MISPEL: A Multi-Crop Spectral Library for Statistical Crop Trait Retrieval and Agricultural Monitoring
Abstract
1. Introduction
2. Materials and Methods
2.1. Methodology
2.2. Data Collection
2.2.1. Hyperspectral Measurements and Reference Data Acquisition
2.2.2. Independent Validation Data
2.3. Spectral Library Establishment
2.4. Model Selection, Implementation, and Accuracy Assessment
2.5. Model Application to Sentinel-2 Data
2.6. SNAP-Based Sentinel-2 LAI Retrieval
2.7. Independent Model Validation
3. Results
3.1. Multi-Crop Spectral Library MISPEL and Model Establishment
3.2. Crop-Specific S2 LAI Model Accuracies
3.3. Independent Model Validation at the DEMMIN Test Site
4. Discussion
4.1. Multi-Crop Spectral Library MISPEL and Model Establishment
4.2. Model Validation
4.3. Practical Use
4.4. Caveats
4.5. Outlook
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Fivefold Cross-Validated Crop-Specific Leaf Area Index (LAI) Model Metrics Based on MISPEL
| Crop | Abbreviation | MISPEL | Model | NRMSE | RMSE | RSQ | N |
| All | ALL | FR | GLMNET | 0.28 | 1.02 | 0.7 | 1411 |
| All | ALL | FR | GP | 0.25 | 0.9 | 0.76 | 1411 |
| All | ALL | FR | PLS | 0.29 | 1.06 | 0.67 | 1411 |
| All | ALL | FR | RF | 0.25 | 0.89 | 0.77 | 1411 |
| All | ALL | S2 | GLMNET | 0.3 | 1.1 | 0.65 | 1411 |
| All | ALL | S2 | GP | 0.3 | 1.1 | 0.65 | 1411 |
| All | ALL | S2 | PLS | 0.31 | 1.12 | 0.63 | 1411 |
| All | ALL | S2 | RF | 0.26 | 0.92 | 0.75 | 1411 |
| Broad bean | BB | FR | GLMNET | 0.24 | 0.9 | 0.78 | 49 |
| Broad bean | BB | FR | GP | 0.26 | 0.99 | 0.76 | 49 |
| Broad bean | BB | FR | PLS | 0.25 | 0.95 | 0.76 | 49 |
| Broad bean | BB | FR | RF | 0.22 | 0.85 | 0.8 | 49 |
| Broad bean | BB | S2 | GLMNET | 0.22 | 0.82 | 0.82 | 49 |
| Broad bean | BB | S2 | GP | 0.24 | 0.91 | 0.78 | 49 |
| Broad bean | BB | S2 | PLS | 0.24 | 0.92 | 0.79 | 49 |
| Broad bean | BB | S2 | RF | 0.22 | 0.84 | 0.8 | 49 |
| Oat | OA | FR | GLMNET | 0.17 | 0.5 | 0.76 | 39 |
| Oat | OA | FR | GP | 0.19 | 0.56 | 0.73 | 39 |
| Oat | OA | FR | PLS | 0.19 | 0.57 | 0.73 | 39 |
| Oat | OA | FR | RF | 0.22 | 0.66 | 0.59 | 39 |
| Oat | OA | S2 | GLMNET | 0.19 | 0.57 | 0.71 | 39 |
| Oat | OA | S2 | GP | 0.19 | 0.57 | 0.71 | 39 |
| Oat | OA | S2 | PLS | 0.19 | 0.57 | 0.72 | 39 |
| Oat | OA | S2 | RF | 0.2 | 0.6 | 0.67 | 39 |
| Potato | PT | FR | GLMNET | 0.32 | 0.93 | 0.68 | 54 |
| Potato | PT | FR | GP | 0.41 | 1.2 | 0.52 | 54 |
| Potato | PT | FR | PLS | 0.35 | 1.02 | 0.63 | 54 |
| Potato | PT | FR | RF | 0.32 | 0.94 | 0.7 | 54 |
| Potato | PT | S2 | GLMNET | 0.36 | 1.04 | 0.58 | 54 |
| Potato | PT | S2 | GP | 0.36 | 1.06 | 0.59 | 54 |
| Potato | PT | S2 | PLS | 0.36 | 1.06 | 0.59 | 54 |
| Potato | PT | S2 | RF | 0.32 | 0.92 | 0.72 | 54 |
| Spring barley | SBA | FR | GLMNET | 0.24 | 0.77 | 0.66 | 75 |
| Spring barley | SBA | FR | GP | 0.23 | 0.75 | 0.66 | 75 |
| Spring barley | SBA | FR | PLS | 0.27 | 0.89 | 0.53 | 75 |
| Spring barley | SBA | FR | RF | 0.19 | 0.6 | 0.81 | 75 |
| Spring barley | SBA | S2 | GLMNET | 0.26 | 0.85 | 0.52 | 75 |
| Spring barley | SBA | S2 | GP | 0.27 | 0.89 | 0.47 | 75 |
| Spring barley | SBA | S2 | PLS | 0.27 | 0.89 | 0.5 | 75 |
| Spring barley | SBA | S2 | RF | 0.22 | 0.71 | 0.72 | 75 |
| Sugar beet | SBE | FR | GLMNET | 0.25 | 0.76 | 0.6 | 106 |
| Sugar beet | SBE | FR | GP | 0.3 | 0.91 | 0.5 | 106 |
| Sugar beet | SBE | FR | PLS | 0.26 | 0.77 | 0.59 | 106 |
| Sugar beet | SBE | FR | RF | 0.27 | 0.81 | 0.55 | 106 |
| Sugar beet | SBE | S2 | GLMNET | 0.26 | 0.77 | 0.59 | 106 |
