46.47 | 76.10 |
| 16 | 90.15 | 97.73 | 100.00 | 90.15 | 93.18 | 87.88 | 65.40 | 88.64 | 72.73 | 100.00 | 83.34 | 88.64 |
| OA (%) | 71.47 ± 10.27 | 69.93 ± 0.09 | 79.36 ± 0.29 | 80.63 ± 0.36 | 85.79 ± 0.67 | 83.51 ± 0.18 | 83.22 ± 0.98 | 81.61 ± 0.71 | 81.90 ± 1.99 | 83.97 ± 0.57 | 84.47 ± 2.00 | 87.75 ± 0.35 |
| AA (%) | 60.01 ± 5.83 | 63.60 ± 0.33 | 68.13 ± 1.76 | 76.63 ± 0.56 | 83.04 ± 1.85 | 72.28 ± 0.68 | 77.11 ± 3.77 | 72.00 ± 1.77 | 79.99 ± 5.33 | 81.62 ± 1.35 | 68.02 ± 2.24 | 88.42 ± 1.37 |
| Κ (%) | 66.97 ± 12.19 | 65.23 ± 0.10 | 76.48 ± 0.38 | 77.95 ± 0.42 | 84.53 ± 0.43 | 81.29 ± 0.20 | 80.91 ± 1.13 | 79.21 ± 0.81 | 80.30 ± 1.52 | 81.79 ± 0.67 | 82.34 ± 2.27 | 86.11 ± 0.41 |
| Class | SVM | RF | 2D CNN | 3D CNN | HybirdSN | SSRN | InternImage | ViT | Next-ViT | SSFTT | SSTN | TransHSI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 66.28 | 79.03 | 90.44 | 86.48 | 92.34 | 96.37 | 94.35 | 87.99 | 89.38 | 92.93 | 88.67 | 89.58 |
| 2 | 83.95 | 56.78 | 70.57 | 75.58 | 75.50 | 86.33 | 78.54 | 71.29 | 84.53 | 77.14 | 72.99 | 88.59 |
| 3 | 35.54 | 43.22 | 69.97 | 69.51 | 65.84 | 68.08 | 73.38 | 68.30 | 63.07 | 82.26 | 56.97 | 76.53 |
| 4 | 93.10 | 95.91 | 81.21 | 72.78 | 86.44 | 84.71 | 87.02 | 85.23 | 87.37 | 87.58 | 78.80 | 92.07 |
| 5 | 99.28 | 99.10 | 99.91 | 99.94 | 100.00 | 99.97 | 99.75 | 100.00 | 100.00 | 100.00 | 99.55 | 99.91 |
| 6 | 33.03 | 77.45 | 94.02 | 89.52 | 90.13 | 78.81 | 97.13 | 96.65 | 91.28 | 88.99 | 97.19 | 80.75 |
| 7 | 90.52 | 79.14 | 98.51 | 99.15 | 99.46 | 98.03 | 97.10 | 95.07 | 97.69 | 98.24 | 97.76 | 97.96 |
| 8 | 91.35 | 88.03 | 97.51 | 97.39 | 98.27 | 99.13 | 97.27 | 97.53 | 96.16 | 96.36 | 97.77 | 97.11 |
| 9 | 99.87 | 99.79 | 98.25 | 99.79 | 99.87 | 99.58 | 96.81 | 94.92 | 98.78 | 99.96 | 97.48 | 99.54 |
| OA (%) | 75.34 ± 0.00 | 70.09 ± 0.09 | 81.45 ± 0.24 | 81.98 ± 1.04 | 83.85 ± 0.91 | 88.11 ± 0.91 | 86.52 ± 0.14 | 81.76 ± 0.03 | 87.32 ± 1.11 | 85.20 ± 0.06 | 81.84 ± 1.16 | 89.03 ± 1.72 |
| AA (%) | 76.99 ± 0.00 | 79.83 ± 0.05 | 88.93 ± 0.57 | 87.79 ± 1.04 | 89.76 ± 0.48 | 90.11 ± 0.71 | 91.26 ± 0.41 | 88.55 ± 0.50 | 89.81 ± 0.56 | 91.49 ± 0.16 | 87.47 ± 1.24 | 91.34 ± 2.05 |