| Sugar beet | SBE | S2 | GP | 0.26 | 0.78 | 0.57 | 106 |
| Sugar beet | SBE | S2 | PLS | 0.27 | 0.81 | 0.56 | 106 |
| Sugar beet | SBE | S2 | RF | 0.26 | 0.78 | 0.57 | 106 |
| Triticale | TR | FR | GLMNET | 0.24 | 0.8 | 0.82 | 66 |
| Triticale | TR | FR | GP | 0.28 | 0.92 | 0.78 | 66 |
| Triticale | TR | FR | PLS | 0.25 | 0.84 | 0.8 | 66 |
| Triticale | TR | FR | RF | 0.27 | 0.89 | 0.78 | 66 |
| Triticale | TR | S2 | GLMNET | 0.23 | 0.78 | 0.83 | 66 |
| Triticale | TR | S2 | GP | 0.24 | 0.79 | 0.83 | 66 |
| Triticale | TR | S2 | PLS | 0.24 | 0.79 | 0.82 | 66 |
| Triticale | TR | S2 | RF | 0.25 | 0.84 | 0.81 | 66 |
| Winter barley | WB | FR | GLMNET | 0.2 | 0.8 | 0.77 | 33 |
| Winter barley | WB | FR | GP | 0.22 | 0.86 | 0.75 | 33 |
| Winter barley | WB | FR | PLS | 0.2 | 0.81 | 0.75 | 33 |
| Winter barley | WB | FR | RF | 0.21 | 0.83 | 0.76 | 33 |
| Winter barley | WB | S2 | GLMNET | 0.18 | 0.74 | 0.83 | 33 |
| Winter barley | WB | S2 | GP | 0.19 | 0.75 | 0.79 | 33 |
| Winter barley | WB | S2 | PLS | 0.19 | 0.75 | 0.79 | 33 |
| Winter barley | WB | S2 | RF | 0.22 | 0.87 | 0.7 | 33 |
| Winter rapeseed | WRA | FR | GLMNET | 0.22 | 0.97 | 0.76 | 426 |
| Winter rapeseed | WRA | FR | GP | 0.2 | 0.88 | 0.81 | 426 |
| Winter rapeseed | WRA | FR | PLS | 0.25 | 1.07 | 0.71 | 426 |
| Winter rapeseed | WRA | FR | RF | 0.21 | 0.92 | 0.79 | 426 |
| Winter rapeseed | WRA | S2 | GLMNET | 0.23 | 0.99 | 0.75 | 426 |
| Winter rapeseed | WRA | S2 | GP | 0.23 | 0.99 | 0.75 | 426 |
| Winter rapeseed | WRA | S2 | PLS | 0.24 | 1.03 | 0.73 | 426 |
| Winter rapeseed | WRA | S2 | RF | 0.22 | 0.96 | 0.77 | 426 |
| Winter rye | WRY | FR | GLMNET | 0.22 | 0.63 | 0.83 | 157 |
| Winter rye | WRY | FR | GP | 0.17 | 0.5 | 0.9 | 157 |
| Winter rye | WRY | FR | PLS | 0.22 | 0.64 | 0.82 | 157 |
| Winter rye | WRY | FR | RF | 0.17 | 0.49 | 0.9 | 157 |
| Winter rye | WRY | S2 | GLMNET | 0.19 | 0.54 | 0.87 | 157 |
| Winter rye | WRY | S2 | GP | 0.21 | 0.62 | 0.84 | 157 |
| Winter rye | WRY | S2 | PLS | 0.22 | 0.63 | 0.83 | 157 |
| Winter rye | WRY | S2 | RF | 0.18 | 0.51 | 0.89 | 157 |
| Winter wheat | WW | FR | GLMNET | 0.22 | 0.78 | 0.83 | 406 |
| Winter wheat | WW | FR | GP | 0.2 | 0.7 | 0.86 | 406 |
| Winter wheat | WW | FR | PLS | 0.23 | 0.82 | 0.81 | 406 |
| Winter wheat | WW | FR | RF | 0.22 | 0.77 | 0.83 | 406 |
| Winter wheat | WW | S2 | GLMNET | 0.22 | 0.79 | 0.83 | 406 |
| Winter wheat | WW | S2 | GP | 0.23 | 0.79 | 0.83 | 406 |
| Winter wheat | WW | S2 | PLS | 0.23 | 0.81 | 0.82 | 406 |
| Winter wheat | WW | S2 | RF | 0.23 | 0.79 | 0.83 | 406 |
References
- Huang, J.; Gómez-Dans, J.L.; Huang, H.; Ma, H.; Wu, Q.; Lewis, P.E.; Liang, S.; Chen, Z.; Xue, J.H.; Wu, Y.; et al. Assimilation of remote sensing into crop growth models: Current status and perspectives. Agric. For. Meteorol. 2019, 276–277, 107609. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C. Inversion of a radiative transfer model for estimating vegetation LAI and chlorophyll in a heterogeneous grassland. Remote Sens. Environ. 2008, 112, 2592–2604. [Google Scholar] [CrossRef]
- Zheng, G.; Moskal, L.M. Retrieving Leaf Area Index (LAI) Using Remote Sensing: Theories, Methods and Sensors. Sensors 2009, 9, 2719–2745. [Google Scholar] [CrossRef] [PubMed]
- Delegido, J.; Verrelst, J.; Alonso, L.; Moreno, J. Evaluation of Sentinel-2 red-edge bands for empirical estimation of green LAI and chlorophyll content. Sensors 2011, 11, 7063–7081. [Google Scholar] [CrossRef] [PubMed]
- Gerighausen, H.; Lilienthal, H.; Jarmer, T.; Siegmann, B. Evaluation of leaf area index and dry matter predictions for crop growth modelling and yield esti-mation based on field reflectance measurements. In Proceedings of the 9th EARSeL Imaging Spectroscopy Workshop, Luxembourg, 14–16 April 2015; Volume 14, pp. 71–90. [Google Scholar]
- Siegmann, B.; Jarmer, T. Comparison of different regression models and validation techniques for the assessment of wheat leaf area index from hyperspectral data. Int. J. Remote Sens. 2015, 36, 4519–4534. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.; Camps-Valls, G.; Moreno, J. Experimental Sentinel-2 LAI estimation using parametric, non-parametric and physical retrieval methods—A comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Upreti, D.; Huang, W.; Kong, W.; Pascucci, S.; Pignatti, S.; Zhou, X.; Ye, H.; Casa, R. A Comparison of Hybrid Machine Learning Algorithms for the Retrieval of Wheat Biophysical Variables from Sentinel-2. Remote Sens. 2019, 11, 481. [Google Scholar] [CrossRef]
- Xie, Q.; Jadu, D.; Huete, A.; Jiang, A.; Yin, G.; Ding, Y.; Peng, D.; C. Hall, C.; Brown, L.; Shi, Y.; et al. Retrieval of crop biophysical parameters from Sentinel-2 remote sensing imagery. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 187–195. [Google Scholar] [CrossRef]
- Kamenova, I.; Dimitrov, P. Evaluation of Sentinel-2 vegetation indices for prediction of LAI, fAPAR and fCover of winter wheat in Bulgaria. Eur. J. Remote Sens. 2021, 54, 89–108. [Google Scholar] [CrossRef]
- Caballero, G.; Pezzola, A.; Winschel, C.; Casella, A.; Angonova, P.S.; Rivera-Caicedo, J.P.; Berger, K.; Verrelst, J.; Delegido, J. Seasonal Mapping of Irrigated Winter Wheat Traits in Argentina with a Hybrid Retrieval Workflow Using Sentinel-2 Imagery. Remote Sens. 2022, 14, 4531. [Google Scholar] [CrossRef]
- Hansen, P.M.; Schjoerring, J.K. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Wocher, M.; Berger, K.; Verrelst, J.; Hank, T. Retrieval of carbon content and biomass from hyperspectral imagery over cultivated areas. ISPRS J. Photogramm. Remote Sens. 2022, 193, 104–114. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Kooistra, L. Using Hyperspectral Remote Sensing Data for Retrieving Canopy Chlorophyll and Nitrogen Content. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 574–583. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Huang, T.; Wang, J.; Lin, L.; Wang, L.; Yang, M. Estimation of Leaf Nitrogen Content in Wheat Using New Hyperspectral Indices and a Random Forest Regression Algorithm. Remote Sens. 2018, 10, 1940. [Google Scholar] [CrossRef]
- Perich, G.; Aasen, H.; Verrelst, J.; Argento, F.; Walter, A.; Liebisch, F. Crop Nitrogen Retrieval Methods for Simulated Sentinel-2 Data Using In-Field Spectrometer Data. Remote Sens. 2021, 13, 2404. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Yang, G.; Li, C.; Li, Z.; Wang, Y.; Feng, H.; Xu, B. Estimation of Winter Wheat Above-Ground Biomass Using Unmanned Aerial Vehicle-Based Snapshot Hyperspectral Sensor and Crop Height Improved Models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef]
- Prey, L.; Schmidhalter, U. Simulation of satellite reflectance data using high-frequency ground based hyperspectral canopy measurements for in-season estimation of grain yield and grain nitrogen status in winter wheat. ISPRS J. Photogramm. Remote Sens. 2019, 149, 176–187. [Google Scholar] [CrossRef]