| κ (%) | 66.88 ± 0.00 | 62.76 ± 0.08 | 76.31 ± 0.20 | 76.71 ± 1.35 | 79.10 ± 1.03 | 84.21 ± 1.04 | 82.51 ± 0.19 | 76.71 ± 0.01 | 83.32 ± 1.38 | 80.78 ± 0.11 | 76.75 ± 1.41 | 85.41 ± 2.16 |
| Class | SVM | RF | 2D CNN | 3D CNN | HybirdSN | SSRN | InternImage | ViT | Next -ViT | SSFTT | SSTN | TransHSI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 92.94 | 91.14 | 85.05 | 82.70 | 87.25 | 87.77 | 76.86 | 80.27 | 80.48 | 90.06 | 81.03 | 83.42 |
| 2 | 88.57 | 87.49 | 87.42 | 88.67 | 85.61 | 84.94 | 82.74 | 82.86 | 89.44 | 83.84 | 85.26 | 88.54 |
| 3 | 100.00 | 100.00 | 97.79 | 98.80 | 99.58 | 99.64 | 97.79 | 96.59 | 97.91 | 97.85 | 94.56 | 99.46 |
| 4 | 97.11 | 96.76 | 98.27 | 98.40 | 98.32 | 97.44 | 95.63 | 96.67 | 98.03 | 97.63 | 95.96 | 96.69 |
| 5 | 83.64 | 83.76 | 93.75 | 93.26 | 97.95 | 94.83 | 94.19 | 94.73 | 97.05 | 95.34 | 85.79 | 97.68 |
| 6 | 91.35 | 89.35 | 93.34 | 92.15 | 93.75 | 94.91 | 91.86 | 93.57 | 95.75 | 92.40 | 94.69 | 92.59 |
| 7 | 98.96 | 98.09 | 98.61 | 99.65 | 99.83 | 100.00 | 93.23 | 98.79 | 99.31 | 97.57 | 96.18 | 100.00 |
| 8 | 80.55 | 79.77 | 82.47 | 86.26 | 87.17 | 86.46 | 84.64 | 87.19 | 87.97 | 82.04 | 87.76 | 88.91 |
| 9 | 88.75 | 93.00 | 89.35 | 91.24 | 91.65 | 92.02 | 91.42 | 89.10 | 92.14 | 91.02 | 91.67 | 92.12 |
| 10 | 41.19 | 47.04 | 59.75 | 62.60 | 62.09 | 64.32 | 62.30 | 60.67 | 65.53 | 61.91 | 64.03 | 68.66 |
| 11 | 48.26 | 58.45 | 68.94 | 67.64 | 76.50 | 78.36 | 73.67 | 67.43 | 69.36 | 72.74 | 61.05 | 74.13 |
| 12 | 10.35 | 29.70 | 38.81 | 42.78 | 62.97 | 79.11 | 73.63 | 75.81 | 60.61 | 65.21 | 14.83 | 72.69 |
| 13 | 52.80 | 52.71 | 67.22 | 65.04 | 72.17 | 69.86 | 66.40 | 68.70 | 72.41 | 67.91 | 72.98 | 72.63 |
| 14 | 60.37 | 59.76 | 71.14 | 78.01 | 77.30 | 81.82 | 77.13 | 88.06 | 75.62 | 67.84 | 70.14 | 80.64 |
| 15 | 96.77 | 94.39 | 98.78 | 98.93 | 99.55 | 98.07 | 99.09 | 95.08 | 99.33 | 97.69 | 99.06 | 99.40 |
| 16 | 61.21 | 71.45 | 91.14 | 91.67 | 93.62 | 94.11 | 91.75 | 87.44 | 91.05 | 90.88 | 90.56 | 93.70 |
| 17 | 84.21 | 99.65 | 90.52 | 94.74 | 100.00 | 99.65 | 98.60 | 100.00 | 98.95 | 98.59 | 98.59 | 100.00 |