- Grote, U.; Fasse, A.; Nguyen, T.T.; Erenstein, O. Food Security and the Dynamics of Wheat and Maize Value Chains in Africa and Asia. Front. Sustain. Food Syst. 2021, 4, 617009. [Google Scholar] [CrossRef]
- FAO. Crops and Livestock Products; FAO: Rome, Italy, 2023. [Google Scholar]
- Bundesministerium für Ernährung und Landwirtschaft. Daten und Fakten: Land-, Forst- und Ernährungswirtschaft mit Fischerei und Wein- und Gartenbau; Bundesministerium für Ernährung und Landwirtschaft: Berlin, Germany, 2022. [Google Scholar]
- Guanter, L.; Kaufmann, H.; Segl, K.; Foerster, S.; Rogass, C.; Chabrillat, S.; Kuester, T.; Hollstein, A.; Rossner, G.; Chlebek, C.; et al. The EnMAP Spaceborne Imaging Spectroscopy Mission for Earth Observation. Remote Sens. 2015, 7, 8830–8857. [Google Scholar] [CrossRef]
- Lee, C.M.; Cable, M.L.; Hook, S.J.; Green, R.O.; Ustin, S.L.; Mandl, D.J.; Middleton, E.M. An introduction to the NASA Hyperspectral InfraRed Imager (HyspIRI) mission and preparatory activities. Remote Sens. Environ. 2015, 167, 6–19. [Google Scholar] [CrossRef]
- Labate, D.; Ceccherini, M.; Cisbani, A.; de Cosmo, V.; Galeazzi, C.; Giunti, L.; Melozzi, M.; Pieraccini, S.; Stagi, M. The PRISMA payload optomechanical design, a high performance instrument for a new hyperspectral mission. Acta Astronaut. 2009, 65, 1429–1436. [Google Scholar] [CrossRef]
- Nieke, J.; Rast, M. Towards the Copernicus Hyperspectral Imaging Mission For The Environment (CHIME). In Proceedings of the IGARSS, Valencia, Spain, 22–27 July 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 157–159. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Teucher, M.; Thürkow, D.; Alb, P.; Conrad, C. Digital In Situ Data Collection in Earth Observation, Monitoring and Agriculture—Progress towards Digital Agriculture. Remote Sens. 2022, 14, 393. [Google Scholar] [CrossRef]
- Rao, N.R. Development of a crop–specific spectral library and discrimination of various agricultural crop varieties using hyperspectral imagery. Int. J. Remote Sens. 2008, 29, 131–144. [Google Scholar] [CrossRef]
- Baldridge, A.M.; Hook, S.J.; Grove, C.I.; Rivera, G. The ASTER spectral library version 2.0. Remote Sens. Environ. 2009, 113, 711–715. [Google Scholar] [CrossRef]
- Hueni, A.; Nieke, J.; Schopfer, J.; Kneubühler, M.; Itten, K.I. The spectral database SPECCHIO for improved long-term usability and data sharing. Comput. Geosci. 2009, 35, 557–565. [Google Scholar] [CrossRef]
- Zomer, R.J.; Trabucco, A.; Ustin, S.L. Building spectral libraries for wetlands land cover classification and hyperspectral remote sensing. J. Environ. Manag. 2009, 90, 2170–2177. [Google Scholar] [CrossRef]
- Nidamanuri, R.R.; Zbell, B. Existence of characteristic spectral signatures for agricultural crops—potential for automated crop mapping by hyperspectral imaging. Geocarto Int. 2012, 27, 103–118. [Google Scholar] [CrossRef]
- Kotthaus, S.; Smith, T.E.; Wooster, M.J.; Grimmond, C. Derivation of an urban materials spectral library through emittance and reflectance spectroscopy. Isprs J. Photogramm. Remote Sens. 2014, 94, 194–212. [Google Scholar] [CrossRef]
- Jiménez, M.; Díaz-Delgado, R. Towards a Standard Plant Species Spectral Library Protocol for Vegetation Mapping: A Case Study in the Shrubland of Doñana National Park. ISPRS Int. J. Geo-Inf. 2015, 4, 2472–2495. [Google Scholar] [CrossRef]