| 18 | 27.14 | 42.29 | 89.74 | 93.70 | 95.80 | 95.10 | 91.95 | 92.90 | 94.72 | 92.88 | 88.17 | 96.75 |
| 19 | 20.23 | 34.91 | 87.41 | 88.45 | 90.19 | 88.54 | 88.03 | 92.36 | 91.96 | 83.17 | 74.62 | 88.53 |
| 20 | 70.15 | 78.82 | 92.00 | 93.76 | 96.89 | 96.76 | 90.03 | 96.89 | 94.20 | 96.28 | 87.88 | 96.08 |
| OA (%) | 74.78 ± 0.00 | 78.44 ± 0.03 | 82.44 ± 0.20 | 83.80 ± 0.24 | 85.40 ± 0.10 | 85.64 ± 0.23 | 83.68 ± 0.22 | 82.81 ± 0.39 | 85.52 ± 0.28 | 83.55 ± 0.38 | 83.62 ± 0.58 | 86.36 ± 0.65 |
| AA (%) | 69.73 ± 0.00 | 74.43 ± 0.04 | 84.07 ± 0.65 | 85.42 ± 0.44 | 88.41 ± 0.58 | 89.19 ± 0.85 | 86.05 ± 0.40 | 87.25 ± 0.40 | 87.59 ± 1.15 | 86.14 ± 0.36 | 81.74 ± 1.37 | 89.13 ± 0.70 |
| κ (%) | 67.32 ± 0.00 | 71.73 ± 0.03 | 77.59 ± 0.23 | 79.26 ± 0.28 | 81.30 ± 0.10 | 81.60 ± 0.20 | 79.07 ± 0.29 | 78.15 ± 0.46 | 81.40 ± 0.34 | 78.90 ± 0.45 | 78.95 ± 0.76 | 82.53 ± 0.77 |
| Experiments | 3D CNNs | Trans_1 | 2D CNNs | Trans_2 | Concat | Trans_3 |
|---|---|---|---|---|---|---|
| (a) | × | × | √ | √ | √ | √ |
| (b) | √ | √ | × | × | √ | √ |
| (c) | √ | √ | √ | √ | × | √ |
| (d) | √ | × | √ | × | √ | × |
| (e) | √ | × | √ | √ | √ | √ |
| (f) | √ | √ | √ | × | √ | √ |
| (g) | √ | √ | √ | √ | √ | × |
| TransHSI | √ | √ | √ | √ | √ | √ |
| Class | (a) | (b) | (c) | (d) | (e) | (f) | (g) | TransHSI |
|---|---|---|---|---|---|---|---|---|
| 1 | 94.67 ± 4.62 | 98.67 ± 2.31 | 61.33 ± 53.12 | 60.00 ± 8.00 | 98.67 ± 2.31 | 100.00 ± 0.00 | 96.00 ± 6.93 | 88.00 ± 6.93 |
| 2 | 88.64 ± 5.71 | 85.73 ± 7.93 | 83.36 ± 5.12 | 81.33 ± 1.43 | 82.62 ± 2.06 | 84.30 ± 3.81 | 79.90 ± 10.33 | 90.82 ± 3.08 |
| 3 | 78.96 ± 1.73 | 87.21 ± 3.97 | 86.22 ± 5.51 | 82.10 ± 4.84 | 82.67 ± 1.87 | 80.45 ± 2.75 | 80.61 ± 5.88 | 79.87 ± 3.62 |
| 4 | 56.57 ± 11.91 | 44.11 ± 8.23 | 45.79 ± 2.54 | 84.18 ± 7.44 | 53.20 ± 23.26 | 40.40 ± 2.67 | 78.17 ± 3.55 | 59.59 ± 30.17 |
| 5 | 92.58 ± 0.56 | 91.24 ± 1.83 | 89.30 ± 2.20 | 89.05 ± 0.97 | 89.17 ± 0.56 | 92.95 ± 1.28 | 88.93 ± 1.12 | 91.61 ± 0.97 |
| 6 | 88.51 ± 0.91 | 98.68 ± 0.99 | 94.73 ± 2.27 | 96.23 ± 0.71 | 91.90 ± 1.88 | 95.29 ± 2.40 | 86.25 ± 2.12 | 92.37 ± 0.29 |