- Fuchs, M.; Awan, A.A.; Akhtar, S.S.; Ahmad, I.; Sadiq, S.; Razzak, A.; Haider, N. Lithological mapping with multispectral data—setup and application of a spectral database for rocks in the Balakot area, Northern Pakistan. J. Mt. Sci. 2017, 14, 948–963. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.; Yuan, L.; Liu, P.; Zhang, Y.; Wu, K. Construction of a plant spectral library based on an optimised feature selection method. Biosyst. Eng. 2020, 195, 1–16. [Google Scholar] [CrossRef]
- de Peppo, M.; Taramelli, A.; Boschetti, M.; Mantino, A.; Volpi, I.; Filipponi, F.; Tornato, A.; Valentini, E.; Ragaglini, G. Non-Parametric Statistical Approaches for Leaf Area Index Estimation from Sentinel-2 Data: A Multi-Crop Assessment. Remote Sens. 2021, 13, 2841. [Google Scholar] [CrossRef]
- Verrelst, J.; Malenovský, Z.; van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.P.; Lewis, p.; North, P.; Moreno, J. Quantifying Vegetation Biophysical Variables from Imaging Spectroscopy Data: A Review on Retrieval Methods. Surv. Geophys. 2019, 40, 589–629. [Google Scholar] [CrossRef] [PubMed]
- Pasqualotto, N.; Delegido, J.; van Wittenberghe, S.; Rinaldi, M.; Moreno, J. Multi-Crop Green LAI Estimation with a New Simple Sentinel-2 LAI Index (SeLI). Sensors 2019, 19, 904. [Google Scholar] [CrossRef]
- Verrelst, J.; Muñoz, J.; Alonso, L.; Delegido, J.; Rivera, J.P.; Camps-Valls, G.; Moreno, J. Machine learning regression algorithms for biophysical parameter retrieval: Opportunities for Sentinel-2 and -3. Remote Sens. Environ. 2012, 118, 127–139. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal Component Analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial Least-Squares Regression: A Tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, G.; Wang, J.; Song, X.; Feng, H. Winter wheat biomass estimation based on spectral indices, band depth analysis and partial least squares regression using hyperspectral measurements. Comput. Electron. Agric. 2014, 100, 51–59. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
- Foster, A.J.; Kakani, V.G.; Mosali, J. Estimation of bioenergy crop yield and N status by hyperspectral canopy reflectance and partial least square regression. Precis. Agric. 2017, 18, 192–209. [Google Scholar] [CrossRef]
- Wang, B.; Chen, J.; Ju, W.; Qiu, F.; Zhang, Q.; Fang, M.; Chen, F. Limited Effects of Water Absorption on Reducing the Accuracy of Leaf Nitrogen Estimation. Remote Sens. 2017, 9, 291. [Google Scholar] [CrossRef]
- Hoerl, A.E.; Kennard, R.W. Ridge Regression: Applications to Nonorthogonal Problems. Technometrics 1970, 12, 69–82. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Lazaridis, D.C.; Verbesselt, J.; Robinson, A.P. Penalized regression techniques for prediction: A case study for predicting tree mortality using remotely sensed vegetation indicesThis article is one of a selection of papers from Extending Forest Inventory and Monitoring over Space and Time. Can. J. For. Res. 2011, 41, 24–34. [Google Scholar] [CrossRef]
- Zandler, H.; Brenning, A.; Samimi, C. Quantifying dwarf shrub biomass in an arid environment: Comparing empirical methods in a high dimensional setting. Remote Sens. Environ. 2015, 158, 140–155. [Google Scholar] [CrossRef]
- Caicedo, J.P.R.; Verrelst, J.; Munoz-Mari, J.; Moreno, J.; Camps-Valls, G. Toward a Semiautomatic Machine Learning Retrieval of Biophysical Parameters. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1249–1259. [Google Scholar] [CrossRef]
- Lázaro-Gredilla, M.; Titsias, M.K.; Verrelst, J.; Camps-Valls, G. Retrieval of Biophysical Parameters with Heteroscedastic Gaussian Processes. IEEE Geosci. Remote Sens. Lett. 2013, 11, 2013. [Google Scholar] [CrossRef]