| 7 | 75.00 ± 0.00 | 86.11 ± 24.06 | 61.11 ± 53.58 | 86.11 ± 12.73 | 91.67 ± 14.43 | 77.78 ± 17.35 | 61.11 ± 52.93 | 97.22 ± 4.81 |
| 8 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 99.07 ± 1.62 | 100.00 ± 0.00 | 96.93 ± 5.31 |
| 9 | 46.67 ± 5.77 | 80.00 ± 34.64 | 46.67 ± 50.33 | 43.33 ± 11.55 | 30.00 ± 43.59 | 96.67 ± 5.77 | 0.00 ± 0.00 | 96.67 ± 5.77 |
| 10 | 70.97 ± 4.83 | 89.53 ± 5.81 | 81.38 ± 4.13 | 85.69 ± 3.81 | 78.99 ± 2.95 | 86.81 ± 5.62 | 93.64 ± 1.39 | 85.15 ± 3.54 |
| 11 | 82.75 ± 1.81 | 83.94 ± 5.77 | 91.67 ± 1.52 | 81.28 ± 2.27 | 82.63 ± 3.10 | 83.29 ± 1.93 | 84.63 ± 0.95 | 83.44 ± 2.16 |
| 12 | 87.71 ± 3.79 | 86.17 ± 4.09 | 91.25 ± 6.07 | 80.14 ± 1.07 | 93.15 ± 4.38 | 92.91 ± 0.62 | 95.98 ± 1.14 | 83.33 ± 5.84 |
| 13 | 93.75 ± 1.25 | 96.67 ± 2.60 | 96.25 ± 4.51 | 95.00 ± 4.33 | 95.83 ± 2.60 | 95.00 ± 1.25 | 90.00 ± 0.00 | 99.17 ± 0.72 |
| 14 | 99.69 ± 0.28 | 99.08 ± 0.66 | 99.45 ± 0.64 | 98.23 ± 2.76 | 97.49 ± 1.87 | 99.21 ± 0.56 | 100.00 ± 0.00 | 98.84 ± 2.01 |
| 15 | 24.24 ± 3.50 | 23.57 ± 16.45 | 14.14 ± 3.03 | 42.76 ± 6.17 | 68.69 ± 7.28 | 30.30 ± 20.28 | 34.68 ± 4.98 | 76.10 ± 2.10 |
| 16 | 95.46 ± 6.01 | 100.00 ± 0.00 | 66.67 ± 57.74 | 88.63 ± 3.94 | 85.61 ± 12.92 | 98.48 ± 2.63 | 76.51 ± 10.50 | 88.64 ± 10.41 |
| OA (%) | 84.68 ± 1.00 | 87.67 ± 0.33 | 87.25 ± 0.10 | 85.65 ± 0.42 | 85.92 ± 1.28 | 86.70 ± 0.23 | 86.86 ± 0.49 | 87.75 ± 0.35 |
| AA (%) | 79.76 ± 0.60 | 84.42 ± 4.26 | 75.58 ± 12.19 | 80.88 ± 0.93 | 82.64 ± 2.54 | 84.56 ± 0.69 | 77.90 ± 2.96 | 88.42 ± 1.37 |
| κ (%) | 82.59 ± 1.12 | 86.02 ± 0.38 | 85.48 ± 0.12 | 83.73 ± 0.49 | 84.02 ± 1.46 | 84.93 ± 0.23 | 85.10 ± 0.53 | 86.11 ± 0.41 |
| Class | (a) | (b) | (c) | (d) | (e) | (f) | (g) | TransHSI |
|---|---|---|---|---|---|---|---|---|
| 1 | 94.37 ± 1.40 | 93.24 ± 1.79 | 94.99 ± 2.30 | 94.22 ± 1.48 | 92.96 ± 4.82 | 93.23 ± 3.35 | 96.14 ± 0.56 | 89.58 ± 3.41 |
| 2 | 76.92 ± 2.98 | 77.88 ± 1.45 | 81.24 ± 0.75 | 85.95 ± 1.82 | 87.68 ± 4.63 | 81.91 ± 0.65 | 80.33 ± 0.17 | 88.59 ± 4.49 |
| 3 | 82.79 ± 1.32 | 73.96 ± 7.11 | 75.68 ± 1.38 | 63.73 ± 11.11 | 72.51 ± 6.12 | 80.88 ± 8.26 | 75.19 ± 2.93 | 76.53 ± 7.10 |