- Chen, B.; Wu, Z.; Wang, J.; Dong, J.; Guan, L.; Chen, J.; Yang, K.; Xie, G. Spatio-temporal prediction of leaf area index of rubber plantation using HJ-1A/1B CCD images and recurrent neural network. ISPRS J. Photogramm. Remote Sens. 2015, 102, 148–160. [Google Scholar] [CrossRef]
- Neinavaz, E.; Skidmore, A.K.; Darvishzadeh, R.; Groen, T.A. Retrieval of leaf area index in different plant species using thermal hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2016, 119, 390–401. [Google Scholar] [CrossRef]
- Combal, B.; Baret, F.; Weiss, M.; Trubuil, A.; Myneni, R.; Knyazikhin, Y.; Wang, L. Retrieval of canopy biophysical variables from bidirectional reflectance: Using prior information to solve the ill-posed inverse problem. Remote Sens. Environ. 2002, 84, 1–15. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L. PROSPECT+SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Berger, K.; Atzberger, C.; Danner, M.; D’Urso, G.; Mauser, W.; Vuolo, F.; Hank, T. Evaluation of the PROSAIL Model Capabilities for Future Hyperspectral Model Environments: A Review Study. Remote Sens. 2018, 10, 85. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F.; Myneni, R.B.; Pragnère, A.; Knyazikhin, Y. Investigation of a model inversion technique to estimate canopy biophysical variables from spectral and directional reflectance data. Agronomie 2000, 20, 3–22. [Google Scholar] [CrossRef]
- Locherer, M.; Hank, T.; Danner, M.; Mauser, W. Retrieval of Seasonal Leaf Area Index from Simulated EnMAP Data through Optimized LUT-Based Inversion of the PROSAIL Model. Remote Sens. 2015, 7, 10321–10346. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F.; Jay, S. S2ToolBox Level 2 Products LAI, FAPAR, FCOVER; HAL Open Science: Lyon, France, 2020. [Google Scholar]
- Borg, E.; Lippert, K.; Zabel, E.; Löpmeier, F.J.; Fichtelmann, B.; Jahncke, D.; Maass, H. DEMMIN—Teststandort zur Kalibrierung und Validierung von Fernerkundungsmissionen; Rebenstorf, R.W., Ed.; Self-published: Neubrandenburg, Germany, 2009; Volume 15, pp. 401–419. [Google Scholar]
- Zacharias, S.; Bogena, H.; Samaniego, L.; Mauder, M.; Fuß, R.; Pütz, T.; Frenzel, M.; Schwank, M.; Baessler, C.; Butterbach-Bahl, K.; et al. A Network of Terrestrial Environmental Observatories in Germany. Vadose Zone J. 2011, 10, 955–973. [Google Scholar] [CrossRef]
- Meier, U.; Bleiholder, H.; Buhr, L.; Feller, C.; Hack, H.; Heß, M.; Lancashire, P.D.; Schock, U.; Strauß, R.; van den Boom, T.; et al. The BBCH system to coding the phenological growth stages of plants–history and publications. J. Kult. 2009, 61, 41–52. [Google Scholar]
- VDLUFA. Bestimmung von organischem Kohlenstoff durch Verbrennung und Gasanalyse (Differenzmethode). In Das VDLUFA Methodenbuch; VDLUFA, Ed.; VDLUFA: Darmstadt, Germany, 1991; Volume 1. [Google Scholar]
- VDLUFA. Bestimmung von Gesamtstickstoff nach trockener Verbrennung (Elementaranalyse). In Das VDLUFA Methodenbuch; VDLUFA, Ed.; VDLUFA: Darmstadt, Germany, 1991; Volume 1. [Google Scholar]
- LI-COR Biosciences. FV2200. 2010. Available online: https://www.licor.com/env/products/leaf_area/LAI-2200C/software (accessed on 14 July 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Lehnert, L.W.; Meyer, H.; Obermeier, W.A.; Silva, B.; Regeling, B.; Bendix, J. Hyperspectral Data Analysis in R: The hsdar Package. J. Stat. Softw. 2019, 89, 1–23. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–13. [Google Scholar] [CrossRef]
- Rasmussen, C.E. Gaussian Processes in Machine Learning. In Advanced Lectures on Machine Learning; Bousquet, O., von Luxburg, U., Rätsch, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 63–71. [Google Scholar]
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab-an S4 package for kernel methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef]
- Liland, K.H.; Mevik, B.H.; Wehrens, R. pls: Partial Least Squares and Principal Component Regression. 2022. Available online: https://cran.r-project.org/web/packages/pls/index.html (accessed on 14 July 2023).
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Wright, M.N.; Ziegler, A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Kuhn, M. caret: Classification and Regression Training. 2021. [Google Scholar]
- CODE-DE. CODE-DE: The German Access to Copernicus Data. 2022. Available online: https://code-de.org/en/ (accessed on 14 July 2023).
- Hijmans, R.J. terra: Spatial Data Analysis. 2021. Available online: https://CRAN.R-project.org/package=terra (accessed on 14 July 2023).
- Robinson, D. fuzzyjoin: Join Tables Together on Inexact Matching. 2020. Available online: https://cran.r-project.org/package=fuzzyjoin (accessed on 14 July 2023).
- Schiefer, F.; Schmidtlein, S.; Kattenborn, T. The retrieval of plant functional traits from canopy spectra through RTM-inversions and statistical models are both critically affected by plant phenology. Ecol. Indic. 2021, 121, 107062. [Google Scholar] [CrossRef]
- Estevez, J.; Vicent, J.; Rivera-Caicedo, J.P.; Morcillo-Pallarés, P.; Vuolo, F.; Sabater, N.; Camps-Valls, G.; Moreno, J.; Verrelst, J. Gaussian Processes Retrieval of LAI from Sentinel-2 Top-of-Atmosphere Radiance Data. ISPRS J. Photogramm. Remote Sens. 2020, 167, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Cai, J.; Fu, P.; Hu, L.; Liu, T. Deep learning-based fusion of Landsat-8 and Sentinel-2 images for a harmonized surface reflectance product. Remote Sens. Environ. 2019, 235, 111425. [Google Scholar] [CrossRef]
- Wang, Q.; Atkinson, P.M. Spatio-temporal fusion for daily Sentinel-2 images. Remote Sens. Environ. 2018, 204, 31–42. [Google Scholar] [CrossRef]
- Bratsch, S.; Epstein, H.; Buchhorn, M.; Walker, D.; Landes, H. Relationships between hyperspectral data and components of vegetation biomass in Low Arctic tundra communities at Ivotuk, Alaska. Environ. Res. Lett. 2017, 12, 025003. [Google Scholar] [CrossRef]
- Frantz, D. FORCE—Landsat + Sentinel-2 Analysis Ready Data and Beyond. Remote Sens. 2019, 11, 1124. [Google Scholar] [CrossRef]
- Y, G.D.; Nair, N.G.; Satpathy, P.; Christopher, J. Covariate Shift: A Review and Analysis on Classifiers. In Proceedings of the Global Conference for Advancement in Technology (GCAT), Bangalore, India, 18–20 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Segarra, J.; Buchaillot, M.L.; Araus, J.L.; Kefauver, S.C. Remote Sensing for Precision Agriculture: Sentinel-2 Improved Features and Applications. Agronomy 2020, 10, 641. [Google Scholar] [CrossRef]
- Kang, Y.; Özdoğan, M. Field-level crop yield mapping with Landsat using a hierarchical data assimilation approach. Remote Sens. Environ. 2019, 228, 144–163. [Google Scholar] [CrossRef]
- Rivera-Caicedo, J.P.; Verrelst, J.; Muñoz-Marí, J.; Camps-Valls, G.; Moreno, J. Hyperspectral dimensionality reduction for biophysical variable statistical retrieval. ISPRS J. Photogramm. Remote Sens. 2017, 132, 88–101. [Google Scholar] [CrossRef]