| 4 | 86.28 ± 3.86 | 89.09 ± 0.79 | 90.54 ± 1.38 | 89.62 ± 2.83 | 87.17 ± 5.22 | 87.48 ± 3.23 | 92.83 ± 0.02 | 92.07 ± 2.04 |
| 5 | 100.00 ± 0.00 | 99.73 ± 0.47 | 99.79 ± 0.36 | 100.00 ± 0.00 | 99.88 ± 0.21 | 99.94 ± 0.10 | 99.58 ± 0.05 | 99.91 ± 0.16 |
| 6 | 85.80 ± 4.31 | 91.06 ± 2.53 | 82.32 ± 2.62 | 80.68 ± 4.55 | 78.84 ± 5.32 | 90.96 ± 2.91 | 91.12 ± 0.83 | 80.75 ± 3.15 |
| 7 | 99.05 ± 0.66 | 99.22 ± 0.42 | 99.56 ± 0.15 | 98.61 ± 0.41 | 98.33 ± 1.05 | 99.39 ± 0.18 | 97.18 ± 0.21 | 97.96 ± 0.47 |
| 8 | 98.26 ± 0.33 | 97.61 ± 0.52 | 96.51 ± 0.74 | 99.62 ± 0.14 | 98.08 ± 0.36 | 95.80 ± 2.77 | 98.67 ± 0.06 | 97.11 ± 0.59 |
| 9 | 98.91 ± 0.70 | 99.54 ± 0.38 | 99.24 ± 0.33 | 99.46 ± 0.32 | 99.20 ± 0.41 | 99.25 ± 0.58 | 99.29 ± 0.14 | 99.54 ± 0.31 |
| OA (%) | 85.05 ± 0.86 | 85.67 ± 0.45 | 86.57 ± 0.66 | 88.03 ± 0.08 | 88.48 ± 0.70 | 87.53 ± 0.29 | 87.60 ± 0.14 | 89.03 ± 1.72 |
| AA (%) | 91.37 ± 1.02 | 91.26 ± 1.02 | 91.10 ± 0.39 | 90.21 ± 1.35 | 90.52 ± 1.29 | 92.09 ± 0.41 | 92.26 ± 0.31 | 91.34 ± 2.05 |
| κ (%) | 80.59 ± 1.02 | 81.39 ± 0.59 | 82.36 ± 0.86 | 84.13 ± 0.11 | 84.66 ± 0.82 | 83.68 ± 0.39 | 83.82 ± 0.16 | 85.41 ± 2.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Yu, H.; Li, P.; Wang, R. TransHSI: A Hybrid CNN-Transformer Method for Disjoint Sample-Based Hyperspectral Image Classification. Remote Sens. 2023, 15, 5331. https://doi.org/10.3390/rs15225331
Zhang P, Yu H, Li P, Wang R. TransHSI: A Hybrid CNN-Transformer Method for Disjoint Sample-Based Hyperspectral Image Classification. Remote Sensing. 2023; 15(22):5331. https://doi.org/10.3390/rs15225331
Chicago/Turabian StyleZhang, Ping, Haiyang Yu, Pengao Li, and Ruili Wang. 2023. "TransHSI: A Hybrid CNN-Transformer Method for Disjoint Sample-Based Hyperspectral Image Classification" Remote Sensing 15, no. 22: 5331. https://doi.org/10.3390/rs15225331
APA StyleZhang, P., Yu, H., Li, P., & Wang, R. (2023). TransHSI: A Hybrid CNN-Transformer Method for Disjoint Sample-Based Hyperspectral Image Classification. Remote Sensing, 15(22), 5331. https://doi.org/10.3390/rs15225331