- Cherif, E.; Feilhauer, H.; Berger, K.; Dao, P.D.; Ewald, M.; Hank, T.B.; He, Y.; Kovach, K.R.; Lu, B.; Townsend, P.A.; et al. From spectra to plant functional traits: Transferable multi-trait models from heterogeneous and sparse data. Remote Sens. Environ. 2023, 292, 113580. [Google Scholar] [CrossRef]
- Preidl, S.; Lange, M.; Doktor, D. Introducing APiC for regionalised land cover mapping on the national scale using Sentinel-2A imagery. Remote Sens. Environ. 2020, 240, 111673. [Google Scholar] [CrossRef]
- Blickensdörfer, L.; Schwieder, M.; Pflugmacher, D.; Nendel, C.; Erasmi, S.; Hostert, P. Mapping of crop types and crop sequences with combined time series of Sentinel-1, Sentinel-2 and Landsat 8 data for Germany. Remote Sens. Environ. 2022, 269, 112831. [Google Scholar] [CrossRef]
- d’Andrimont, R.; Verhegghen, A.; Lemoine, G.; Kempeneers, P.; Meroni, M.; van der Velde, M. From parcel to continental scale—A first European crop type map based on Sentinel-1 and LUCAS Copernicus in-situ observations. Remote Sens. Environ. 2021, 266, 112708. [Google Scholar] [CrossRef]
- Dhillon, M.S.; Dahms, T.; Kübert-Flock, C.; Liepa, A.; Rummler, T.; Arnault, J.; Steffan-Dewenter, I.; Ullmann, T. Impact of STARFM on Crop Yield Predictions: Fusing MODIS with Landsat 5, 7, and 8 NDVIs in Bavaria Germany. Remote Sens. 2023, 15, 1651. [Google Scholar] [CrossRef]
- Tewes, A.; Hoffmann, H.; Krauss, G.; Schäfer, F.; Kerkhoff, C.; Gaiser, T. New Approaches for the Assimilation of LAI Measurements into a Crop Model Ensemble to Improve Wheat Biomass Estimations. Agronomy 2020, 10, 446. [Google Scholar] [CrossRef]

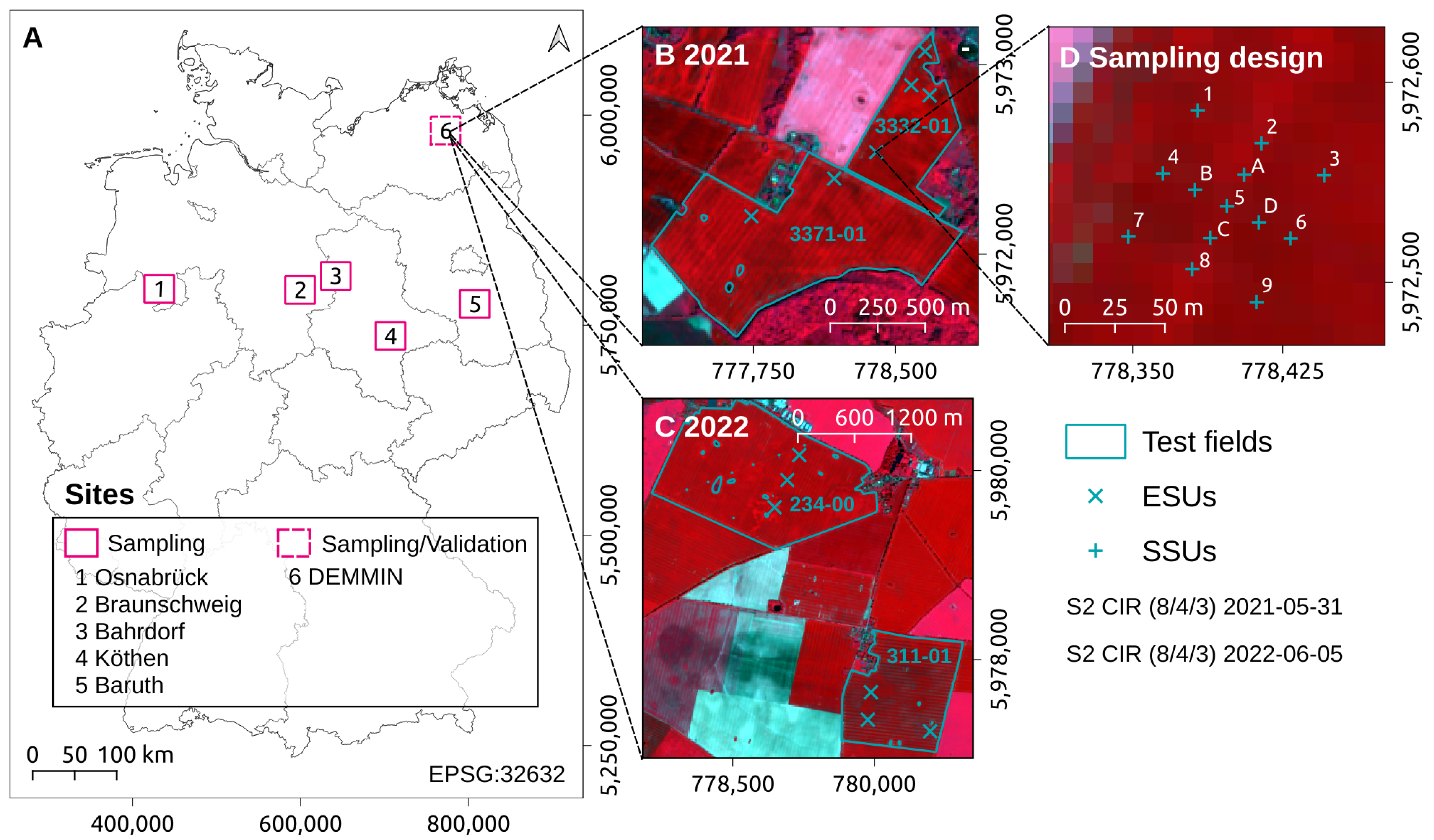
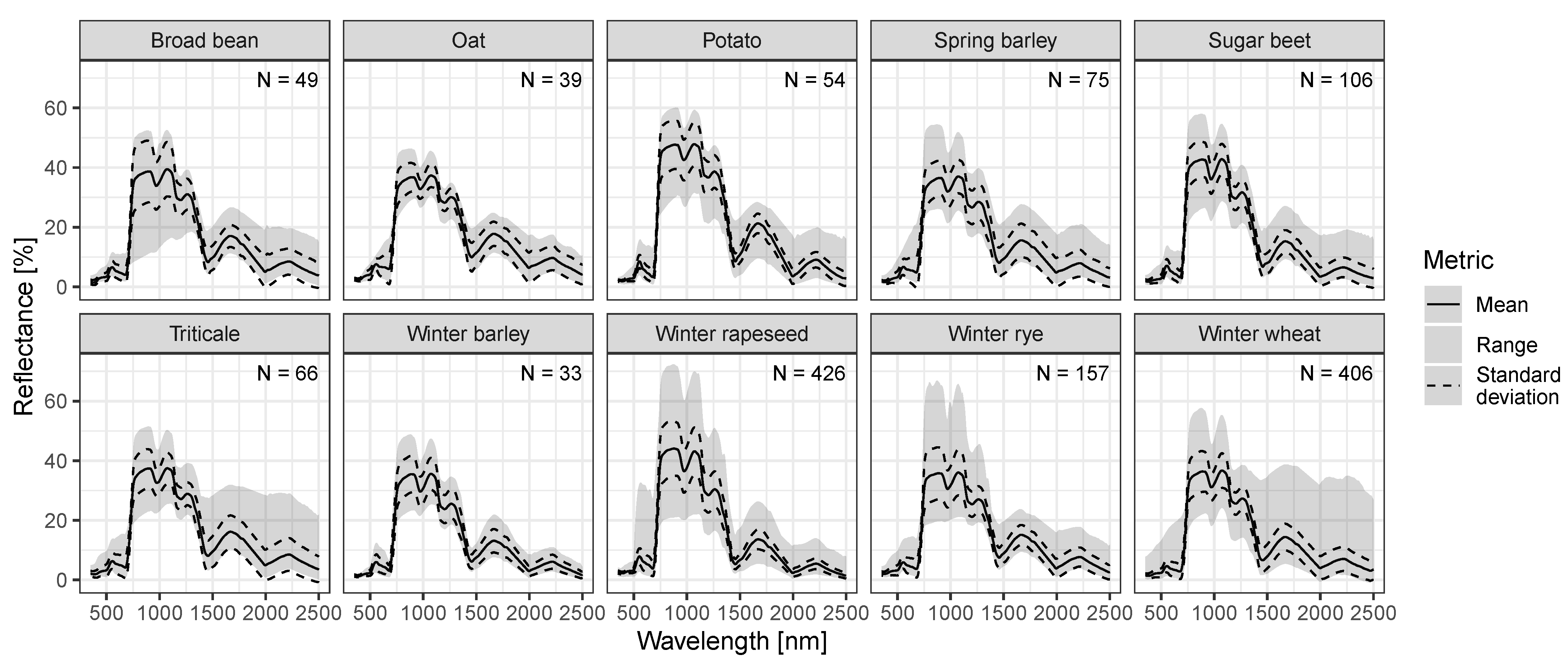
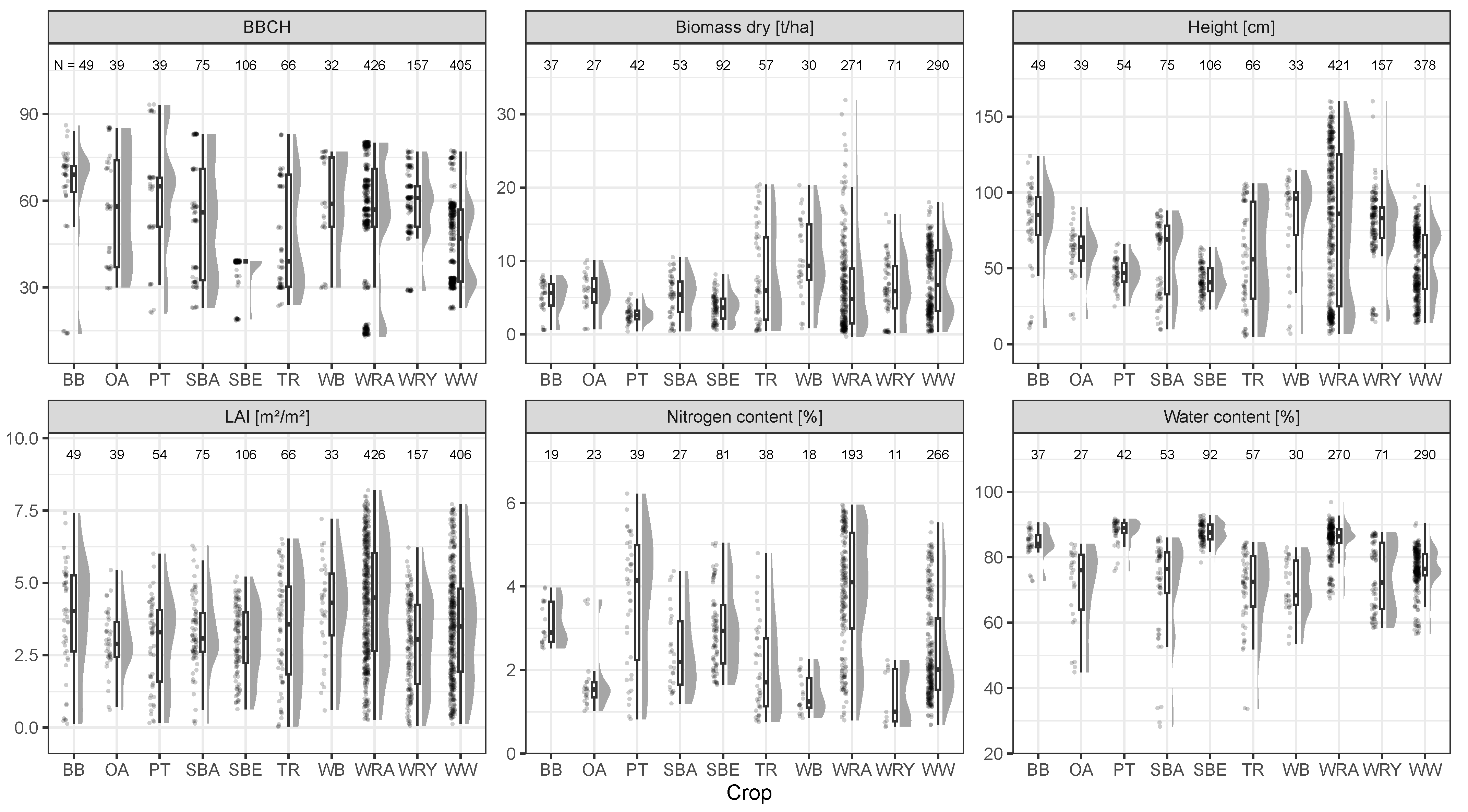
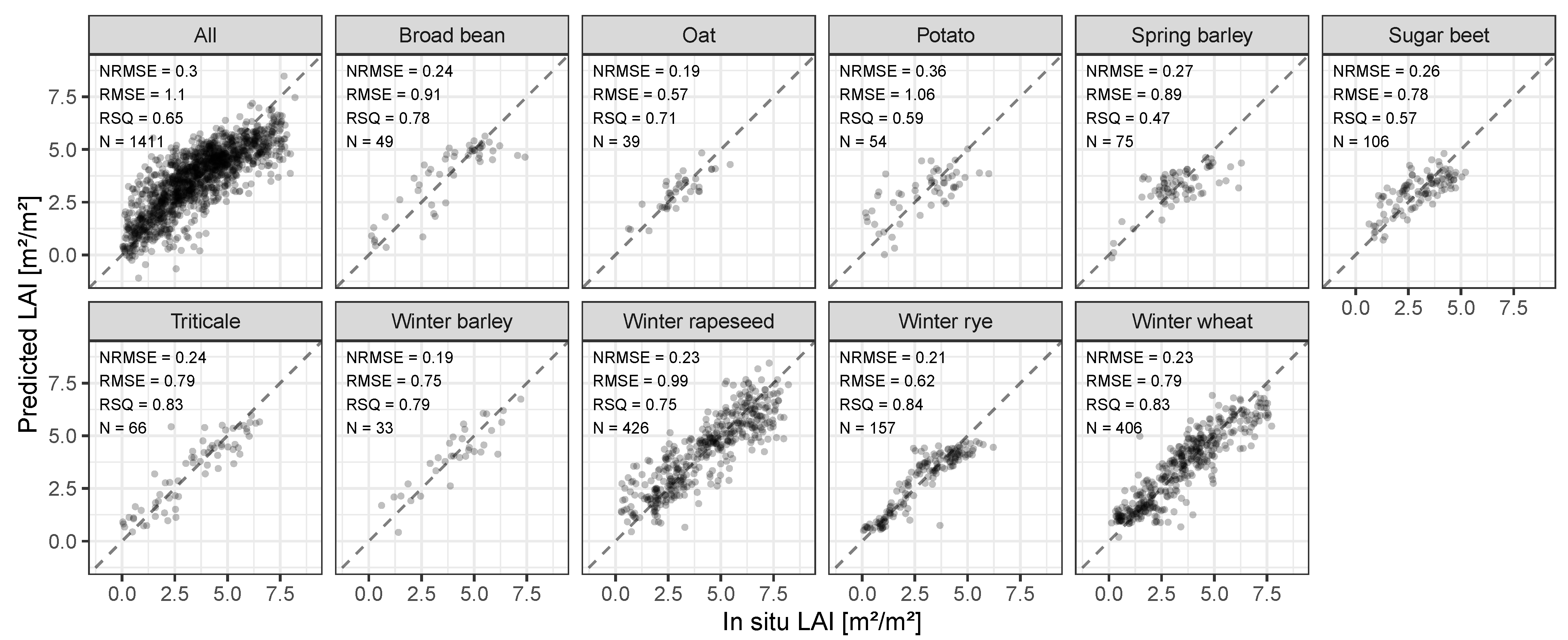


| Crop | Botanical Name | Abbreviation | N |
|---|---|---|---|
| Broad bean | Vicia faba | BB | 49 |
| Oat | Avena sativa | OA | 39 |
| Potato | Solanum tuberosum | PT | 54 |
| Spring barley | Hordeum vulgare | SBA | 75 |
| Sugar beet | Beta vulgaris | SBE | 106 |
| Triticale | xTriticosecale | TR | 66 |
| Winter barley | Hordeum vulgare | WB | 33 |
| Winter rapeseed | Brassica napus | WRA | 426 |
| Winter rye | Secale cereale | WRY | 157 |
| Winter wheat | Triticum aestivum | WW | 406 |
| MISPEL | Model | NRMSE | RMSE | RSQ | N |
|---|---|---|---|---|---|
| FR | GLMNET | 0.22 | 0.78 | 0.83 | 406 |
| FR | GP | 0.20 | 0.70 | 0.86 | 406 |
| FR | PLS | 0.23 | 0.82 | 0.81 | 406 |
| FR | RF | 0.22 | 0.77 | 0.83 | 406 |
| S2 | GLMNET | 0.22 | 0.79 | 0.83 | 406 |
| S2 | GP | 0.23 | 0.79 | 0.83 | 406 |
| S2 | PLS | 0.23 | 0.81 | 0.82 | 406 |
| S2 | RF | 0.23 | 0.79 | 0.83 | 406 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borrmann, P.; Brandt, P.; Gerighausen, H. MISPEL: A Multi-Crop Spectral Library for Statistical Crop Trait Retrieval and Agricultural Monitoring. Remote Sens. 2023, 15, 3664. https://doi.org/10.3390/rs15143664
Borrmann P, Brandt P, Gerighausen H. MISPEL: A Multi-Crop Spectral Library for Statistical Crop Trait Retrieval and Agricultural Monitoring. Remote Sensing. 2023; 15(14):3664. https://doi.org/10.3390/rs15143664
Chicago/Turabian StyleBorrmann, Peter, Patric Brandt, and Heike Gerighausen. 2023. "MISPEL: A Multi-Crop Spectral Library for Statistical Crop Trait Retrieval and Agricultural Monitoring" Remote Sensing 15, no. 14: 3664. https://doi.org/10.3390/rs15143664
APA StyleBorrmann, P., Brandt, P., & Gerighausen, H. (2023). MISPEL: A Multi-Crop Spectral Library for Statistical Crop Trait Retrieval and Agricultural Monitoring. Remote Sensing, 15(14), 3664. https://doi.org/10.3390/rs15143664








